Abstract
Background
Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) is a member of the tumour necrosis factor cytokine family that induces apoptosis upon binding to its death domain containing receptors, TRAIL receptor 1 (DR4) and TRAIL receptor 2 (DR5). Expression of TRAIL receptors is higher in colorectal carcinoma (CRC) as compared to normal colorectal mucosa and targeted therapy with TRAIL leads to preferential killing of tumor cells sparing normal cells.
Methods
We investigated the expression of TRAIL and its receptors in a tissue microarray cohort of 448 Middle Eastern CRC. We also studied the correlation between TRAIL receptors and various clinico-pathological features including key molecular alterations and overall survival.
Results
CRC subset with TRAIL-R1 expression was associated with a less aggressive phenotype characterized by early stage (p = 0.0251) and a histology subtype of adenocarcinomas (p = 0.0355). Similarly CRC subset with TRAIL-R2 expression was associated with a well-differentiated tumors (p < 0.0001), histology subtype of adenocarcinomas (p = 0.0010) and tumors in left colon (p = 0.0009). Over expression of pro apoptotic markers: p27KIP1 and KRAS4A isoforms was significantly higher in CRC subset with TRAIL-R1 and TRAIL-R2 expression; TRAIL-R1 expression was also associated with cleaved caspase-3(p = 0.0011). Interestingly, TRAIL-R2 expression was associated with a microsatellite stable (MS--S/L) phenotype (p = 0.0003) and with absence of KRAS mutations (p = 0.0481).
Conclusion
TRAIL-R1 expression was an independent prognostic marker for better survival in all CRC samples and even in the CRC group that received adjuvant therapy. The biological effects of TRAIL in CRC models, its enhancement of chemosensitivity towards standard chemotherapeutic agents and the effect of endogenous TRAIL receptor levels on survival make TRAIL an extremely attractive therapeutic target.
Similar content being viewed by others
Introduction
Colorectal Cancer (CRC) is a major cause of mortality and morbidity worldwide. In Saudi Arabia, the incidence of CRC is increasing. According to the latest statistics, CRC is the second most common cancer among Saudi males and the third most common among Saudi females [1]. Currently available therapeutic approaches for CRC are less effective, and thus the prognosis is poor. Despite a growing number of publications about biomarkers that give information on disease outcome, the best prognostic factors are still simple clinical parameters like number of lymph nodal metastasis, presence of distant metastasis, tumour grade and AJCC stage. Prognostic biomarkers might especially be useful for hypothesis testing for their relevance as predictive markers, as targets for therapy and for the selection of patients for adjuvant treatment [2].
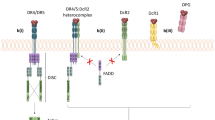
Apoptosis, or programmed cell death, is a major control mechanism by which cells die if DNA damage is not repaired [3]. Apoptosis is an essential biochemical pathway for normal tissue homeostasis, cellular differentiation, and development. Derangements of normal apoptotic mechanisms provide a growth advantage to cancer cells [4]. The understanding of apoptosis has provided the basis for novel targeted therapies that can induce death in cancer cells or sensitize them to established cytotoxic agents and radiation therapy [5]. In addition, as apoptosis usually does not elicit host inflammatory or immune response, this type of cell death is the preferred way of cancer cell killing by various treatments. Accordingly, selectively inducing apoptosis in tumour cells is gaining recognition as a promising therapeutic approach for many cancers [6]. Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL or Apo2 ligand) is a member of the tumour necrosis factor (TNF) cytokine family that induces apoptosis upon binding to its death domain containing receptors, TRAIL receptor 1 (death receptor 4, DR4) and TRAIL receptor 2 (death receptor 5, DR5) [7]. The TRAIL receptors, TRAIL-R1 and TRAIL-R2, are highly expressed in many cancer cells including CRC [8–10]. A further three TRAIL receptors exist, which are unable to induce apoptosis and act as decoys. Decoy receptors 1 (DcR1) and 2 (DcR2), similar to TRAIL-R1 and TRAIL-R2, are expressed on the cell surface. Thus, overexpression of either DcR1 or DcR2 confers protection against TRAIL-induced apoptosis [11, 12]. The fifth TRAIL receptor is osteoprotegerin (OPG), a secreted, low affinity receptor for TRAIL [11, 12]. Binding of TRAIL to TRAIL-R1 and TRAIL-R2 induces trimerization of TRAIL-R1 and TRAIL-R2 [13]. The trimerized TRAIL-R1 and TRAIL-R2 bind to FADD, which recruits caspase 8 and initiates a proteolysis cascade that eventually leads to cell death by apoptosis. Many cancer cells are resistant to death receptor induced apoptosis [4]. The mechanisms of resistance include the presence of decoy receptors for TRAIL [12], the loss of TRAIL receptor expression [14], the overexpression of inhibitory proteins in signal transduction pathways such as FLICE-inhibitory protein [14], and the mutation of TRAIL-R2 gene [15–18].
Oncogenic mutations such as ras may enhance expression of TRAIL receptors; potentially sensitizing these tumors to TRAIL based therapies [19–21]. Constitutively activated Ras increases the tumorigenic potential of cells because it causes deregulation of important intracellular signaling pathways [22]. Activated RAS mediates its biological activity through interaction with various downstream effector targets, thus activating pathways like MEK, PI3K, and Rho GTPases [22, 23]. RAS regulates a RAF-MEK-ERK1/2 kinase cascade and this pathway is found to be active in human colon adenocarcinomas cells [24] as well as in human colorectal tumors [25]. Drosopoulos et al. [21] have shown transformation of the colon cell line Caco-2 by ras oncogenes sensitizes these cells to TRAIL induced apoptosis by causing specific MEK-dependent up-regulation of TRAIL-R1 and TRAIL-R2. Nesterov A et al. [20] have demonstrated that normal cells are sensitized to TRAIL when TRAIL-R2 is up regulated by overexpression of c-myc or oncogenic ras mutants. Thus, RAS-MEK-ERK1/2 signaling pathway can sensitize cells to TRAIL-induced apoptosis by up-regulating TRAIL-R1, TRAIL-R2 and TRAIL-based therapeutic strategies using TRAIL agonists could be used in cases of human colon cancers bearing RAS mutations. Therefore, we also sought to explore the potential link between expression of TRAIL and its receptors with KRAS alterations in CRC.
The aims of the present study were: (a) to determine the TRAIL/TRAIL receptor expression pattern in normal and neoplastic colon epithelium; (b) to correlate immunohistochemical expression patterns with KRAS alterations, microsatellite instability and pro apoptotic markers; (c) to correlate immunohistochemical expression patterns with overall survival.
Results
Expression of TRAIL and its receptors TRAIL-R1 and TRAIL-R2
Incidence of TRAIL-R1, TRAIL-R2 and TRAIL ligand expression in CRC was 85.5% (331/387), 59.4(217/365) and 31.5% (127/403) respectively [Figure 1]. These incidences are within the wide ranges reported earlier - TRAIL: 37.5% to 83%, TRAIL-R1:58.1% to 100.0% and TRAIL-R2: 40.3% to 100% [26–31]. Incidence of non-interpretable tumor spots for TRAIL, TRAIL-R1 and TRAIL R2 ranged from 10 to 18%. Tumor spots were deemed not interpretable if they had insufficient tumor cells, loss of tissue in the spot, or an abundance of necrotic tissue. Expression of TRAIL and its receptors was also evaluated in colorectal adenomas and adjacent colorectal mucosa [Figure 2]. Both TRAIL-R1 and TRAIL-R2 expression was significantly higher in both colorectal adenomas [TRAIL-R1 96.11 ± 78.82(p = 0.0312); TRAIL-R2 59.17 ± 49.69(p = 0.0027)] and carcinoma [TRAIL-R1 173.91 ± 61.20(P < 0.00001); TRAIL-R2 115.63 ± 95.76(p = < 0.0001)] as compared to normal colorectal mucosa (TRAIL-R1 52.13 ± 42.48; TRAIL-R2 24.57 ± 38.77). In addition, there was a significant difference in expression of both TRAIL-R1 (p = 0.0006) and TRAIL-R2 (p = < 0.0001) between colorectal adenomas and carcinoma [Figure 3B and 3C]. Similarly, TRAIL expression was significantly higher in carcinoma (132.87 ± 64.23; p < 0.0001) and adenomas (129.01 ± 38.59; p < 0.0001) as compared to normal colorectal mucosa (48.59 ± 66.07). However, there was no difference in TRAIL expression between adenomas and carcinomas (p = 0.6822; Figure 3A). Thus the TRAIL system may play a key role in colorectal carcinogenesis.
Tissue microarray-based immunohistochemical analysis of TRAIL receptors TRAIL-R1 and TRAIL-R2 in CRC patients. CRC array spots showing overexpression of TRAIL-R1 (A) and TRAIL-R2 (C). In contrast, other CRC tissue array spots showing low expression of TRAIL-R1 (B) and TRAIL-R2 (D). 20 ×/0.70 objective on an Olympus BX 51 microscope (Olympus America Inc, Center Valley, PA, USA), with the inset showing a 40 ×/0.85 aperture magnified view of the same
Tissue microarray-based immunohistochemical analyses of TRAIL receptors TRAIL-R1 and TRAIL-R2 in CRC carcinogenesis. A progressive increase on TRAIL-R1 expression was seen from normal colorectal mucosa (A) to adenoma (B) to carcinoma(C). Similarly, a progressive increase on TRAIL-R2 expression was seen from normal colorectal mucosa (D) to adenoma (E) to carcinoma (F).
Box plot of TRAIL, TRAIL-R1 and TRAILR2 expression in cancer, adenoma, and normal samples of colorectal patients. (A) Using Student's t-test, the mean ± SD of TRAIL expression in normal colorectal mucosa (48.59 ± 66.07), adenoma (129.01 ± 38.59) and cancer (132.87 ± 64.23) and showed a significant association with cancer versus normal (p < 0.0001) and adenoma versus normal (p < 0.0001). (B) Using Student's t-test, the mean ± SD of TRAIL-R1 expression in normal colorectal mucosa (52.13 ± 42.48), adenoma (96.11 ± 78.82), cancer (173.91 ± 61.20) showed a significant association with cancer versus normal (p < 0.0001), adenoma versus normal(p = 0.0312) and adenoma versus cancer(p = 0.0006). (C) Using Student's t-test, the mean ± SD of TRAIL-R2 expression in normal colorectal mucosa (24.57 ± 38.77), adenoma (59.17 ± 49.69) and cancer (115.63 ± 95.76) showed a significant association with cancer versus normal (p < 0.0001), adenoma versus normal(p = 0.0027) and adenoma versus cancer(p < 0.0001).
Association of TRAIL, TRAIL-R1 and TRAIL-R2 with clinico-pathological parameters
TRAIL-R1 was associated with histology subtype of adenocarcinomas (p = 0.0355), early AJCC stage (p = 0.0251) and a trend of higher expression was noted with well-differentiated tumors (p = 0.0887). No association was seen with age, gender and tumor site (Table 1). Similarly, TRAIL-R2 was associated with histology subtype of adenocarcinomas (p = 0.0010, tumors in the left colon (p = 0.0009) and a significantly higher expression was noted with well-differentiated tumors (p < 0.0001). No associations were seen with age, gender and tumor stage (Table 2). TRAIL ligand expression was not associated with any of the clinico-pathological parameters (see Additional File 1 Table S1).
Association of TRAIL, TRAIL-R1 and TRAIL-R2 with KRAS mutations and KRAS splice variants KRAS4A and KRAS4B
TRAIL-R2 expression was significantly higher in the CRC subset lacking KRAS mutations as compared to CRC with KRAS mutations (p = 0.0481; Figure 4). Interestingly, both TRAIL-R1(p < 0.0001) and TRAIL-R2(p < 0.0001) showed a highly significant association with the pro-apoptotic KRAS4A isoform. However, TRAIL-R1 expression did not show any correlations with KRAS mutations and KRAS4B isoform (Table 1 & 2). TRAIL expression did not show any associations with KRAS mutations or expression of KRAS splice variants (see Additional File 1 Table S1).
Box plot analysis of TRAIL-R1 and TRAIL-R2 expression in KRAS mutated versus non-mutated colorectal patients. (A) Using Student's t-test, the mean ± SD of TRAIL-R1 expression in KRAS mutated (181.28 ± 61.33)and non mutated (181.25 ± 63.92).However, this difference in expression was not statistically significant (p = 0.9824). (B Using Student's t-test, the mean ± SD of TRAIL-R2 expression in KRAS mutated (113.36 ± 92.45) and not mutated (140.00 ± 92.26).There was statistically significant difference in expression (p = 0.0466).
Associations of TRAIL, TRAIL-R1 and TRAIL-R2 with microsatellite instability, cleaved caspase 3 and p27kip1
p27kip1 expression was significantly associated with both TRAIL-R1 (p = 0.0024) and TRAIL-R2 (p < 0.0001; Table 1 & 2). CRC with expression of TRAIL-R1 but not TRAIL-R2 or TRAIL also showed expression of cleaved caspase3 (p = 0.0011). Although TRAIL-R2 was associated with a phenotype of microsatellite stable (MSI-S/L) tumors (p = 0.0003), no associations were seen between TRAIL-R1 or TRAIL and microsatellite instability status.
Overall survival in all patients, selected stage subgroups and combination groups of TRAIL receptors
CRC with low TRAIL-R1 expression also showed a poor 5 year overall survival of 53.9% as compared to 68.1% with high TRAIL-R1 expression (p = 0.0124; Figure 5A). Similarly, CRC with low TRAIL-R2 expression also showed a poor 5 year overall survival of 57.6% as compared to 67.3% with high TRAIL-R2 expression (p = 0.0211; Figure 5B). TRAIL expression did not show any prognostic significance (p = 0.2901). To exclude that the observed prognostic difference were caused by classical prognostic factors of CRC, we performed a multivariate analysis (Cox proportional hazards) with histological subtype, tumor grade, tumor stage, age, gender and microsatellite instability status as variables (Table 3). In the multivariate analysis, only TRAIL-R1 expression retained its significance. The relative risk was 1.84 (for low TRAIL-R1 expression (95% CI 1.10 - 3.02; p = 0.0273) and 6.56 for high stage group III-IV (95% CI 3.67- 12.78; p = < 0.0001). Thus, TRAIL-R1 was an independent prognostic marker in Middle Eastern Colorectal Carcinoma. To exclude that TRAIL-R1 is not a readout of KRAS-4A or p27 we reanalyzed our data and did a Cox proportional hazards model where we included age, gender, Stage, Grade, KRAS-4A, p27 and TRAIL-R1 expression (see Additional File 1 Table S2). In a Cox proportional Hazards model, the independent prognostic significance of TRAIL-R1 was weakened (p = 0.0883). However, AJCC stage, p27 and KRAS4A still remained independent prognostic markers.
(A) Kaplan Meier survival analysis in CRC patients with over expression of Trail R1 had a better overall survival of 68.1% at 5 years as compared to 53.9% with reduced TRAIL-R1 expression (p = 0.0124). (B) CRC patients Trail R2 also showed a better overall survival as of 67.3% compared to 57.6% with reduced TRAIL-R2 expression ( p = 0.0211).
Although TRAIL-R1 expression was significantly more in early stage tumors, a vast majority of Stage III & IV tumors (82%) also showed TRAIL-R1 expression. Both TRAIL-R1 (p = 0.0060) and TRAIL-R2 (p = 0.0263) were associated with better outcome only in the advanced Stage group (III and IV; see Additional File 2). When stage II and III were taken together only TRAIL-R2 expression was associated with better overall survival (p = 0.0088); TRAIL-R1 expression was not significant (p = 0.2508; see Additional File 3). Co-expression of TRAIL-R1 and TRAIL-R2 was seen in 56.85% of the CRC( 191/336) and was associated with a good survival (p = 0.0107; see Additional File 4) which remained significant in multivariate analysis (Cox proportional hazards) with TRAIL-R1/R2 co-expression, tumor grade, tumor stage, age and gender as variables (see Additional File 1 Table S3).
TRAIL death receptors and response to adjuvant therapy
The availability of 220 CRC from affected individuals who had undergone adjuvant therapy: chemotherapy and/or radiotherapy, allowed us to investigate the possible impact of TRAIL-R1 on response to adjuvant therapy. For this analysis, we first stratified the individuals into two groups: (i) CRC patient who have received adjuvant therapy (n = 220), and (ii) CRC patient who have been treated by surgical resection only and have not received adjuvant therapy (n = 90). There was a statistically significant difference in survival between individuals with tumors with TRAIL-R1 overexpression versus those with reduced expression (p = 0.0033; see Additional File 5). To exclude that the observed prognostic difference was caused by classical prognostic factors of CRC we performed a multivariate analysis (Cox proportional hazards) with TRAIL-R1 expression, tumor grade, tumor stage, age and gender as variables (see Additional File 1 Table S4). We found that the prognostic value of TRAIL-R1 expression in adjuvant treated individuals was independent of these factors. Similarly, TRAIL-R2 expression was also associated with trend towards better outcome in the adjuvant treated CRC subgroup (p = 0.0998) but no association with outcome was seen in the group which did not receive adjuvant therapy.
Discussion
We conducted this study to examine the relations of TRAIL and it receptors: TRAIL-R1 and TRAIL-R2 with clinical, pathologic, molecular characteristics and patient survival in Saudi colorectal cancers. Expression of TRAIL-R1 or TRAIL-R2 was associated with a less aggressive phenotype characterized by an early AJCC stage and well-differentiated tumors. TRAIL-R2 expression was associated with microsatellite stable phenotype and with absence of KRAS mutations. TRAIL-R1 but not TRAIL-R2 was an independent prognostic marker for better survival.
Using immunohistochemistry, we have studied the expression of TRAIL and its receptors in Saudi CRC; incidence of TRAIL R1, TRAIL-R2 and TRAIL expression was 85.5%, 59.4% and 31.5% respectively. In agreement with earlier studies, we have also observed a progressive increase in expression of TRAIL and its receptors: TRAIL-R1 and TRAIL-R2 in colorectal carcinoma and noted a strong association of TRAIL -R1 or TRAIL-R2 expression with differentiation and an early stage. The prognostic implication of TRAIL receptor expression is the subject of intensive investigation as malignant cells are more sensitive to TRAIL- induced apoptosis than their benign counterparts are and this potentially affects the future management of patients [32–34]. Furthermore, our data indicates that high TRAIL-R1 expression was an independent prognostic marker for better survival in Saudi CRC patients. TRAIL-R2 was also associated significantly with better outcome but failed to remain significant in multivariate analysis. TRAIL-R1 expression was also associated with better outcome in the following subgroups: Stage III and IV (p = 0.0060) and CRC subgroup who received adjuvant therapy(p = 0.0033). To elucidate the role of TRAIL expression further analysis was done in the following subgroup: CRC subgroup with high co-expression of TRAIL and TRAIL-R1 and CRC subgroup with high co-expression of TRAIL and TRAIL-R2. Both these combination groups were not associated with outcome (data not shown). Thus, TRAIL ligand co-expression with TRAIL receptors does not influence the outcome.
These findings are in agreement with earlier studies by Starter et al [26] where TRAIL-R1 expression was associated with a better disease free survival in a cohort of 129 Stage II and III CRC [26]. Granci et al. [28] studied the TRAIL receptors TRAIL-R 1, -2, -3 and -4 expression by immunohistochemistry in metastatic stage IV CRC and found that concomitant low/mediumTRAIL-R1 and high TRAIL-R3 expression in primary CRC is significantly associated with a poor response to 5-FU-based first-line chemotherapy and with a shorter progression-free survival. Surprisingly, high TRAIL R1 was associated with worse disease free survival and overall survival in 376 CRC patients with Stage III [30]. Ullenhag et al. [29] analyzed FLICE inhibitory protein (c-FLIP) and TRAIL receptors(TRAIL-R1 and R-2) in 476 CRC of all Stage groups(I to IV): Overexpression of FLIPL, (the long form of FLICE inhibitory protein) but not TRAIL-R1 or TRAIL-R2, was an independent prognostic factor for shorter disease free survival. In an attempt to explain these conflicting results of TRAIL and its pro-apoptotic receptors in CRC, we offer the following explanations: a) differences and heterogeneity in samples studied: sample size, ethnic differences, different Stage groups, tumor site- colon or rectal tumors, type of treatment- surgery and/or chemo/radiotherapy; b) differences in scoring system could be another important reason for this difference. The varied effects of TRAIL signaling could be also attributed to the following factors: TRAIL resistance due to presence of decoy receptors [11], number, type and functionality of TRAIL receptors [7, 12] and intracellular anti apoptotic molecules like c-FLIP [35], IAP [36], Mcl-1 [37] and bcl2 [38].
Although TRAIL-R1 lost its statistical significance when included as a prognostic marker in multivariate analysis with p27 and KRAS4A (see Additional File 1 Table S2), this does not argue against the biological role of TRAIL-R1 in CRC as much as it reflects that p27 and KRAS4A are a more powerful predictor of clinical outcome of CRC than TRAIL-R1 expression. We can hypothesize that the TRAIL-R1 functions most effectively in the cells which show co-expression of p27kip1 in concordance with an earlier study [39]. Despite some studies that show a role of Ras signaling pathway in modulating the TRAIL system, studies on the KRAS isoforms - KRAS4A and KRAS 4B are lacking. Alternate approaches to modulate the expression of KRAS isoforms, a greater understanding of the role(s) that each oncoprotein plays in malignant transformation, including the signal transduction pathways affected, is crucial in the development of therapeutic approaches in cancer treatment, which include the use of drugs that target isoform-specific post-translational modifications [40] and of antisense oligonucleotides to modulate alternative splicing [41].
Oncogenic mutations such as ras may enhance expression of TRAIL receptors; potentially sensitizing these tumors to TRAIL based therapies [19–21]. TRAIL-based therapeutic strategies using TRAIL agonists could be used in cases of human colon cancers bearing RAS mutations. In a small cohort of 51 CRC, Oikonomou E et al.[42] have reported a much lower incidence of KRASG12/13 mutations(10%) and have concluded that there is clear correlation between these mutations(KRAS and BRAF) and upregulation of TRAIL-R1 and TRAIL-R2. Despite lack of statistical significance they have concluded that CRC with mutations in KRAS or BRAF gene had significantly upregulated both TRAIL death receptors. In our earlier study [43] KRAS gene mutations were seen in 80/285 CRC (28.1%) and were an independent prognostic marker for poor survival. Interestingly we have observed a significantly higher expression of TRAIL-R2 (p = 0.0481) in CRC subgroup lacking KRAS mutations(75.5%) as compared to the CRC subgroup with KRAS mutations(61.1%). In view of the recent findings of KRAS mutations and PIK3CA mutations contributing to resistance to EGFR inhibitors like Cetuximab,[44, 45] a better understanding of the TRAIL system with context to KRAS mutations might be useful. The KRAS gene has two alternative fourth exon variants that result from differential splicing and activating mutations affect both isoforms [46–48]. Studies in animals indicate that KRAS4A promotes apoptosis while KRAS4B inhibits it, and KRAS4B promotes differentiation [49, 50]. In our study [43], KRAS 4A a pro apoptotic isoform, in particular was found to be an independent prognostic marker for better survival in all CRC patients. Even in the CRC subgroup lacking KRAS mutations KRAS4A was associated with better survival. Furthermore, we have observed a highly significant association of KRAS4A and both the TRAIL receptors: TRAIL-R1(p < 0.0001) and TRAIL-R2(p < 0.0001). Considering the tight linkage between TRAIL-R1 and KRAS4A future studies should be conducted to understand the association between these markers.
In summary, our study shows high TRAIL-R1 expression to be an independent prognostic marker for better survival in colorectal cancer. High TRAIL-R1 or TRAIL-R2 expression was associated with a less aggressive phenotype characterized by early AJCC stage, well-differentiated tumors, microsatellite stable cancers, absence of KRAS mutations and expression of pro apoptotic molecules: KRAS4A, p27kip1 and cleaved caspase 3. Further work is needed to elucidate the biological significance of high TRAIL-R1 expression and better outcome, and to establish the association between TRAIL-R1 expression and response to therapy that targets this receptor. The biological effects of TRAIL in CRC models, its enhancement of chemosensitivity with standard chemotherapeutic agents and the effect of endogenous TRAIL receptor levels on survival make TRAIL an extremely attractive therapeutic target.
Materials and methods
Patient selection and tissue microarray construction
Four hundred forty eight patients with CRC diagnosed between 1990 and 2006 were selected from King Faisal Specialist Hospital and Research Centre. All CRC, 24 adenomas and 229 adjacent normal colorectal mucosa were analyzed in a tissue microarray format. Clinical and histopathological data were available for all these patients. Colorectal Unit, Department of Surgery, provided long-term follow-up data. From our cohort of 448 patients treatment details were available for 310 patients:220 patients received adjuvant therapy; 90 were treated by surgery alone and 138 patients were excluded as we could not retrieve treatment details. Patients with colon cancer underwent surgical colonic resection and those with rectal cancer underwent anterior resection or abdominoperineal resection. All node-positive colon cancers received 5-fluorouracil-based adjuvant chemotherapy. A vast majority of the rectal cancers received radiotherapy alone or chemoradiotherapy prior to surgery, followed by adjuvant chemotherapy after surgery. Fixation of tissues was done overnight with 10% neutral buffered formalin at the Pathology Laboratory of King Faisal Specialist Hospital and Research Centre, Riyadh. Tissue microarrays were constructed from formalin-fixed, paraffin-embedded colorectal carcinoma specimens as described previously [51]. One pathologist (PB) reviewed all tumors for grade and histological subtype. Institutional Review Board (IRB) of the King Faisal Specialist Hospital & Research Centre approved the study.
Immunohistochemistry (IHC)
Tissue microarray slides were processed and stained manually. The streptavidin-biotin peroxidase technique with diaminobenzidine as chromogen was applied. For antigen retrieval, Dako Target Retrieval Solution was used at a pH of 6 for TRAIL-R1 and pH of 9 for TRAIL-R2 was used, and the slides were microwaved at 750W for 5 minutes and then at 250W for 30 minutes. Primary antibodies used, their dilutions, and incidences are listed in Additional File 1 Table S5. The specificity of these antibodies for TRAIL and its receptors has been previously assessed by immunohistochemistry [28, 52], or by Western blot [53]. Endogenous peroxidase activity was quenched using 3% hydrogen peroxidase. Endogenous biotin was blocked and all slides were counterstained with hematoxylin, dehydrated, cleared, and cover slipped with premount. Only fresh cut slides were stained simultaneously to minimize the influence of slide ageing and maximize repeatability and reproducibility of the experiment. As controls, we used a tissue microarray control block comprising multiple cores from normal tissue from various sites, common epithelial cancers and colon cancer cell lines. Omission of the primary antibody also served as a negative control for TRAIL, TRAIL-R1 and -R2 staining.
Immunohistochemistry Assessment
TRAIL-R1, TRAIL-R2 and TRAIL expression was categorized by doing an H score [54, 55]. Each tissue microarray spot was assigned an intensity score from 0-3(I0, I1-3) and proportion of the tumor staining for that intensity was recorded as 5% increments from a range of 0-100(P0, P1-3). A final H score (range 0-300) was obtained by adding the sum of scores obtained for each intensity and proportion of area stained (H score = I1XP1+I2XP2+I3XP3). CRCs were grouped into two groups based on X-tile plots for TRAIL-R1: one with complete absence or reduced staining (H score = 0-110) and the other group showed over expression (H score > 110) depending on the H score. Similarly, X-tile plots were used to stratify the CRC cases into two groups for TRAIL-R2 and TRAIL. X-tile plots were constructed for assessment of biomarker and optimization of cut off points based on outcome as has been described earlier [56, 57]. For cleaved caspase-3 expression, we used the antibody clone C5A-1 from Cell signalling technologies as described previously [43]. CRCs were grouped into two groups based on X-tile plots: one with complete absence or reduced staining (H score = 0-9 for low cleaved caspase3); and the other group showed over expression (H score >9 for high cleaved caspase 3). Grading of p27 nuclear protein staining was based on proportion or percentage of cell nuclei staining and was semi quantified as high or low. Nuclear protein expression of epithelial cells only was scored as high if 50% or more of the nuclei were stained or low if < 50% were stained as described previously [58]. This scoring criteria has been used earlier [59].
Mutational analysis of the KRAS gene
KRAS mutations were done as described earlier [43]. Briefly the step-down cycling condition was used for the detection of exon 1 mutation of the KRAS gene. After 10 minutes denaturing at 95°C, the PCR was run with each temperature for 1 min at five step-down steps, for two cycles each. The denaturing temperature was 95°C and the extension temperature was 72°C for each step, with an annealing temperature of 66°C, 64°C, 62°C, 60°C, and 58°C from the first to the last step. The PCR was finally run at 95°C, 58°C, and 72°C, each for 1 min for 35 cycles, followed by an elongation at 72°C for 5 min. The PCR products were subsequently subjected to direct sequencing PCR with BigDye terminator V 3.0 cycle sequencing reagents (Applied Biosystems, Foster City, CA, USA). The samples were finally analysed on an ABI PRISM 3100xl Genetic Analyzer (Applied Biosystems).
Microsatellite instability
Allelic imbalances were measured by performing microsatellite analysis on all matched normal and tumor tissue by PCR amplification as described previously [60]. A reference panel of five pairs of microsatellite primers, comprising two mononucleotide microsatellites (BAT25, BAT26) and three dinucleotide microsatellites (DS123, D5S346 and D17S250) were used to determine tumor MSI status. Multiplex PCR was performed in a total volume of 25 ml using 50 ng of genomic DNA, 2.5 ml 10 Taq buffer, 1.5 ml MgCl2 (25 mM), 10 pmol of fluorescent-labeled primers, 0.05 ml dNTP (10 mM) and 0.2 ml Taq polymerase (1 U ml 1 ) (all reagents were from Qiagen Inc., Valencia, CA, USA). PCR was performed using an MJ Research PTC-200 thermocycler. The PCR conditions were as follows: after an initial 10 min denaturation step at 95 1C, 40 amplification cycles were performed consisting of 40 s at 95 1C, 40 s at 54 1C and a 1 min elongation step at 72 1C. Amplification was completed with a final extension step at 72 1C for 7 min. The fluorescent-labeled products were finally analysed on an ABI PRISM 3100 l Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Tumors were classified as MSI if at least two or more markers out of the five were unstable and as MSS if only one or none of the markers was unstable.
Statistical Analysis
The JMP8 (SAS Institute, Inc., Cary, NC) software package was used for data analyses. Survival curves were generated using the Kaplan-Meier method, with significance evaluated using the Mantel-Cox log-rank test. Risk ratio (relative risk for death) was calculated using the Cox Proportional Hazard model in both univariate and multivariate analyses. Comparisons between groups were made with the paired Student's t-test. Chi-square tests were used to examine relationship between nominal variables. The limit of significance for all analyses was defined as a p-value of 0.05.
References
Al-Eid HS AS: Cancer Incidence Report Saudi Arabia 1999-200. 2004, May 2004, King Faisal Specialist Hospital and Research Centre,
Oldenhuis CN, Oosting SF, Gietema JA, de Vries EG: Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer. 2008, 44: 946-953. 10.1016/j.ejca.2008.03.006
Lowe SW, Lin AW: Apoptosis in cancer. Carcinogenesis. 2000, 21: 485-495. 10.1093/carcin/21.3.485
Nagata S: Apoptosis by death factor. Cell. 1997, 88: 355-365. 10.1016/S0092-8674(00)81874-7
Ghobrial IM, Witzig TE, Adjei AA: Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin. 2005, 55: 178-194. 10.3322/canjclin.55.3.178
Qiao L, Wong BC: Targeting apoptosis as an approach for gastrointestinal cancer therapy. Drug Resist Updat. 2009, 12: 55-64. 10.1016/j.drup.2009.02.002
LeBlanc HN, Ashkenazi A: Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003, 10: 66-75. 10.1038/sj.cdd.4401187
Pukac L, Kanakaraj P, Humphreys R, Alderson R, Bloom M, Sung C, Riccobene T, Johnson R, Fiscella M, Mahoney A: HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody, induces cell death in multiple tumour types in vitro and in vivo. Br J Cancer. 2005, 92: 1430-1441. 10.1038/sj.bjc.6602487
Martin JH, Potthoff A, Ledig S, Cornberg M, Jandl O, Manns MP, Kubicka S, Flemming P, Athmann C, Beil W, Wagner S: Effect of H. pylori on the expression of TRAIL, FasL and their receptor subtypes in human gastric epithelial cells and their role in apoptosis. Helicobacter. 2004, 9: 371-386. 10.1111/j.1083-4389.2004.00269.x
Kuraoka K, Matsumura S, Sanada Y, Nakachi K, Imai K, Eguchi H, Matsusaki K, Oue N, Nakayama H, Yasui W: A single nucleotide polymorphism in the extracellular domain of TRAIL receptor DR4 at nucleotide 626 in gastric cancer patients in Japan. Oncol Rep. 2005, 14: 465-470.
Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM: An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997, 277: 815-818. 10.1126/science.277.5327.815
Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI: Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997, 277: 818-821. 10.1126/science.277.5327.818
Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, Blenis J, Tschopp J: TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000, 2: 241-243. 10.1038/35008667
Zhang XD, Franco A, Myers K, Gray C, Nguyen T, Hersey P: Relation of TNF-related apoptosis-inducing ligand (TRAIL) receptor and FLICE-inhibitory protein expression to TRAIL-induced apoptosis of melanoma. Cancer Res. 1999, 59: 2747-2753.
Pai SI, Wu GS, Ozoren N, Wu L, Jen J, Sidransky D, El-Deiry WS: Rare loss-of-function mutation of a death receptor gene in head and neck cancer. Cancer Res. 1998, 58: 3513-3518.
Lee SH, Shin MS, Kim HS, Lee HK, Park WS, Kim SY, Lee JH, Han SY, Park JY, Oh RR: Alterations of the DR5/TRAIL receptor 2 gene in non-small cell lung cancers. Cancer Res. 1999, 59: 5683-5686.
Lee SH, Shin MS, Kim HS, Lee HK, Park WS, Kim SY, Lee JH, Han SY, Park JY, Oh RR: Somatic mutations of TRAIL-receptor 1 and TRAIL-receptor 2 genes in non-Hodgkin's lymphoma. Oncogene. 2001, 20: 399-403. 10.1038/sj.onc.1204103
Shin MS, Kim HS, Lee SH, Park WS, Kim SY, Park JY, Lee JH, Lee SK, Lee SN, Jung SS: Mutations of tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and receptor 2 (TRAIL-R2) genes in metastatic breast cancers. Cancer Res. 2001, 61: 4942-4946.
Wang Y, Engels IH, Knee DA, Nasoff M, Deveraux QL, Quon KC: Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell. 2004, 5: 501-512. 10.1016/S1535-6108(04)00113-8
Nesterov A, Nikrad M, Johnson T, Kraft AS: Oncogenic Ras sensitizes normal human cells to tumor necrosis factor-alpha-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2004, 64: 3922-3927. 10.1158/0008-5472.CAN-03-2219
Drosopoulos KG, Roberts ML, Cermak L, Sasazuki T, Shirasawa S, Andera L, Pintzas A: Transformation by oncogenic RAS sensitizes human colon cells to TRAIL-induced apoptosis by up-regulating death receptor 4 and death receptor 5 through a MEK-dependent pathway. J Biol Chem. 2005, 280: 22856-22867. 10.1074/jbc.M412483200
Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O'Hagan R, Pantginis J, Zhou H: Essential role for oncogenic Ras in tumour maintenance. Nature. 1999, 400: 468-472. 10.1038/22788
Li W, Zhu T, Guan KL: Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J Biol Chem. 2004, 279: 37398-37406. 10.1074/jbc.M405730200
Plattner R, Gupta S, Khosravi-Far R, Sato KY, Perucho M, Der CJ, Stanbridge EJ: Differential contribution of the ERK and JNK mitogen-activated protein kinase cascades to Ras transformation of HT1080 fibrosarcoma and DLD-1 colon carcinoma cells. Oncogene. 1999, 18: 1807-1817. 10.1038/sj.onc.1202482
Hoshino R, Chatani Y, Yamori T, Tsuruo T, Oka H, Yoshida O, Shimada Y, Ari-i S, Wada H, Fujimoto J, Kohno M: Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene. 1999, 18: 813-822. 10.1038/sj.onc.1202367
Strater J, Hinz U, Walczak H, Mechtersheimer G, Koretz K, Herfarth C, Moller P, Lehnert T: Expression of TRAIL and TRAIL receptors in colon carcinoma: TRAIL-R1 is an independent prognostic parameter. Clin Cancer Res. 2002, 8: 3734-3740.
Koornstra JJ, Jalving M, Rijcken FE, Westra J, Zwart N, Hollema H, de Vries EG, Hofstra RW, Plukker JT, de Jong S, Kleibeuker JH: Expression of tumour necrosis factor-related apoptosis-inducing ligand death receptors in sporadic and hereditary colorectal tumours: potential targets for apoptosis induction. European Journal of Cancer. 2005, 41: 1195-1202. 10.1016/j.ejca.2005.02.018
Granci V, Bibeau F, Kramar A, Boissiere-Michot F, Thezenas S, Thirion A, Gongora C, Martineau P, Del Rio M, Ychou M: Prognostic significance of TRAIL-R1 and TRAIL-R3 expression in metastatic colorectal carcinomas. Eur J Cancer. 2008, 44: 2312-2318. 10.1016/j.ejca.2008.06.042
Ullenhag GJ, Mukherjee A, Watson NF, Al-Attar AH, Scholefield JH, Durrant LG: Overexpression of FLIPL is an independent marker of poor prognosis in colorectal cancer patients. Clin Cancer Res. 2007, 13: 5070-5075. 10.1158/1078-0432.CCR-06-2547
van Geelen CM, Westra JL, de Vries EG, Boersma-van Ek W, Zwart N, Hollema H, Boezen HM, Mulder NH, Plukker JT, de Jong S: Prognostic significance of tumor necrosis factor-related apoptosis-inducing ligand and its receptors in adjuvantly treated stage III colon cancer patients. J Clin Oncol. 2006, 24: 4998-5004. 10.1200/JCO.2006.06.8809
Koornstra JJ, Kleibeuker JH, van Geelen CM, Rijcken FE, Hollema H, de Vries EG, de Jong S: Expression of TRAIL (TNF-related apoptosis-inducing ligand) and its receptors in normal colonic mucosa, adenomas, and carcinomas. J Pathol. 2003, 200: 327-335. 10.1002/path.1364
Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA: Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995, 3: 673-682. 10.1016/1074-7613(95)90057-8
Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A: Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996, 271: 12687-12690. 10.1074/jbc.271.22.12687
Hague A, Hicks DJ, Hasan F, Smartt H, Cohen GM, Paraskeva C, MacFarlane M: Increased sensitivity to TRAIL-induced apoptosis occurs during the adenoma to carcinoma transition of colorectal carcinogenesis. Br J Cancer. 2005, 92: 736-742. 10.1038/sj.bjc.6602387
Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C: Inhibition of death receptor signals by cellular FLIP. Nature. 1997, 388: 190-195. 10.1038/40657
Deveraux QL, Takahashi R, Salvesen GS, Reed JC: X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997, 388: 300-304. 10.1038/40901
Wirth T, Kuhnel F, Fleischmann-Mundt B, Woller N, Djojosubroto M, Rudolph KL, Manns M, Zender L, Kubicka S: Telomerase-dependent virotherapy overcomes resistance of hepatocellular carcinomas against chemotherapy and tumor necrosis factor-related apoptosis-inducing ligand by elimination of Mcl-1. Cancer Res. 2005, 65: 7393-7402. 10.1158/0008-5472.CAN-04-3664
Fulda S, Meyer E, Debatin KM: Inhibition of TRAIL-induced apoptosis by Bcl-2 overexpression. Oncogene. 2002, 21: 2283-2294. 10.1038/sj.onc.1205258
Garofalo M, Quintavalle C, Di Leva G, Zanca C, Romano G, Taccioli C, Liu CG, Croce CM, Condorelli G: MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene. 2008, 27: 3845-3855. 10.1038/onc.2008.6
Adjei AA: Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001, 93: 1062-1074. 10.1093/jnci/93.14.1062
Sazani P, Kole R: Therapeutic potential of antisense oligonucleotides as modulators of alternative splicing. J Clin Invest. 2003, 112: 481-486.
Oikonomou E, Kosmidou V, Katseli A, Kothonidis K, Mourtzoukou D, Kontogeorgos G, Andera L, Zografos G, Pintzas A: TRAIL receptor upregulation and the implication of KRAS/BRAF mutations in human colon cancer tumors. Int J Cancer. 2009, 125: 2127-2135. 10.1002/ijc.24613
Abubaker J, Bavi P, Al-Haqawi W, Sultana M, Al-Harbi S, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F: Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009, 219: 435-445. 10.1002/path.2625
Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M: KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006, 66: 3992-3995. 10.1158/0008-5472.CAN-06-0191
Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L: PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009, 69: 1851-1857. 10.1158/0008-5472.CAN-08-2466
Malumbres M, Barbacid M: RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003, 3: 459-465. 10.1038/nrc1097
Capon DJ, Seeburg PH, McGrath JP, Hayflick JS, Edman U, Levinson AD, Goeddel DV: Activation of Ki-ras2 gene in human colon and lung carcinomas by two different point mutations. Nature. 1983, 304: 507-513. 10.1038/304507a0
Welman A, Burger MM, Hagmann J: Structure and function of the C-terminal hypervariable region of K-Ras4B in plasma membrane targetting and transformation. Oncogene. 2000, 19: 4582-4591. 10.1038/sj.onc.1203818
Plowman SJ, Arends MJ, Brownstein DG, Luo F, Devenney PS, Rose L, Ritchie AM, Berry RL, Harrison DJ, Hooper ML, Patek CE: The K-Ras 4A isoform promotes apoptosis but does not affect either lifespan or spontaneous tumor incidence in aging mice. Exp Cell Res. 2006, 312: 16-26. 10.1016/j.yexcr.2005.10.004
Patek CE, Arends MJ, Rose L, Luo F, Walker M, Devenney PS, Berry RL, Lawrence NJ, Ridgway RA, Sansom OJ, Hooper ML: The pro-apoptotic K-Ras 4A proto-oncoprotein does not affect tumorigenesis in the ApcMin/+ mouse small intestine. BMC Gastroenterol. 2008, 8: 24- 10.1186/1471-230X-8-24
Bavi P, Jehan Z, Atizado V, Al-Dossari H, Al-Dayel F, Tulbah A, Amr SS, Sheikh SS, Ezzat A, El-Solh H: Prevalence of fragile histidine triad expression in tumors from saudi arabia: a tissue microarray analysis. Cancer Epidemiol Biomarkers Prev. 2006, 15: 1708-1718. 10.1158/1055-9965.EPI-05-0972
Sanlioglu AD, Dirice E, Elpek O, Korcum AF, Balci MK, Omer A, Griffith TS, Sanlioglu S: High levels of endogenous tumor necrosis factor-related apoptosis-inducing ligand expression correlate with increased cell death in human pancreas. Pancreas. 2008, 36: 385-393. 10.1097/MPA.0b013e318158a4e5
Tomek S, Koestler W, Horak P, Grunt T, Brodowicz T, Pribill I, Halaschek J, Haller G, Wiltschke C, Zielinski CC, Krainer M: Trail-induced apoptosis and interaction with cytotoxic agents in soft tissue sarcoma cell lines. Eur J Cancer. 2003, 39: 1318-1329. 10.1016/S0959-8049(03)00227-2
McCarty KS, Miller LS, Cox EB, Konrath J, McCarty KS: Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985, 109: 716-721.
Uddin S, Siraj AK, Al-Rasheed M, Ahmed M, Bu R, Myers JN, Al-Nuaim A, Al-Sobhi S, Al-Dayel F, Bavi P: Fatty acid synthase and AKT pathway signaling in a subset of papillary thyroid cancers. J Clin Endocrinol Metab. 2008, 93: 4088-4097. 10.1210/jc.2008-0503
Camp RL, Dolled-Filhart M, Rimm DL: X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004, 10: 7252-7259. 10.1158/1078-0432.CCR-04-0713
McCarthy MM, Sznol M, DiVito KA, Camp RL, Rimm DL, Kluger HM: Evaluating the expression and prognostic value of TRAIL-R1 and TRAIL-R2 in breast cancer. Clin Cancer Res. 2005, 11: 5188-5194. 10.1158/1078-0432.CCR-05-0158
Uddin S, Ahmed M, Bavi P, El-Sayed R, Al-Sanea N, AbdulJabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Hussain AR, Al-Kuraya KS: Bortezomib (Velcade) induces p27Kip1 expression through S-phase kinase protein 2 degradation in colorectal cancer. Cancer Res. 2008, 68: 3379-3388. 10.1158/0008-5472.CAN-07-6109
Shapira M, Ben-Izhak O, Bishara B, Futerman B, Minkov I, Krausz MM, Pagano M, Hershko DD: Alterations in the expression of the cell cycle regulatory protein cyclin kinase subunit 1 in colorectal carcinoma. Cancer. 2004, 100: 1615-1621. 10.1002/cncr.20172
Abubaker J, Bavi P, Al-Harbi S, Ibrahim M, Siraj AK, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F: Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. Oncogene. 2008, 27: 3539-3545. 10.1038/sj.onc.1211013
Acknowledgements
We thank Valorie Balde and Hassan Al Dossarie for their skilful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PB designed research, performed experiments, analyzed data, and wrote the paper; SP performed validation experiments and helped in writing the paper; JA performed experiments and analyzed data; ZQ & TG collected and analyzed data; performed statistical analysis FA provided archival pathology material, review of histopathology slides; NA, AA, LAH and SA provided fresh tissue samples, clinical data and reviewed the manuscript, SU analyzed data and helped in writing the paper; KSA designed research, analyzed data, and wrote the paper.
All authors read and approved the final manuscript.
Electronic supplementary material
12943_2010_718_MOESM1_ESM.DOC
Additional file 1: Additional file 1. Table S1. Clinico-pathological characteristics and TRAIL expression in patients with colorectal carcinoma. Table S2. Cox regression analysis for overall survival of colorectal carcinoma patients: TRAIL-R1, KRAS4A and p27kip1. Table S3. Trail-R1 & R2 co-expression: Cox regression analysis for overall survival of patients with colorectal carcinoma. Table S4. Trail R1 in Adjuvant treated Group: Cox regression analysis for overall survival of patients with colorectal carcinoma. Table S5. Antibodies used for tissue micro array Immunohistochemical analysis. (DOC 130 KB)
12943_2010_718_MOESM2_ESM.TIFF
Additional file 2: Additional file 2. Prognostic significance of TRAIL-R1 & TRAIL-R2 in early and late stage CRC and Kaplan Meier survival analysis. [A] In the early Stage subgroup (I and II) CRC patients TRAIL-R1 expression was not associated with prognostic outcome (p = 0.4703). [B] In the advanced Stage subgroup(III and IV) CRC patients with over expression of TRAIL-R1(n = 157) had a better overall survival of 48.8% at 5 years as compared to 26.0% with reduced TRAIL-R1 expression (n = 30; p = 0.0060). [C] In the early Stage subgroup (I and II) CRC patients TRAIL-R2 expression was not associated with prognostic outcome (p = 0.5613). [D] In the advanced Stage subgroup(III and IV) CRC patients with over expression of TRAIL-R2 (n = 112) had a better overall survival of 50.0% at 5 years as compared to 35.2% with reduced TRAIL-R2 expression (n = 70; p = 0.0263). (TIFF 1 MB)
12943_2010_718_MOESM3_ESM.TIFF
Additional file 3: Additional file 3. Prognostic significance of TRAIL-R1 & R2 in CRC with Stage II and III and Kaplan Meier survival analysis. [A] In the Stage subgroup(II and III) CRC patients with over expression of TRAIL-R1(n = 226) had a better overall survival of 67.6% at 5 years as compared to 63.1% with reduced TRAIL-R1 expression (n = 41; p = 0.2508). [B] In the Stage subgroup(II and III) CRC patients with over expression of TRAIL-R2(n = 149) had a better overall survival of 71.3% at 5 years as compared to 57.3% with reduced TRAIL-R2 expression (n = 105; p = 0.0088). (TIFF 1 MB)
12943_2010_718_MOESM4_ESM.TIFF
Additional file 4: Additional file 4. Prognostic significance of co-expression of TRAIL receptors: TRAIL-R1 and TRAIL-R2 in CRC and Kaplan Meier survival analysis. The CRC subgroup with overexpression of TRAIL-R1 and TRAIL-R2 (n = 191) had a better overall survival of 69.5% at 5 years as compared to Reduced TRAIL-R1 and TRAIL-R2 expression of 57.9% (n = 145; p = 0.0107). (TIFF 1 MB)
12943_2010_718_MOESM5_ESM.TIFF
Additional file 5: Additional file 5. Prognostic significance of TRAIL-R1 & TRAIL-R2 in CRC based on adjuvant therapy and Kaplan Meier survival analysis. [A] CRC patients with over expression of TRAIL-R1 in adjuvant treated group had a better overall survival of 70.6% at 5 years as compared to 48.4% with reduced TRAIL-R1 expression (n = 203; p = 0.0033). [B] In the non-adjuvant treated group TRAIL-R1 expression was not associated with prognostic outcome (p = 0.8801). [C] CRC patients with over expression of TRAIL-R2 in adjuvant treated had a better overall survival of 73.5% at 5 years as compared to 46.5% with reduced TRAIL-R2 expression (n = 168; p = 0.0998). [D] In the non-adjuvant treated group TRAIL-R2 expression was not associated with prognostic outcome (p = 0.8069). (TIFF 1 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bavi, P., Prabhakaran, S.E., Abubaker, J. et al. Prognostic significance of TRAIL death receptors in Middle Eastern colorectal carcinomas and their correlation to oncogenic KRAS alterations. Mol Cancer 9, 203 (2010). https://doi.org/10.1186/1476-4598-9-203
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-4598-9-203