Abstract
Background
Hypoxia is a condition of low oxygen tension occurring in the tumor microenvironment and it is related to poor prognosis in human cancer. To examine the relationship between hypoxia and neuroblastoma, we generated and tested an in vitro derived hypoxia gene signature for its ability to predict patients' outcome.
Results
We obtained the gene expression profile of 11 hypoxic neuroblastoma cell lines and we derived a robust 62 probesets signature (NB-hypo) taking advantage of the strong discriminating power of the l1-l2 feature selection technique combined with the analysis of differential gene expression. We profiled gene expression of the tumors of 88 neuroblastoma patients and divided them according to the NB-hypo expression values by K-means clustering. The NB-hypo successfully stratifies the neuroblastoma patients into good and poor prognosis groups. Multivariate Cox analysis revealed that the NB-hypo is a significant independent predictor after controlling for commonly used risk factors including the amplification of MYCN oncogene. NB-hypo increases the resolution of the MYCN stratification by dividing patients with MYCN not amplified tumors in good and poor outcome suggesting that hypoxia is associated with the aggressiveness of neuroblastoma tumor independently from MYCN amplification.
Conclusions
Our results demonstrate that the NB-hypo is a novel and independent prognostic factor for neuroblastoma and support the view that hypoxia is negatively correlated with tumors' outcome. We show the power of the biology-driven approach in defining hypoxia as a critical molecular program in neuroblastoma and the potential for improvement in the current criteria for risk stratification.
Similar content being viewed by others
Background
Neuroblastoma is the most common pediatric solid tumor, deriving from ganglionic lineage precursors of the sympathetic nervous system [1, 2]. It is diagnosed during infancy and shows notable heterogeneity with regard to histology and clinical behavior [3], ranging from rapid progression associated with metastatic spread and poor clinical outcome to spontaneous, or therapy-induced regression into benign ganglioneuroma [4]. Clinical and molecular risk factors which correlate with prognosis include age at diagnosis, stage, histology, chromosomal aberrations, and amplification of the N-myc proto-oncogene (MYCN), which is the most typical genetic feature of advanced-stage neuroblastoma [3, 5, 6]. MYCN amplification correlates with a more malignant course of the disease, angiogenesis, resistance to therapy, and poor clinical outcome [5, 7–9], suggesting that it may be a progression-related event and a potential therapeutic target [3].
In particular, high expression of the proangiogenic cytokine, vascular endothelial growth factor (VEGF), is a marker of poor prognosis in neuroblastoma tumors [10, 11].
Tumor microenvironment is intimately connected with the evolution of the disease. In particular, hypoxia, a condition of low oxygen tension occurring in poorly vascularized areas, has a profound effects on tumor cell growth, susceptibility to apoptosis, and resistance to radio- and chemotherapy [12, 13]. The response to hypoxia is associated with changes in gene expression [14–16]. Hypoxia activates, among others, hypoxia-inducible transcription factors (HIF-1α and HIF-2α) [12, 17, 18], which transactivate the hypoxia-responsive element (HRE) present in the promoter of many genes encoding angiogenic, metabolic and metastatic factors [13, 19, 20] and contribute to the acquisition of the tumor aggressive phenotype [13, 14, 21].
There is little information on the relationship among hypoxia, tumor phenotypes and clinical parameters in neuroblastoma. Rapidly expanding neuroblastoma tumors present hypoxic areas and metastasize to bone marrow [22] and it has been reported that HIF-2α is significantly correlated with a poor prognosis [23]. Neuroblastoma cells adaptation to hypoxia activates a gene expression program consistent with the pro-metastatic events [24]. Furthermore, hypoxia causes dedifferentiation in vitro and in vivo suggesting a novel mechanism for the selection of highly malignant neuroblastoma cells with stem-cell characteristics [25].
Every cell type is bound to respond to hypoxia and the gene expression profile of the tumor will have, to some extent, the footprint of hypoxia which, in turn, might be a prognostic indicator depending on the proportion of the hypoxic tissue. Therefore, clues to the prognosis of neuroblastoma might be reflected at the time of surgical removal in the pattern of hypoxic gene expression in the primary tumor. Definition, detection and analysis of hypoxia induced gene expression have the potential of leading to interesting and useful molecular predictors of neuroblastoma progression.
We investigated the prognostic potential of hypoxia induced genes in neuroblastoma tumors. A biology-driven approach was chosen to define the hypoxia signature. We performed a systematic analysis of the transcriptome of neuroblastoma cell lines cultured under hypoxic or normoxic conditions and applied a rigorous framework to derive a robust 62 probesets neuroblastoma hypoxia signature (NB-hypo). We show that the NB-hypo have a strong predictive power in the multivariate Cox regression model that includes the classical prognostic factors. This signature can also stratify a heterogeneous subgroup of patients with MYCN not amplified tumors. In conclusion, we demonstrate that NB-hypo is an independent risk factor and provide evidences of the power of the biology-driven approach to study the role of molecular programs in human tumors.
Materials and methods
Patients
The clinical characteristics of the 88 patients used in this study are listed in Table 1. The tumor samples were obtained at the time of diagnosis. The median follow-up time for patients in this study is 3.5 years (range = 0-16 years) and the median age at the diagnosis is 1 year (range = 0-14). Analysis of the predictive value of the current European risk factors MYCN status, International Neuroblastoma Staging System (INSS), and age was performed by Kaplan-Meier curves and log-rank test for overall survival (OS). All the prognostic factors stratified the patients in our cohort (p < 0.0001). Overall survival was 72.6% for the MYCN not amplified patients, compared with 10.4% for the MYCN amplified patients (HR = 4.30, 95% CI 3.66-30.69). For INSS, OS curves were characterized by a survival rate of 93.6% for the stage 1, 2, 3, and 4S patients, compared with a survival rate of 25.4% for the stage 4 patients (HR = 16.65, 95% CI 5.37-23.74). Overall survival was 100% for the patients with age at the diagnosis < 1 year, compared with 46.6% for the patients with age at the diagnosis > 1 year (HR and 95% CI undefined).
Cell lines and culture conditions
The human neuroblastoma cell lines GI-LI-N, ACN, GI-ME-N, IMR-32, LAN-1, SK-N-BE(2)C, SK-N-F1, and SK-N-SH were purchased from the Interlab Cell Line Collection while SHEP-2, SHEP-21N over-expressing MYCN, and SHEP-21N not over-expressing MYCN were kindly provided by Dr. M. Schwab (Division of Tumour Genetics, German Cancer Research Centre, Heidelberg, Germany). The cell lines were cultured in RPMI 1640 (Euroclone Ltd., Celbio, Milan, Italy), supplemented with 10% heat-inactivated fetal bovine serum (Sigma, Milan Italy), 2 mmol/L L-glutamine, 10 mM Hepes, 100 units/mL penicillin, and 100 μg/mL streptomycin (Euroclone Ltd), at 37°C in a humidified incubator containing 20% O2, 5% CO2, and 75% N2. Hypoxic conditions (1% O2) were achieved by culturing the cells in an anaerobic workstation incubator (BUG BOX, Jouan, ALC International S.r.l., Cologno Monzese, Milan, Italy) flushed with a gas mixture containing 1% O2, 5% CO2, and balanced N2 at 37°C in a humidified atmosphere. Oxygen tension in the medium was measured with a portable, trace oxygen analyzer (Oxi 315i/set, WTW; VWR International, Milan, Italy).
RNA extraction and microarray experiments
Total RNA was extracted using Trizol (Invitrogen Life technologies, Irvine, CA) according to the manufacturer's instructions. RNA was resuspended in diethyl pyrocarbonate-treated H2O (DEPC water), the physical quality control of RNA integrity was carried out by electrophoresis using Agilent Bioanalyzer 2100 (Agilent Technologies Waldbronn, Germany) and quantified by NanoDrop (NanoDrop Technologies Wilmington, Delawere USA). Total RNA was reverse transcribed into cDNA and biotin labeled according to the Affymetrix instructions (Affymetrix, SantaClara, CA). Biotin-labeled cRNA was cleaned up with the Qiagen RNeasy Mini kit and ethanol precipitation, checked for quality with Agilent Bioanalyzer 2100, and fragmented by incubation at 94°C for 35 min in 40 mmol/L Tris-acetate (pH 8.1), 100 mmol/L potassium acetate, and 30 mmol/L magnesium acetate. Fragmented cRNA was used for hybridization to Affymetrix HG-U133 Plus 2.0 arrays. GeneChips were scanned using an Affymetrix GeneChip Scanner 3000. The microarray data were released into the GEO-database (accession number GSE17714). Expression values were quantified, and array quality control was performed using the statistical algorithms implemented in Affymetrix Microarray Suite 5.0. All microarrays were examined for surface defects, grid placement, and background intensity. All the global microarray quality metrics that are summarized in Affymetrix report files were within normal ranges for all the samples. The scale factors (SF) for all the hybridizations were within 1 SD of the mean (SF 1-3). All samples had 3'/5' Gapdh and 3'/5' Actb ratios below the maximum threshold of 3. To asses RNA integrity, "RNA digestion plot" was performed. Quality control and RNA digestion plot were used as implemented in the R package affy. Array signals were scaled to an average intensity of 500, and the resulting data were normalized as follows: 1) per microarray sample, dividing the raw data by 50th percentile of all measurements; and 2) per gene, by dividing the normalized data by the median of the expression level for the gene in all samples.
Data analysis and statistics of gene expression in cell lines
To define the hypoxia gene signature, we independently applied the l1-l2 regularization with double optimization and the evaluation of differential gene expression to the dataset of 11 neuroblastoma cell lines cultured in hypoxic or normoxic conditions. The signature was defined as the intersection of the probesets selected by the two methods.
l1-l2 regularization technique was proposed in the context of statistical learning as a feature selection method [26] and successfully applied in the field of computational biology [27]. l1-l2 regularization is a supervised multivariate analysis which statistical significance and model selection is performed within double selection bias free cross-validation loops. Detailed descriptions of the application of l1-l2 algorithm and the method applied to select optimal values of the parameters for signature definition were previously reported [28]. Briefly, the output of the l1-l2 regularization algorithm depends on one free parameter εthat governs the amount of correlation allowed among the selected variables (probesets); the higher the ε, the more probesets are taken into account. The algorithm analyzed all the probesets on the chip simultaneously, thereby dealing with 54675-dim vectors. The system was characterized by a leave-one-out error of 10% and performed the validation loop producing 22 lists of probesets for each εvalue. A common list was obtained as the union of the 22 lists, with a frequency score counting how many times each probeset was selected by the algorithm in the 22 cross validation loops. In order to define the frequency threshold, the behavior of each ε curve was analyzed. The minimal list is obtained for values of ε equal to or lower than 1, whereas the largest list, which is correlation aware, is obtained for ε equal to 100. Thus, to include every probesets concurring in the identification of the hypoxia status, we set ε equal to 100.
In order to define the differentially expressed probesets, a fold change was calculated as the ratio between the expression level in the hypoxic and normoxic conditions for each individual cell line. The probesets that were modulated of at least 2-fold were selected and a t-test with Benjamini and Hochberg multiple hypothesis correction was applied. Only those probesets that were selected by both l1-l2 regularization algorithm and differentially expressed method were included in the NB-hypo.
Data analysis and statistics of gene expression in neuroblastoma tumors
The predictive value of the signature was assessed on the tumor expression dataset. The patients were divided into two clusters by applying K-means clustering, with euclidean distance and 100 iterations, on the normalized expression values of the NB-hypo.
We performed a t-test to assess the significance of the obtained clustering versus the random clustering distribution. A permutation test was performed in order to measure the stability of the obtained clusters. We sampled 300 times a random signature and repeated the clustering procedure, each time evaluating the misclassification distance between the obtained and random clusters. Hierarchical clustering analysis with complete linkage was performed by Cluster 3.0 software (Michael Eisen, Stanford University, California, USA). The dendrogram was visualized using TreeView 1.6 (Michael Eisen, Stanford University, California, USA).
The probability of overall survival (OS) and event-free survival (EFS) was calculated using Kaplan-Meier method, and the significance of the difference between Kaplan-Meier curves was calculated by the log-rank test using Prism 4.03 (GraphPad Software, Inc.).
Multivariate Cox proportional regression analysis was performed to evaluate the prognostic significance of NB-hypo and the currently used risk factors such as age at diagnosis (> 1 year vs. < 1 year), International Neuroblastoma Staging System (INSS) stage (stage 4 vs. not stage 4), and MYCN status (amplified vs. not amplified). Hazard ratios and 95% confidence interval for survival were calculated (StatPlus 4.7, AnalystSoft).
Results
The hypoxia signature of neuroblastoma cell lines
The neuroblastoma hypoxia signature was defined by the biology-driven approach outlined in Figure 1A. We utilized an in vitro experimental model consisting of 11 neuroblastoma cell lines, cultured under normoxic or hypoxic conditions for 18 hrs. RNA was extracted, processed and the gene expression profile was determined. To derive the hypoxia signature we applied the framework detailed in the Materials and Methods' section, which combines the discriminatory power of the l1 - l2 regularization algorithm and the biological strength of differentially expressed genes. The application of l1 - l2 algorithm to the neuroblastoma cell lines dataset defined a list of 137 probesets able to discriminate hypoxic from normoxic cell lines (Figure 1B). In the figure the multi dimensional model is projected on its 3 principal components. The differential expression analysis of hypoxic vs. normoxic cell lines identified 174 significant modulated probesets. We intersected the cluster of probesets originated from the l1 - l2 regularization with that created by the analysis of the differential expression, obtaining a 62 probesets signature that is referred to as NB-hypo. The 62 probesets correspond to 32 genes (Table 2) comprising mainly known hypoxia inducible genes providing indirect validation of our signature. The NB-hypo, which was generated only by objective criteria, was then applied to the gene expression profiles of tumors from neuroblastoma patients.
Definition of neuroblastoma hypoxia signature (NB-hypo). A, workflow of the biology-driven approach applied to define the hypoxia signature. The workflow is divided into three main blocks representing, from top to bottom, the experimental data collection, the functional genomics data analysis, and the evaluation of the NB-hypo as prognostic factor in neuroblastoma patients. B, principal components representation of the multivariate analysis performed on the 11 cell lines by l1 - l2 algorithm. This figure illustrates a 3-dimensional visualization of the dataset restricted to the selected probesets projected on their 3 principal components. Open squares (H) represent the cell lines in hypoxic status and the black circles (N) the corresponding cell lines in normoxic status.
Discriminating power of the NB-hypo
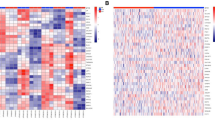
To study the prognostic value of the NB-hypo, we classified 88 neuroblastoma patients into two groups by applying a k-means clustering to the gene expression values of the 62 probesets (Figure 2). The significance of the clustering performance was assessed by permutation tests of 62 random probesets. The misclassification distance was calculated each time and the t-test (p < 0.001, data not shown) indicated that the NB-hypo significantly stratified patients into two clusters of 21 (cluster 1) and 67 (cluster 2) patients respectively. The expression levels of the probesets were grouped by hierarchical clustering and are represented in the heatmap in Figure 2. Cluster 1 consists of tumors in which the hypoxia probesets are highly expressed and stable whereas cluster 2 consists of tumors in which the expression levels are lower and less stable.
Heatmap of the 62 probesets in the 88 neuroblastoma tumors. The expression data for each individual probeset have been scaled and are represented by pseudo-colors in the heatmap. Red color corresponds to high level of expression and green color corresponds to low level of expression. The 88 patients (columns) were divided into two groups by k-means clustering. Cluster 1 consists of 21 patients and cluster 2 consists of 67 patients. The expression values of the 62 probesets were grouped by hierarchical clustering (rows). Hierarchical clustering dendrogram is on the left and the corresponding probesets on the right.
The survival in the resulting groups was analyzed by Kaplan-Meier curves, log-rank test for overall survival (OS) (Figure 3A) and event-free survival (EFS) (Figure 3B). The results show a significant separation (p < 0.001) of the two curves in both OS and EFS demonstrating that the NB-hypo can stratify patients into poor (cluster 1) and good prognosis (cluster 2) groups. OS curves are characterized by a survival rate of 73.2% for the patients with good prognosis, compared with a survival rate of 25.5% for the patients with poor prognosis (HR = 4.26, 95% CI 3.33-22.81). EFS curves are characterized by a survival rate of 67.7% for the patients with good prognosis, compared with a survival rate of 27.7% for the patients with poor prognosis (HR = 3.08, 95% CI 1.94-11.09). The latter belong to the class with highly expressed hypoxia-related probesets (Figure 2), suggesting that the hypoxic microenvironment takes part in the definition of the neuroblastoma aggressiveness.
Kaplan-Meier and log-rank analysis for 88 neuroblastoma patients. OS (A) and EFS (B) of patients classified according to the NB-hypo. OS (C) of patients classified according to 4 VEGF probesets. OS (D) of patients classified according to dendritic cells hypoxia signature. Solid and dashed curves represent good and poor prognosis patients, respectively. The p-value of the log-rank test is shown.
As a control, we tested the discriminating power of the four probesets characterizing VEGF, a gene represented in the NB-hypo and broadly up-regulated in hypoxic cells. The expression values of the four probesets were used to classify the 88 patients into two groups by k-means clustering (Figure 3C). VEGF alone was not capable of dividing the patients in statistically significant groups demonstrating the need for a complex signature.
To address the question of the relationship between effectiveness of the hypoxia signature and cell lineage, we tested the hypoxia signature of human dendritic cells [29]. The latter signature did not stratify the patients (Figure 3D) indicating that the prognostic value of the NB-hypo was dependent on the nature of the cell lines used for its definition.
To determine whether the NB-hypo is an independent risk predictor over the currently used risk factors, we created a multivariate Cox proportional hazard regression model using NB-hypo, age, INSS stage, and MYCN status. The results are shown in Table 3. INSS stage and NB-hypo have been found to be significant predictors (p < 0.01), with Hazard Ratios (HR) of 7.15 (95% CI, 2.08 to 24.64) and 2.73 (95% CI, 1.31 to 5.69) respectively, thus providing the evidence that hypoxia is a prognostic factor in neuroblastoma patients. There is no a significant correlation between INSS stage and NB-hypo. However, every stage 2 and stage 3 patients who survived and every stage 1 patients were classified by NB-hypo in the good prognosis group, while in the poor prognosis group 13 out of 21 (62%) were stage 4 patients (data not shown).
We studied the relationship between NB-hypo and other hypoxia signatures generated in different tumor systems in terms of overlapping, neuroblastoma patients' stratification and risk assessment. We considered the following hypoxia signatures: the hypoxia metagene described by Winter et al. [30] and redefined by Buffa et al. [31]; the VEGF signature by Hu et al. [32]; two early hypoxia signatures by Seigneuric et al. [33]. First, we investigated whether the 32 genes composing our signature are part of the considered hypoxia signatures. Table 4 shows the result of the overlap analysis. Few genes belonging to the NB-hypo are found in each of the other hypoxia signatures. Moreover, a total of 12 out of 32 genes are represented in at least one of the other signatures (genes in bold in Table 2). We then tested the predictive performance of the hypoxia signatures in stratifying neuroblastoma patients by performing survival curves analysis and multivariate Cox regression. The results are reported in Table 5 and demonstrate that most of these signatures fail to stratify the patients, with the exception of Winter's hypoxia signature. However it is not a significant predictor in the multivariate Cox analysis. These results demonstrate the effectiveness of the biology-driven approach applied to derive the neuroblastoma hypoxia signature and that hypoxia signatures derived from different tumor types are less effective than the one derived from the same tumor type for which they are conceived. In conclusion, we show that the NB-hypo is an independent and significant risk factor that can predict neuroblastoma patients' outcome.
NB-hypo as a prognostic factor in MYCN not amplified patients
MYCN amplification is a very important risk factor in neuroblastoma and correlates with unfavorable prognosis. However, within patients lacking MYCN amplification, the commonly used prognostic factors give little information regarding outcome [34]. Thus, it was important to determine the predictive power of the NB-hypo in MYCN not amplified tumors found in 72 out of 88 patients. We classified the neuroblastoma patients into two classes by applying k-means clustering to the gene expression values of the 62 probesets. The Kaplan-Meier curves and log-rank test for overall survival (OS) (Figure 4A) and event-free survival (EFS) (Figure 4B) are shown. The results demonstrate a significant separation of the two curves in both OS (p < 0.001) and EFS (p < 0.001) thereby identifying good and poor prognosis MYCN not amplified patients. The good prognosis cluster consists of 61 patients, whereas 11 patients belong to the poor prognostic cluster. OS curves are characterized by a survival rate of 81.4% for the patients with good prognosis, compared with a survival rate of 24.2% for the patients with poor prognosis (HR = 6.71, 95% CI 8.74-182.30). EFS curves are characterized by a survival rate of 74.8% for the patients with good prognosis, compared with a survival rate of 27.3% for the patients with poor prognosis (HR = 4.53, 95% CI 3.49-50.25).
We created a multivariate Cox proportional hazard regression model of age, INSS stage and NB-hypo (Table 6). The NB-hypo (p = 0.001; HR = 5.04; 95% CI, 2.00 to 12.69) is a significant independent predictor of outcome and it is equivalent to the INSS stage (p = 0.002; HR = 7.35; 95% CI, 2.13 to 25.40). Although the limited number of patients did not allow further sub grouping with statistical significance, it is interesting to analyze the association of the patients' risk factors profile with poor and good prognosis groups (Figure 5). All stage 1 and stage 4S patients and every stage 2 and stage 3 patients who survived were correctly classified by NB-hypo as good prognosis. Interestingly, NB-hypo correctly classified in the poor prognosis group the only stage 3, age>1 year intermediate risk and stage 2, age>1 year low risk patients who died.
Risk factors and survival of 72 MYCN not amplified patients. Patients are divided in poor and good prognosis groups according to the NB-hypo. Columns represent individual patients. The first line represents the patients according to the International Neuroblastoma Staging System (INSS). The second line represents the patients according to the age at diagnosis (> 1 year vs. < 1 year). In the last line, the patients are divided in deceased, black squares, and alive, gray squares.
Our results demonstrate that NB-hypo increases the resolution of the MYCN stratification by dividing patients with MYCN not amplified tumors in good and poor outcome groups suggesting that the neuroblastoma hypoxia is associated with the aggressiveness of neuroblastoma tumor independently from MYCN amplification.
Discussion
Identification of new markers for outcome prediction will improve the effectiveness of risk related therapy for neuroblastoma patients. Furthermore, definition of the molecular programs linked to the prognostic markers is important for targeted therapy and specific drug discovery. Utilizing an innovative biology-driven approach, we selected 62 probesets representing a robust hypoxia signature of neuroblastoma cell lines and we tested it on a cohort of 88 neuroblastoma patients for outcome prediction. We found that NB-hypo is a strong, independent risk predictor also for patients with MYCN not amplified tumors. These results demonstrate the power of the biology-driven approach to identify molecular programs related to tumor progression and point to hypoxia and NB-hypo as important prognostic indicators in neuroblastoma.
The precise definition of the relationship between hypoxia and human cancer is complicated by the difficulty in measuring hypoxia at the tumor site. The use of an independent hypoxia signature to analyze tumor gene expression profile and related outcome is a powerful way to probe the hypoxic status of tumors and its implications [30, 35]. We previously identified an 11 probesets hypoxia signature in 9 neuroblastoma cell lines by using l1 - l2 feature selection technique, a multivariate method specifically adopted to detect the subtle hypoxia response masked by strong transcriptional pattern [28]. The present study is based on the analysis of 11 neuroblastoma cell lines and on the modification of the l1 - l2 algorithm parameters in order to consider additional discriminative features. Furthermore, we combined the unique discriminating power of l1-l2 algorithm with the biological weight of the differential expression analysis to develop a robust method to identify a hypoxia signature that could better account for the biology of the tumor. This framework was carefully tested in its components and it is based on rigorous statistics and verification.
The definition of NB-hypo as a risk factor was obtained by estimating the Kaplan-Meier survival curves and the Cox proportional hazard regression model. We found that the NB-hypo stratifies neuroblastoma patients into good and poor prognosis groups with a significant separation of the patients for both OS and EFS. Risk assessment in neuroblastoma is based upon a number of factors which include, among others, age at diagnosis, International Neuroblastoma Staging System (INSS) stage, and MYCN status [36]. However, despite elaborate risk estimation strategies, outcome prediction for patients with neuroblastoma is still imperfect, as suggested particularly by current low- and intermediate risk patients with adverse outcome [34]. Our finding that NB-hypo is a true independent prognostic factor may help improving the risk assessment in neuroblastoma patients.
We and other have observed that the response to hypoxia is highly heterogeneous in different cell lines [28, 35] raising the question of the effectiveness of the hypoxia signature generated in other cell types in stratifying neuroblastoma tumors. We found that the hypoxia gene set derived from dendritic cells did not stratify the patients. Furthermore, the single VEGF gene did not divide the patients in significant risk related groups even if it is almost universally induced by hypoxia, indicating that a more complex gene set was needed. Furthermore, we studied the relationship between NB-hypo and other hypoxia signatures generated in different tumor systems in terms of overlapping, neuroblastoma patients' stratification and risk assessment. We found that only a limited overlapping consistent with the notion that hypoxia modulates different genes in different cells. Furthermore, only the Winter's hypoxia signature [30] out of the 5 tested was able to stratify neuroblastoma patients but without reaching a level of significance in the multivariate Cox analysis. These results demonstrated that NB-hypo is the only independent risk factor, among the signatures tested, capable of producing a significant patients' stratification. This property of NB-hypo could be due to the match between cellular system used to derive the signature and tumor, to the rigorous computational framework utilized or, more likely, to a combination of these factors.
NB-hypo probesets are highly expressed in aggressive tumors indicating that neuroblastoma hypoxia leads to growth, metastasis and poor outcome when reaching the levels that make it measurable by microarray. These results provide evidence that hypoxia is a sizable component of progressing neuroblastoma tumor. Hypoxia is a common characteristic of many aggressive tumors [37] and there are several reports associating HIF-1α/2α expression with the patients' outcome in a broad range of cancers [38]. There is evidence that hypoxia plays a role in causing the dedifferentiation of neuroblastoma cells in vitro [39, 40]. Furthermore, Forristal et al. [41] reported that HIF-2α is important in maintaining the pluripotency of human embryonic stem cells in hypoxic condition and Pietras et al. [42] demonstrated that HIF-2α maintains bone marrow- derived neuroblastoma tumor cells at a neural crest-like stage of differentiation in vitro and in vivo. Thus, the hypoxia-HIF-2α system promotes the undifferentiated phenotype either by dedifferentiation or inhibition of differentiation and may contribute to the aggressiveness through these mechanisms. The relationship between HIFs system and hypoxia is complicated by the fact that different environmental signals, such as genetic alterations, transition metals, chelating agents, hormones, and growth factors, share with hypoxia the property of inducing HIF-1α/2α and HIF-dependent gene transcription under normal pO2 [38, 43, 45]. Several reports associate HIF expression with the outcome of a broad range of cancers [38]. Correlation between HIF-2α, VEGF expression and poor prognosis [23] or pro-angiogenic activity was reported in neuroblastoma [46]. Recently, Noguera et al. [47] demonstrated an independence between HIF-1α and HIF-2α expression in neuroblastoma specimens and a correlation between HIF-1α and favorable outcome. These results show that the expression of HIF-1α/2α is not a specific and universal indicator of tissue hypoxia and the prognostic significance of HIFs may be unrelated to the hypoxic status of the neuroblastoma tumor. Thus, the assessment of tissue hypoxia requires measurement of multiple markers and NB-hypo could be a relevant tool for this purpose.
A computational approach using a priori biological knowledge to analyze the data of clinical neuroblastoma studies was applied successfully to the analysis of MYCN transcriptional targets whose over-expression contributes to the prediction of relapses and death from neuroblastoma [48]. The importance of MYCN in risk assessment is shown by the fact that MYCN gene is amplified, and often over-expressed, in about 22% of all neuroblastoma patients and it is an independent predictor for poor prognosis [49]. All metastatic tumors with amplified MYCN genes are aggressive, whereas metastatic tumors with non amplified MYCN genes have variable clinical behaviors influenced by the patient's age at diagnosis [50]. A 55-gene signature derived from the expression profile of metastatic neuroblastoma lacking MYCN amplification provided a new definition of high and low risk of disease progression [51]. We pursued the study of MYCN not amplified patients and tested the NB-hypo for its ability to predict the outcome of the disease. We analyzed the 72 MYCN not amplified patients of our cohort and we found that the NB-hypo significantly stratified them into good and poor prognosis groups and it was an independent risk factor relative to age and INSS.
Among MYCN not amplified tumors, stage 4 age>1 year patients are classified as high risk whereas the remaining are classified as low or intermediate risk based, among other factors, on age at the diagnosis and stage. We observed situations in which NB-hypo distinguished the clinical outcome of MYCN not amplified tumors more accurately than age or stage. In fact, NB-hypo correctly classified not only the deceased stage 4 age>1 year patients in the poor prognosis group, but included also in this category the deceased two stage 3 and the one stage 2 patients of the cohort belonging to the intermediate and low risk categories respectively. These results point to a high resolution of our signature. However, validation in larger cohort is needed to asses the effectiveness of NB-hypo in the above mentioned groups of patients. Although MYCN amplification is associated to the progression of the disease [5, 8], our results indicate that hypoxia is related to the aggressiveness of neuroblastoma independently from MYCN amplification and it might be responsible for increasing the risk of poor outcome in patients with a more favorable biology.
Several microarray studies on neuroblastoma gene expression were published using different platforms and analysis methods [51–55]. Platform heterogeneity complicates the comparison of the signatures, although useful information can be obtained also from these datasets by appropriate algorithms [56]. The analysis of neuroblastoma tumor gene expression profiles, pioneered by Wei et al. [52], was followed by studies confirming the forecast of individual survival from gene expression data analyzed by the CASPAR algorithm [57, 58]. Differences in the specific pattern of gene expression were described in spontaneous regressing, progressing [59] and intermediate risk [54] neuroblastoma. However, the overlap in identified expressed genes with prognostic information is low. Recent work by Vermeulen et al. [60] showed that a 59-gene signature, originated from the re-analysis of published gene expression studies, is an accurate and robust predictor of neuroblastoma outcome. These gene expression-based predictors were derived from the supervised association between the gene expression profile and clinical outcome. One shortcoming of this approach is that it is difficult to relate the signatures to the pathophysiology of the tumor; the second is that the results must be validated on an independent cohort of patients different from that from which the signature was originated. One advantage of the biology-driven approach is the immediate appreciation of the molecular program related to the prognostic indication. The second advantage is that independent validation of the results is not needed because the signature is not derived from the tumor profiles. Utilizing a biology-driven approach Chi et al. [35] demonstrated the prognostic value of hypoxia gene signature in breast and ovarian cancer. These results, together with our findings, show the power of controlled ex vivo studies in defining hypoxia as critical molecular programs in cancers and the potential for improvement in the current criteria for risk stratification of cancer patients.
The characterization of the tumor at diagnosis is indispensable for deciding the treatment and includes the evaluation of risk factors such as MYCN amplification status, hystotype, tissue markers, and chromosomal rearrangements [36]. The gene expression signatures are novel tools that may improve the stratification of patients thereby conditioning the choice of treatment. The hypoxic status detected by NB-hypo may be important to identify the tumors that may have high genetic instability [61] and high content of undifferentiated cells [25] at the time of excision as result of hypoxic phenotype. These characteristics of the primary tumor may be those that promote the aggressiveness of the disease and could be targeted by individualized therapies. There is a keen interest in biomarkers contained in body fluids because they are easy obtainable and can give indications about the response to therapy. These biomarkers includes catecholamines, ferritin, LDH [62], and other factors such as midkine [63], RANKL and OPG [64] but they do not define all risk groups and they can not substitute the analysis of the tumor mass. For the time being, the combination of tumor and body fluid characterization is needed for the optimal assessment of the treatment. It is reasonable to foresee that the study of the tumor mass will lead to the choice of individualized therapy and the longitudinal assessment of body fluids' biomarkers will monitor the response to therapy.
The prognostic molecular signatures linked to in vitro experimental models provide a more direct route to the development of targeted therapeutics. Many therapeutic agents are already under development to specifically target HIF pathways [65, 66] or to target cells under hypoxic environments by hypoxia dependent gene therapy [17]. An alternative approach is to target the proangiogenic factors induced by hypoxia such as VEGF and its receptors [67]. The clinical results obtained so far are not very promising (reviewed in [68]) because anti-VEGF therapy causes vascular regression, with concomitant increase of intratumor hypoxia, activation of HIF target genes and increases in aggressiveness and metastatic spread [69, 70]. These results suggest that the hypoxia, rather than of angiogenesis, may be the critical target for neuroblastoma therapy.
Conclusions
Neuroblastoma shows notable heterogeneity with regard to histology and clinical behavior, ranging from rapid progression and poor clinical outcome to spontaneous or therapy-induced regression. Despite elaborate risk estimation strategies, outcome prediction for patients with neuroblastoma is still imperfect and requires new molecular indicators. We utilized an innovative biology driven approach to study the prognostic power of the molecular signature of hypoxia, a condition of low oxygen tension developing in the tumor mass. We demonstrated that the hypoxia signature is a strong, independent risk predictor in neuroblastoma patients. Our signature will improve the effectiveness of risk related therapy by helping patients' stratification. Furthermore, our result point to the transcriptional response to hypoxia as a negative event leading to poor outcome, suggesting that those neuroblastoma patients classified as high risk by our signature may benefit from therapeutic protocols targeting hypoxia pathway.
References
De Preter K, Vandesompele J, Heimann P, Yigit N, Beckman S, Schramm A, Eggert A, Stallings R, Benoit Y, Renard M: Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biology. 2006, 7: R84- 10.1186/gb-2006-7-9-r84
Thiele CJ: Neuroblastoma. Human Cell Culture. Edited by: Master JRW, Palsson B. 1999, 21-22. London: Kluwer Academic,
Maris J, Hogarty M, Bagatell R, Cohn S: Neuroblastoma. Lancet. 2007, 369: 2106-2120. 10.1016/S0140-6736(07)60983-0
Weinstein J, Katzenstein H, Cohn S: Advances in the diagnosis and treatment of neuroblastoma. Oncologist. 2003, 8: 278-292. 10.1634/theoncologist.8-3-278
Bordow S, Norris M, Haber P, Marshall G, Haber M: Prognostic significance of MYCN oncogene expression in childhood neuroblastoma. Journal of clinical oncology. 1998, 16: 3286-3294.
van Noesel MM, Versteeg R: Pediatric neuroblastomas: genetic and epigenetic 'danse macabre'. Gene. 2004, 325: 1-15. 10.1016/j.gene.2003.09.042
Boon K, Caron H, van Asperen R, Valentijn L, Hermus M, van Sluis P, Roobeek I, Weis I, Voute P, Schwab M: N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001, 20: 1383-1393. 10.1093/emboj/20.6.1383
Meitar D, Crawford S, Rademaker A, Cohn S: Tumor angiogenesis correlates with metastatic disease, N-myc amplification, and poor outcome in human neuroblastoma. Journal of clinical oncology. 1996, 14: 405-414.
Puppo M, Pastorino S, Melillo G, Pezzolo A, Varesio L, Bosco MC: Induction of apoptosis by flavopiridol in human neuroblastoma cells is enhanced under hypoxia and associated with N-myc proto-oncogene down-regulation. Clin Cancer Res. 2004, 10: 8704-8719. 10.1158/1078-0432.CCR-03-0422
Puppo M, Battaglia F, Ottaviano C, Delfino S, Ribatti D, Varesio L, Bosco M: Topotecan inhibits vascular endothelial growth factor production and angiogenic activity induced by hypoxia in human neuroblastoma by targeting hypoxia-inducible factor-1alpha and -2alpha. Mol Cancer Ther. 2008, 7: 1974-1984. 10.1158/1535-7163.MCT-07-2059
Kim ES, Serur A, Huang J, Manley CA, McCrudden KW, Frischer JS, Soffer SZ, Ring L, New T, Zabski S: Potent VEGF blockade causes regression of coopted vessels in a model of neuroblastoma. Proc Natl Acad Sci USA. 2002, 99: 11399-11404. 10.1073/pnas.172398399
Semenza GL: Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009, 19: 12-16. 10.1016/j.semcancer.2008.11.009
Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P: Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998, 394: 485-490. 10.1038/28867
Harris AL: Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002, 2: 38-47. 10.1038/nrc704
Bosco MC, Puppo M, Santangelo C, Anfosso L, Pfeffer U, Fardin P, Battaglia F, Varesio L: Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J Immunol. 2006, 177: 1941-1955.
Bosco MC, Puppo M, Blengio F, Fraone T, Cappello P, Giovarelli M, Varesio L: Monocytes and dendritic cells in a hypoxic environment: Spotlights on chemotaxis and migration. Immunobiology. 2008, 213: 733-749. 10.1016/j.imbio.2008.07.031
Carta L, Pastorino S, Melillo G, Bosco M, Massazza S, Varesio L: Engineering of macrophages to produce IFN-gamma in response to hypoxia. J Immunol. 2001, 166: 5374-5380.
Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL: The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000, 157: 411-421.
Melillo G, Musso T, Sica A, Taylor L, Cox G, Varesio L: A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. J Exp Med. 1995, 182: 1683-1693. 10.1084/jem.182.6.1683
Chan DA, Giaccia AJ: Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 2007, 26: 333-339. 10.1007/s10555-007-9063-1
Rankin EB, Giaccia AJ: The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008, 15: 678-685. 10.1038/cdd.2008.21
Matthay K, Villablanca J, Seeger R, Stram D, Harris R, Ramsay N, Swift P, Shimada H, Black C, Brodeur GM: Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children's Cancer Group. N Engl J Med. 1999, 341: 1165-1173. 10.1056/NEJM199910143411601
Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K: Recruitment of HIF-1 alpha and HIF-2 alpha to common target genes is differentially regulated in neuroblastoma: HIF-2 alpha promotes an aggressive phenotype. Cancer Cell. 2006, 10: 413-423. 10.1016/j.ccr.2006.08.026
Poomthavorn P, Wong SH, Higgins S, Werther GA, Russo VC: Activation of a prometastatic gene expression program in hypoxic neuroblastoma cells. Endocr Relat Cancer. 2009, 16: 991-1004. 10.1677/ERC-08-0340
Edsjo A, Holmquist L, Pahlman S: Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin Cancer Biol. 2007, 17: 248-256. 10.1016/j.semcancer.2006.04.005
Zou H, Hastie T: Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2005, 67: 301-320. 10.1111/j.1467-9868.2005.00503.x.
De Mol C, Mosci S, Traskine M, Verri A: A Regularized Method for Selecting Nested Groups of Relevant Genes from Microarray Data. Journal of Computational Biology. 2009, 16: 677-690. 10.1089/cmb.2008.0171
Fardin P, Barla A, Mosci S, Rosasco L, Verri A, Varesio L: The l1-l2 regularization framework unmasks the hypoxia signature hidden in the transcriptome of a set of heterogeneous neuroblastoma cell lines. BMC Genomics. 2009, 10: 474- 10.1186/1471-2164-10-474
Ricciardi A, Elia AR, Cappello P, Puppo M, Vanni C, Fardin P, Eva A, Munroe D, Wu X, Giovarelli M: Transcriptome of hypoxic immature dendritic cells: modulation of chemokine/receptor expression. Mol Cancer Res. 2008, 6: 175-185. 10.1158/1541-7786.MCR-07-0391
Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ: Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007, 67: 3441-3449. 10.1158/0008-5472.CAN-06-3322
Buffa FM, Harris AL, West CM, Miller CJ: Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. British Journal of Cancer. 2010, 102: 428-435. 10.1038/sj.bjc.6605450
Hu ZY, Fan C, Livasy C, He XP, Oh DS, Ewend MG, Carey LA, Subramanian S, West R, Ikpatt F: A compact VEGF signature associated with distant metastases and poor outcomes. Bmc Medicine. 2009, 7: 10.1186/1741-7015-7-9.
Seigneuric R, Starmans MHW, Fung G, Krishnapuram B, Nuyten DSA, van Erk A, Magagnin MG, Rouschop KM, Krishnan S, Rao RB: Impact of supervised gene signatures of early hypoxia on patient survival. Radiotherapy and Oncology. 2007, 83: 374-382. 10.1016/j.radonc.2007.05.002
Berthold F, Hero B, Breu H, Christiansen H, Erttmann R, Gnekow A, Herrmann F, Klingebiel T, Lampert F, Muller-Weihrich S: The recurrence patterns of stages I, II and III neuroblastoma: experience with 77 relapsing patients. Ann Oncol. 1996, 7: 183-187.
Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Borresen-Dale AL: Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006, 3: e47- 10.1371/journal.pmed.0030047
Ambros PF, Ambros IM, Brodeur GM, Haber M, Khan J, Nakagawara A, Schleiermacher G, Speleman F, Spitz R, London WB: International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. British Journal of Cancer. 2009, 100: 1471-1482. 10.1038/sj.bjc.6605014
Brown JM, William WR: Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004, 4: 437-447. 10.1038/nrc1367
Semenza G: Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003, 3: 721-732. 10.1038/nrc1187
Jogi A, Ora I, Nilsson H, Lindeheim A, Makino Y, Poellinger L, Axelson H, Pahlman S: Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. PNAS. 2002, 99: 7021-7026. 10.1073/pnas.102660199
Pietras A, Gisselsson D, Ora I, Noguera R, Beckman S, Navarro S, Pahlman S: High levels of HIF-2 alpha highlight an immature neural crest-like neuroblastoma cell cohort located in a perivascular niche. Journal of Pathology. 2008, 214: 482-488. 10.1002/path.2304
Forristal CE, Wright KL, Hanley NA, Oreffo ROC, Houghton FD: Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction. 2010, 139: 85-97. 10.1530/REP-09-0300
Pietras A, Hansford LM, Johnsson AS, Bridges E, Sjolund J, Gisselsson D, Rehn M, Beckman S, Noguera R, Navarro S: HIF-2 alpha maintains an undifferentiated state in neural crest-like human neuroblastoma tumor-initiating cells. PNAS. 2009, 106: 16805-16810. 10.1073/pnas.0904606106
Wenger R, Stiehl D, Camenisch G: Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005, 2005: re12- 10.1126/stke.3062005re12
Lofstedt T, Fredlund E, Holmquist-Mengelbier L, Pietras A, Ovenberger M, Poellinger L, Pahlman S: Hypoxia inducible factor-2 alpha in cancer. Cell Cycle. 2007, 6: 919-926.
Carroll VA, Ashcroft M: Role of hypoxia-inducible factor (HIF)-1 alpha-versus HIF-2 alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-1, or loss of von Hippel-Lindau function: Implications for targeting the HIF pathway. Cancer Res. 2006, 66: 6264-6270. 10.1158/0008-5472.CAN-05-2519
Favier J, Lapointe S, Maliba R, Sirois MG: HIF2 alpha reduces growth rate but promotes angiogenesis in a mouse model of neuroblastoma. BMC Cancer. 2007, 7:
Noguera R, Fredlund E, Piqueras M, Pietras A, Beckman S, Navarro S, Pahlman S: HIF-1 alpha and HIF-2 alpha Are Differentially Regulated In vivo in Neuroblastoma: High HIF-1 alpha Correlates Negatively to Advanced Clinical Stage and Tumor Vascularization. Clinical Cancer Research. 2009, 15: 7130-7136. 10.1158/1078-0432.CCR-09-0223
Fredlund E, Ringner M, Maris JM, Pahlman S: High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. PNAS. 2008, 105: 14094-14099. 10.1073/pnas.0804455105
Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D: Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985, 313: 1111-1116. 10.1056/NEJM198510313131802
Schmidt ML, Lukens JN, Seeger RC, Brodeur GM, Shimada H, Gerbing RB, Stram DO, Perez C, Haase GM, Matthay KK: Biologic factors determine prognosis in infants with stage IV neuroblastoma: A prospective Children's Cancer Group study. J Clin Oncol. 2000, 18: 1260-1268.
Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, Matthay K, Buckley J, Ortega A, Seeger R: Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006, 98: 1193-1203. 10.1093/jnci/djj330
Wei J, Greer B, Westermann F, Steinberg S, Son C, Chen Q, Whiteford C, Bilke S, Krasnoselsky A, Cenacchi N: Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res. 2004, 64: 6883-6891. 10.1158/0008-5472.CAN-04-0695
Schramm A, Vandesompele J, Schulte JH, Dreesmann S, Kaderali L, Brors B, Eils R, Speleman F, Eggert A: Translating expression profiling into a clinically feasible test to predict neuroblastoma outcome. Clinical Cancer Research. 2007, 13: 1459-1465. 10.1158/1078-0432.CCR-06-2032
Ohira M, Oba S, Nakamura Y, Isogai E, Kaneko S, Nakagawa A, Hirata T, Kubo H, Goto T, Yamada S: Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005, 7: 337-350. 10.1016/j.ccr.2005.03.019
Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, Ernestus K, Konig R, Haas S, Eils R: Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. Journal of clinical oncology. 2006, 24: 5070-5078. 10.1200/JCO.2006.06.1879
Chen QR, Song YK, Wei JS, Bilke S, Asgharzadeh S, Seeger RC, Khan J: An integrated cross-platform prognosis study on neuroblastoma patients. Genomics. 2008, 92: 195-203. 10.1016/j.ygeno.2008.05.014
Oberthuer A, Kaderali L, Kahlert Y, Hero B, Westermann F, Berthold F, Brors B, Eils R, Fischer M: Subclassification and individual survival time prediction from gene expression data of neuroblastoma patients by using CASPAR. Clin Cancer Res. 2008, 14: 6590-6601. 10.1158/1078-0432.CCR-07-4377
Schramm A, Mierswa I, Kaderali L, Morik K, Eggert A, Schulte JH: Reanalysis of neuroblastoma expression profiling data using improved methodology and extended follow-up increases validity of outcome prediction. Cancer Lett. 2009, 282: 55-62. 10.1016/j.canlet.2009.02.052
Fischer M, Oberthuer A, Brors B, Kahlert Y, Skowron M, Voth H, Warnat P, Ernestus K, Hero B, Berthold F: Differential expression of neuronal genes defines subtypes of disseminated neuroblastoma with favorable and unfavorable outcome. Clinical Cancer Research. 2006, 12: 5118-5128. 10.1158/1078-0432.CCR-06-0985
Vermeulen J, De Preter K, Naranjo A, Vercruysse L, Van Roy N, Hellemans J, Swerts K, Bravo S, Scaruffi P, Tonini GP: Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009, 10: 663-671. 10.1016/S1470-2045(09)70154-8
Vaupel P: The role of hypoxia-induced factors in tumor progression. Oncologist. 2004, 9 (Suppl 5): 10-17. 10.1634/theoncologist.9-90005-10
Haupt R, Garaventa A, Gambini C, Parodi S, Cangemi G, Casale F, Viscardi E, Bianchi M, Prete A, Jenkner A: Improved survival of children with neuroblastoma between 1979 and 2005: a report of the Italian Neuroblastoma Registry. J Clin Oncol. 2010, 28: 2331-2338. 10.1200/JCO.2009.24.8351
Ikematsu S, Nakagawara A, Nakamura Y, Ohira M, Shinjo M, Kishida S, Kadomatsu K: Plasma midkine level is a prognostic factor for human neuroblastoma. Cancer Sci. 2008, 99: 2070-2074. 10.1111/j.1349-7006.2008.00957.x
Granchi D, Garaventa A, Amato I, Paolucci P, Baldini N: Plasma levels of receptor activator of nuclear factor-kappaB ligand and osteoprotegerin in patients with neuroblastoma. Int J Cancer. 2006, 119: 146-151. 10.1002/ijc.21783
Hewitson KS, Schofield CJ: The HIF pathway as a therapeutic target. Drug Discov Today. 2004, 9: 704-711. 10.1016/S1359-6446(04)03202-7
Melillo G: Targeting hypoxia cell signaling for cancer therapy. Cancer Metastasis Rev. 2007, 26: 341-352. 10.1007/s10555-007-9059-x
Zhang LB, Smith K, Chong AL, Stempak D, Yeger H, Marrano P, Thorner PS, Irwin MS, Kaplan DR, Baruchel S: In Vivo Antitumor and Antimetastatic Activity of Sunitinib in Preclinical Neuroblastoma Mouse Model. Neoplasia. 2009, 11: 426-435.
Schmidt C: Why do tumors become resistant to antiangiogenesis drugs?. J Natl Cancer Inst. 2009, 101: 1530-1532. 10.1093/jnci/djp425
Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O: Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009, 15: 220-231. 10.1016/j.ccr.2009.01.027
Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS: Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009, 15: 232-239. 10.1016/j.ccr.2009.01.021
Semenza G: Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med. 2001, 7: 345-350. 10.1016/S1471-4914(01)02090-1
Murata M, Yudo K, Nakamura H, Chiha J, Okamoto K, Suematsu N, Nishioka K, Beppu M, Inoue K, Kato T: Hypoxia Upregulates the Expression of Angiopoietin-Like-4 in Human Articular Chondrocytes: Role of Angiopoietin-Like-4 in the Expression of Matrix Metalloproteinases and Cartilage Degradation. Journal of Orthopaedic Research. 2009, 27: 50-57. 10.1002/jor.20703
Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ: An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009, 37: 4587-4602. 10.1093/nar/gkp425
Yoon DY, Buchler P, Saarikoski ST, Hines OJ, Reber HA, Hankinson O: Identification of genes differentially induced by hypoxia in pancreatic cancer cells. Biochem Biophys Res Commun. 2001, 288: 882-886. 10.1006/bbrc.2001.5867
Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM: Hypoxia-Induced Autophagy Is Mediated through Hypoxia-Inducible Factor Induction of BNIP3 and BNIP3L via Their BH3 Domains. Molecular and Cellular Biology. 2009, 29: 2570-2581. 10.1128/MCB.00166-09
Zhong XP, Wang D, Zhang YB, Gui JF: Identification and characterization of hypoxia-induced genes in Carassius auratus blastulae embryonic cells using suppression subtractive hybridization. Comp Biochem Physiol B Biochem Mol Biol. 2009, 152: 161-170. 10.1016/j.cbpb.2008.10.013
Lee M, Kim J, Suk K, Park J: Identification of the hypoxia-inducible factor 1 alpha-responsive HGTD-P gene as a mediator in the mitochondrial apoptotic pathway. Mol Cell Biol. 2004, 24: 3918-3927. 10.1128/MCB.24.9.3918-3927.2004
Patiar S, Harris AL: Role of hypoxia-inducible factor-1alpha as a cancer therapy target. Endocr Relat Cancer. 2006, 13 (Suppl 1): S61-S75. 10.1677/erc.1.01290
Metzen E: Enzyme substrate recognition in oxygen sensing: how the HIF trap snaps. Biochem J. 2007, 408: e5-e6. 10.1042/BJ20071306
Kang SG, Brown AL, Chung JH: Oxygen tension regulates the stability of insulin receptor substrate-1 (IRS-1) through caspase-mediated cleavage. J Biol Chem. 2007, 282: 6090-6097. 10.1074/jbc.M610659200
Le Jan S, Le Meur N, Cazes A, Philippe J, Le Cunff M, Leger J, Corvol P, Germain S: Characterization of the expression of the hypoxia-induced genes neuritin, TXNIP and IGFBP3 in cancer. FEBS Lett. 2006, 580: 3395-3400. 10.1016/j.febslet.2006.05.011
Wellmann S, Bettkober M, Zelmer A, Seeger K, Faigle M, Eltzschig HK, Buhrer C: Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun. 2008, 372: 892-897. 10.1016/j.bbrc.2008.05.150
Corn PG, Ricci MS, Scata KA, Arsham AM, Simon MC, Dicker DT, El Deiry WS: Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Biol Ther. 2005, 4: 1285-1294.
Sibold S, Roh V, Keogh A, Studer P, Tiffon C, Angst E, Vorburger SA, Weimann R, Candinas D, Stroka D: Hypoxia increases cytoplasmic expression of NDRG1, but is insufficient for its membrane localization in human hepatocellular carcinoma. FEBS Lett. 2007, 581: 989-994. 10.1016/j.febslet.2007.01.080
Hofbauer KH, Gess B, Lohaus C, Meyer HE, Katschinski D, Kurtz A: Oxygen tension regulates the expression of a group of procollagen hydroxylases. Eur J Biochem. 2003, 270: 4515-4522. 10.1046/j.1432-1033.2003.03846.x
Minchenko O, Opentanova I, Minchenko D, Ogura T, Esumi H: Hypoxia induces transcription of 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase-4 gene via hypoxia-inducible factor-1alpha activation. FEBS Lett. 2004, 576: 14-20. 10.1016/j.febslet.2004.08.053
Li H, Ko HP, Whitlock JP: Induction of phosphoglycerate kinase 1 gene expression by hypoxia. Roles of Arnt and HIF1alpha. J Biol Chem. 1996, 271: 21262-21267. 10.1074/jbc.271.35.21262
Semenza GL, Roth PH, Fang HM, Wang GL: Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994, 269: 23757-23763.
Zhang JZ, Behrooz A, Ismail-Beigi F: Regulation of glucose transport by hypoxia. Am J Kidney Dis. 1999, 34: 189-202. 10.1016/S0272-6386(99)70131-9
Niitsu Y, Hori O, Yamaguchi A, Bando Y, Ozawa K, Tamatani M, Ogawa S, Tohyama M: Exposure of cultured primary rat astrocytes to hypoxia results in intracellular glucose depletion and induction of glycolytic enzymes. Brain Res Mol Brain Res. 1999, 74: 26-34. 10.1016/S0169-328X(99)00245-4
Murat A, Migliavacca E, Hussain SF, Heimberger AB, Desbaillets I, Hamou MF, Ruegg C, Stupp R, Delorenzi M, Hegi ME: Modulation of angiogenic and inflammatory response in glioblastoma by hypoxia. PLoS ONE. 2009, 4: e5947- 10.1371/journal.pone.0005947
Acknowledgements
The authors would like to thank Ms. Daniela Segalerba for technical assistance with cell culturing and RNA extraction, and Dr. Sara Barzaghi for the editorial assistance. This work was supported by the Fondazione Italiana per la Lotta al Neuroblastoma (Italy), the KiKa foundation (the Netherlands), the SKK foundation and the Dutch Cancer Society.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PF and LV conceived the initial idea, the experimental design, supervised the work, and wrote the manuscript. PF performed microarray experiments and interpreted results. AB contributed with the development of MATLAB and R scripts for data processing, normalization and analysis, performed the supervised analysis, and visualized the results. SM wrote the core code for l1 - l2 regularization and contributed with the development of MATLAB and R scripts for data processing, normalization and analysis. LR performed the unsupervised analysis and helped in the design and implementation of the supervised analysis. AV supervised the entire statistical data analysis. RV organized tumor samples experiments and supervised the work. HC and JM organized tumor series collection. IØ collected all survival data, sectioned tumors for expression arraying and reviewed the tumor samples. MP performed hypoxia experiment on cell lines. AE interpreted the results and supervised the work. All authors read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Fardin, P., Barla, A., Mosci, S. et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol Cancer 9, 185 (2010). https://doi.org/10.1186/1476-4598-9-185
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-4598-9-185