Abstract
Modelling studies on the spatial distribution and spread of infectious diseases are becoming increasingly detailed and sophisticated, with global risk mapping and epidemic modelling studies now popular. Yet, in deriving populations at risk of disease estimates, these spatial models must rely on existing global and regional datasets on population distribution, which are often based on outdated and coarse resolution data. Moreover, a variety of different methods have been used to model population distribution at large spatial scales. In this review we describe the main global gridded population datasets that are freely available for health researchers and compare their construction methods, and highlight the uncertainties inherent in these population datasets. We review their application in past studies on disease risk and dynamics, and discuss how the choice of dataset can affect results. Moreover, we highlight how the lack of contemporary, detailed and reliable data on human population distribution in low income countries is proving a barrier to obtaining accurate large-scale estimates of population at risk and constructing reliable models of disease spread, and suggest research directions required to further reduce these barriers.
Similar content being viewed by others
Introduction
Mapping and modelling methods used to study the spatial distribution and spread of vector-borne and directly transmitted infectious diseases are becoming increasingly widespread and sophisticated as the field of spatial epidemiology grows. Spatial epidemiology is defined as "the study of spatial variation in disease risk or incidence" [1], and its aims are both to describe and to understand these variations [2], with the ultimate objective being to assist public health decision making. Interactions between pathogens, vectors and hosts, and between these agents and their environment determine spatial variations in disease risk and make the transmission of vector-borne and other infectious diseases an intrinsically spatial process [1, 3].
Most studies on infectious disease dynamics are not spatially-explicit, i.e. elements are not explicitly localized in space. Models are typically based on the metapopulation concept, which considers isolated subpopulations subject to colonization and extinction dynamics [4–6]. If the species of interest is a parasite, colonization means infection and a local extinction occurs when the host dies or recovers [5]. This approach is spatially-implicit, as it avoids the use of geographical maps to locate elements. In the majority of non-spatial mathematical models of infectious diseases, the total population is assumed to be constant [7], but population data have been included, for instance, in non-spatial models of HIV [8], pertussis [9], malaria [7], or in global burden of disease calculations [10–16]. However, the spatial nature of infectious diseases, and particularly spatial heterogeneities in transmission and spread, make risk maps and spatially-explicit models of disease incidence valuable tools for understanding disease dynamics and planning public health interventions [1, 2, 17]. Defining the extent of infectious diseases as a public health burden and their distribution and dynamics in time and space are critical to scoping the financial requirements, for setting a control agenda and for monitoring.
The emergence of spatially-explicit studies in infectious disease research has been supported by improvements in spatial data and tools such as remote sensing and geographical information systems (GIS) [18–23], as well as advances in spatially-explicit modelling methods [17, 24]. GIS are commonly used to combine spatial data from different sources, for mapping disease and for performing spatial analyses to identify the causal factors of observed spatial patterns such as cluster detection or landscape fragmentation analyses [20, 25]. In addition, the growth in computing, data collection and the centralization of epidemiological data, has lead to an increase in the sophistication and complexity in the mapping and modelling of infectious disease risks.
Among the agents involved in the disease transmission process, human hosts play a crucial role as their density [26], spatial location, demographic characteristics (e.g. age-risk profiles [27–30]) and behaviour [31–33] determine their exposure to infection. Any approach that requires the use of modelled disease rates or dynamics requires reasonable information on the resident population for the time period one is intending to estimate risk. Where risks and spread of diseases are heterogeneous in space, population distributions and counts should ideally be resolved to higher levels of spatial detail than large regional estimates. Accurate and detailed information on population size and distribution are therefore of significant importance for deriving populations at risk and infection movement estimates in spatial epidemiological studies [34]. For many low-income countries of the World, where disease burden is greatest, however, spatially detailed, contemporary census data do not exist. This is especially true for much of Africa, where currently available census data are often over a decade old, and at administrative boundary levels just below national-level [35, 36].
Modelling techniques for the spatial reallocation of populations within census units have been developed in an attempt to overcome the difficulties caused by input census data of varying resolutions. National census population data can be represented as continuous gridded population distribution (or count) datasets through the use of spatial interpolation algorithms. Here, we firstly review and compare the methods used in the construction of existing large-scale population datasets, and secondly review applications of these datasets in past studies of disease risk and dynamics.
Mapping humans
Spatial demographic data
Our knowledge of human distribution in many areas of the World remains surprisingly poor. A growing interest in the global mapping of human populations emerged in the 1990s [37, 38]. Until then, the only information on the spatial distribution of people came from maps showing the location of towns, cities and administrative boundaries on one hand and sparse, inconsistent population data coming from national censuses or demographic surveys on the other [39]. Wright (1936) provided one of the first examples of the combination of demographic and spatial data to build a population density map of the Cape Cod region in the United States [40]. Improvements in demographic and spatial data availability and the development of methods to combine them led to the creation of global population density datasets.
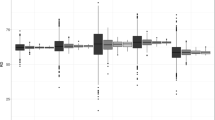
Demographic data come from different sources: censuses, civil registration systems, governmental or non-governmental administrative data or sample surveys [37]. Civil registration systems provide the most reliable and useful demographic data as they continuously record information on the population of a country, including their spatial distribution. However, up-to-date registration systems only exist in a small number of countries. Instead, censuses are conducted approximately every 10 years by national statistical offices in order to provide consistent and geo-referenced population data. The accuracy and amount of data supplied by national censuses vary considerably from one country to the other. From a temporal point of view, (at the time of writing) the most recent census is more than 25 years old in some sub-Saharan countries such as Angola, Eritrea and the Demographic Republic of Congo [41] (Figure 1a). Large variations also exist in the spatial resolution of available census data, as the ways in which national territories are divided and the administrative level at which population data are collected and summarized vary by country. Figure 1b shows the spatial resolution of census data used in the construction of the Gridded Population of the World version 3 (GPW3) [42] and the Global Rural Urban Mapping Project (GRUMP) [43, 44] spatial population databases (both described below).
Spatial and temporal characteristics of available census data. a) Year of the last national census data available (data source: GeoHive [41]) and b) average spatial resolution (ASR) of census data used in the construction of Gridded Population of the World version 3 (GPW3) and the Global Rural Urban Mapping Project (GRUMP). The ASR measures the effective resolution of administrative units in kilometers. It is calculated as the square root of the land area divided by the number of administrative units [42]. It can be thought of as the "cell size" if all units in a country were square and of equal size.
The link between demographic data and a spatial reference system is essential for geographical analyses. Census data collected at the administrative unit level must be related to an accurate boundary dataset [37]. This has in the past often been neglected, mainly due to a lack of GIS technology, knowledge, resources and methods, as well as computing infrastructure [37, 39, 42], but efforts are now being made across the world to link census data with digital administrative boundaries.
Population distribution modelling methods
A variety of methods for converting population count data from irregular administrative units to regular grids have been developed since the 1990s [43, 44] and have led to the emergence of differing global gridded population datasets. The quality and accessibility of population and spatial data have been improving, making collaborations between demographers and geographers stronger [39, 42, 43]. In addition, GIS technologies are becoming increasingly available and accessible and computer power is continuously improving, allowing the processing of larger and more detailed datasets [37, 39, 42].
Population distribution modelling methods over large spatial scales rely on redistributing populations within census units to obtain continuous population surfaces, i.e. gridded datasets with a number of inhabitants per grid cell. Different interpolation methods have been typically used to reallocate populations within administrative units (Figure 2):
Schematic illustrations of population distribution modelling methods. The population of two administrative units A and B (with total population in A = 8 and total population in B = 16) are redistributed according to different population distribution modelling approaches (areal weighted, pycnophylactic and dasymetric). In the dasymetric method, a higher weight was attributed to the red hatched area.
Areal weighting assumes that the population is uniformly distributed within each administrative unit. The population assigned to a grid cell is simply the total population of the administrative unit divided by the number of cells in the administrative unit. Every grid cells of an administrative unit has therefore the same population value [45]. This method was used to construct the Gridded Population of the World (GPW) database, versions 2 and 3 [42, 46]
Pycnophylactic interpolation starts with the areal weighted method, but smoothes population values using the weighted average of nearest neighbours, while preserving the summation of population data to the original population per areal unit [47]. Pycnophylactic interpolation was used to generate GPW version 1 [48, 49].
Dasymetric modelling involves using ancillary data - often this may include satellite derived land cover data - to redistribute populations within administrative units [45, 50]. Weightings are attributed to the different land cover classes and the population is redistributed accordingly. For example, the Global Rural Urban Mapping Project (GRUMP) uses a similar approach to GPW, but incorporates urban-rural extents and their corresponding populations in the spatial reallocation of census counts [43, 44]. The urban-rural extent information is generated by a variety of input data that include census data, online web sources and National Imagery and Mapping Agency (NIMA) database of populated places [46]. The recently produced AfriPop dataset, which covers the African continent at a fine spatial resolution, also used land cover data to redistribute populations [51, 52]. Other kinds of ancillary data such as the slope or roads can be used for dasymetric mapping.
More sophisticated modelling approaches - called smart interpolation - involve modelling the finescale distribution of populations using a range of satellite and other ancillary data. For example, an accessibility surface developed from road networks and populated places can be used to redistribute people, as was done in the construction of the UNEP database [53–56]. The LandScan dataset is another example of smart interpolation, where various ancillary data such as roads, slope, land cover and nighttime lights are used to determine the probability of population occurrence in cells. Populations are spatially reallocated within each areal unit using modelling approaches based on these probability coefficients [57–59].
Features of each dataset are outlined in Table 1. All of these existing datasets show the spatial distribution of nighttime residential population, except LandScan that maps the 'ambient' population, i.e. the average location of people across time. AfriPop is the only project that also freely provides demographic sub-group gridded datasets, i.e. age composition by 5-years groupings and gender [52]. The most recently updated datasets are LandScan and AfriPop, updated in 2010 and 2011, respectively. However, given its commercial status, the LandScan 2010 dataset is not available in the public domain. LandScan and AfriPop are also the two datasets for which we can expect the most frequent updates in the future. Different levels of transparency in the methodologies are observed. Most of the datasets are fully documented, with methods clearly described and all data sources mentioned (e.g. GPW, GRUMP, UNEP, AfriPop), whereas datasets using more sophisticated interpolation methods are sometimes less transparent. For example, the available documentation of LandScan only enables a general understanding of the methodologies used. These global population distribution datasets that have been created at spatial resolutions of finer than 1 degree have been used in various epidemiological studies, and these are reviewed below.
Uncertainty and error
Given the different input data and the different modelling methods used, the existing gridded population datasets described above clearly differ. Different sources of error and uncertainty are associated with these population datasets, which generally arise from (i) the input data, (ii) temporal projections and (iii) the modelling procedure used.
Uncertainties associated with input data, such as census data, can be important, especially in low income regions where misreporting errors may be frequent [60, 61]. In addition to errors in population counts, inaccuracies in the spatial positioning of administrative unit boundaries can lead to population mapping uncertainties. Population movements also make counts not entirely representative of the long-term residential population. However, censuses are often the only consistent and exhaustive population databases available in countries where registration systems do not exist, and quantifying such uncertainties remains difficult. Figure 3 shows population distribution as mapped by existing global population datasets (LandScan 2008, GRUMP beta, GPW3, AfriPop and UNEP Africa) for a region of Kenya, where census data are available at a high administrative unit level, and a region of Angola, where the spatial resolution of census data is coarse. This figure highlights how the differing approaches to the spatial interpolation of census data produce very different spatial configurations of population distribution when census data are aggregated in large administrative units, as in the case of Angola. Recent studies used Kenya data at different administrative levels to show quantitatively that population map accuracies significantly improve with finer resolution input census data [35, 62, 63]. With fine resolution census data, populations are already distributed in small spatial units, so that the margin of possible errors due to the population distribution modelling is reduced, relative to the output resolution of the modelled surfaces. These studies demonstrate that obtaining as fine a spatial resolution of census data as possible must be the priority starting point in population distribution modelling [35, 62, 63]. This calls for regular updating of input census data, as and when it becomes available. The cost of population censuses is however large and most countries undertake full censuses only once per decade. Therefore, models also help provide estimates in the intervening years. Given the positional uncertainties that may be associated with detailed boundaries, some dataset producers (e.g. LandScan) have prioritised smart modelling methods over obtaining fine resolution census data. Deciding upon which solution produces the more accurate results here is difficult. However, if the methodologies used to construct the fine resolution census data boundaries are well documented and known to match accurately with census enumeration units, then the use of detailed census data likely produces consistently more accurate results in mapping [35].
To adjust population counts to one target year, either inter-censal growth rates or national-level growth rates - often from the United Nations [64] - are used. These national estimates are derived from fertility, mortality and international migration numbers, a method that is inevitably associated with uncertainties [65]. In addition, growth rates can vary substantially within countries, introducing uncertainties when using national-level estimates, and are dependent upon the urban-rural definition used when using urban and rural growth rates.
Modelling approaches using ancillary datasets only increase population distribution model accuracies over the simple gridding (areal weighting) of census data if the ancillary data is more detailed and complete spatially than the input census data, and can be detrimental to modelling accuracies otherwise [35, 62]. The extent to which ancillary data can improve population model accuracies depends on the resolution of census data, and decreases when the resolution of census data becomes finer.
The validation of large-scale population distribution datasets is problematic as no independent source exists that could serve this purpose globally. Map accuracies can be tested in target regions where reference data are available at a finer spatial resolution than the map produced [57, 62, 66, 67]. Recent studies have shown that using certain methods of downscaling increases consistently the mapping accuracy over simple areal weighting of administrative unit census data [35, 63]. Until now, validation efforts at the global level have been limited to comparing results with population totals reported by the UN (in the case of GPW and GRUMP) [39]. Another way to evaluate the accuracy of population datasets is to use geospatial metrics to compare spatial datasets [68]. These metrics quantify differences in the spatial structure of datasets and analyse properties such as spatial correlations within and between datasets [68, 69]. The accuracy of urban extent datasets can be more easily assessed using expert opinion [70] or a set of independent test sites derived from medium or high resolution remote sensing imagery [71, 72]. In any case, a future priority should be to design methods for the incorporation of uncertainty explicitly.
Application of global population distribution data in studies of disease risk and dynamics
Gridded population data have been commonly used to estimate populations at risk of infectious disease and to simulate disease dynamics. These raster datasets have the advantage of being global and consistent in terms of spatial resolution and are particularly useful for population at risk (PAR) assessments and infectious disease risk mapping and modelling. In this section, we will first review studies that have used gridded population datasets as input data for spatial transmission models. Secondly, studies that used existing gridded population data to calculate infectious disease health metrics, such as populations at risk, will be covered. An overview of the literature cited is available in Table 2.
Input data for spatial transmission modelling
Population density and growth are significant drivers for the emergence of different categories of infectious diseases [73]. Jones and colleagues (2008) examined the relationship between the spatial distribution of emerging infectious disease events of different kinds - including vector-borne and non-vector-borne - and population data from GPW3 [73]. For directly-transmitted diseases, the spatial distribution of people is determinant as it controls person-to-person contacts and therefore the spread of the disease. Gridded population data have been used in spatio-temporal models that simulate contacts between infectious and susceptible people and the spatial spread of the disease [74–76]. Even if less obvious, the link between indirectly-transmitted diseases - i.e. infections that require an external agent for transmission to occur, such as a vector, an animal host or the environment - and population density can also be determinant. The effect however varies according to the disease considered, mainly because population size and distribution can modify the habitat of disease vectors or hosts and hence increase or decrease disease incidence. For example, trypanosomiasis is expected to decline in large parts of Africa because of the growth in human population and the expansion of agriculture at the expense of tsetse fly habitat [77]. Urbanization in Africa is also expected to reduce malaria transmission risk [78]. Other transmitting agents can live in urbanized or highly populated places, making the disease transmission risk higher in these areas, such as poultry responsible for avian influenza [79].
Gridded population datasets have been widely used to examine the relationships between infectious disease incidence and population size and distribution. Globally, population density has an opposed impact on dengue and yellow fever: vector preferences mean that dengue risk is higher in highly populated urban areas, whereas yellow fever risk is higher in rural areas [80]. Several authors included gridded population data as a risk factor in disease mapping and modelling [79, 81–85]. When the objective is to study the impact of factors other than population (e.g. climate, ecological or socio-economic variables) on disease transmission, gridded population data can be used as an offset variable in statistical models, i.e. to control for population count differences by analysing rates instead of absolute values [86, 87].
Besides spatial models that simply study the statistical association between disease risk or incidence and population density in order to map disease risk, more sophisticated spatially-explicit models have been developed to study the spatial diffusion of infectious diseases. Several of such spatially-explicit models have successfully used gridded population datasets as input data, for example for creating risk maps [88, 89], for the calculation of a global malaria transmission stability index [90], or to study the potential economic impacts of avian influenza in Nigeria [91]. All of these models were developed at the grid cell level, making gridded population datasets particularly useful. Gridded population data have also been used to develop agent-based simulation models at the regional level [92, 93] and at the global level [94]. Whatever the spatial approach for modelling - patch, distance, group or network - population data are essential, as these models generally require the generation of a virtual society with an appropriate distribution of people. Population distribution datasets have been used to randomly distribute households in study areas according to local population densities [92–94]. In these models, gridded population data provide valuable input datasets mainly because of their wide coverage, consistent spatial detail and availability in the public domain. Moreover, most spatially-explicit models are grid cell-based, meaning that the gridded population datasets are ready to use without any further processing.
Human interactions and movements are crucial for disease spread. However, the complexity of human mobility and its multiscale nature make comprehensive data on movements difficult to obtain [95]. A recurrent issue emerging from large-scale modelling of infectious diseases is therefore the mobility patterns and the spatial details required for transport network data [95]. Gridded population data provide a useful baseline for developing mobility networks. Several authors have combined gridded population data with transport networks to simulate the large-scale spread of infectious diseases [95–100]. In low-income countries, data on population flows are rare and a recent study on the risk of introducing malaria to areas targeted for elimination used a database of bilateral migrant stocks and the GRUMP dataset to evaluate international population movements [101]. Even if population datasets help in the construction of large-scale mobility databases, significant further work is required to fill the data gaps.
Endemic disease health metrics
Population distribution datasets constitute an essential denominator required for many infectious disease studies. It is well known that disease transmission is focal and heterogeneous [102–105], partially due to the clustered nature of human population distribution. As the precision and detail of disease risk mapping improves, spatial population datasets that capture these patterns are therefore required if the sizes of populations at risk (PAR) are to be more accurately quantified.
PAR of some infectious diseases have been estimated based on gridded population datasets, principally malaria [78, 106–115], helminth infections [116–119], human African trypanosomiasis [120] and dengue and yellow fevers [80, 121]. Typically, disease risk maps are spatially overlaid onto population distribution datasets to quantify the number of people residing in specific risk zones or classes, and thus derive PAR numbers. This commonly used method of combining population and prevalence data to assess PAR was also used to study co-infection of diseases that show a clear geographic overlap, such as malaria and helminth infections [122, 123]. This method is simple to implement and provides a useful assessment of PAR. However, PAR assessments generally use the different existing population datasets interchangeably to provide such estimates. Moreover, uncertainties inherent in the population datasets are rarely acknowledged in such calculations. As already described above, existing gridded population datasets clearly differ, especially where census population data are spatially coarse, and both input-based and process-based uncertainties contribute to great variations in mapping precision (Figure 3). As a consequence, large variations in PAR estimates can result from the choice of population dataset, particularly in low-income countries where census data are often spatially and temporally poor [34]. Specific estimates for pregnant women and children have been also derived from population distribution datasets, by combining gridded population data with age, sex and fertility data from the United Nations [124, 125]. However, given that the demographic composition of populations varies substantially within countries, using such national-level demographic estimates introduces additional uncertainties, as already mentioned before.
The combination of endemic disease risk maps with human population counts in the 'at risk' regions presents opportunities for designing the targeting of interventions such as resource allocation, vaccine campaigns or epidemic prevention measures to regions where they will have most cost-effective or burden-reducing impact [80, 108, 111]. For example, PAR estimates of malaria enabled the derivation of intervention coverage estimates [126], the intervention costs for reducing the malaria burden [127], malaria elimination feasibility [128], and the actual funding coverage [129]. The use of gridded population datasets in these studies facilitated more precise estimates of PAR than estimates based on aggregated population data by administrative units. An accurate assessment of 'at risk' populations also presents opportunities for designing disease surveillance and early warning systems for epidemics in the populations [130, 131].
The size of PAR is expected to vary in the future as a response to environmental and demographic changes. Some authors have attempted to assess the future PAR of malaria according to different scenarios [132–135]. While climate factors are predicted to cause limited changes in PAR of malaria for the year 2050 [134] and even a reduction in the size of PAR in Africa in the coming decades [133], demographic changes could significantly increase PAR. Applying forecasted population growth rates to gridded population data enables the derivation of estimates of future global population distribution [132, 135]. The combination of such projected data based on GPW2 with climate scenarios showed that population growth likely will have a larger effect than climate change on future global PAR of malaria estimates [135]. Moroever, a study examining the combined effects of climate, population and urbanisation changes confirmed the likely dominant effect of population growth in the increasing size of malaria PAR estimates in Africa [132]. Dengue fever risk in the future was also estimated based on population and climate projections for 2055 and 2085 [121].
Conclusions
Spatial methods and tools are now widely used in infectious disease research and have led to significant advances in our understanding of disease dynamics, surveillance and control [1, 2, 17, 18]. Population distribution datasets are becoming increasingly important inputs to these models. During the past decade, a number of advances have been made in GIS technologies that have allowed demographers and GIS specialists to begin to map the spatial distribution of human populations globally at an unprecedented level of detail. We have shown in this paper how useful large-scale gridded population datasets are for the calculation of PAR of infectious diseases and for disease risk mapping and modelling. Gridded population datasets allow the user to select geographic boundaries of interest independently from administrative boundaries. Population datasets capture spatial heterogeneities observed in disease transmission risks, making PAR calculations significantly more accurate than can be reached with aggregated population data.
Nevertheless, a number of issues and challenges remain, that, if resolved, would permit a more refined analysis of the spatial distribution of human population around the globe, and reduced uncertainty resulting from their use in epidemiological studies. Population distribution modelling methods have raised several issues and challenges such as the lack of comparability of statistical data from different countries and sources [43], the lack of standard definitions for what constitutes an urban area [136], the need for extensive source and metadata information, and the difficulties in validating the existing population datasets or measuring which existing dataset is the most accurate. The construction of contemporary, well-validated and well-documented spatial demographic datasets should be a priority in order to reduce uncertainties in spatial epidemiological studies [34]. In the absence of a more institutionalized mechanism to generate updated and freely available population datasets, data sharing should be encouraged between projects. Each dataset is for instance built upon similar population data linked to administrative boundaries and a standardized database framework that would encourage sharing of new and improved datasets between projects would greatly facilitate the data production and benefit the users.
Several extensions of population datasets would be particularly useful for infectious disease research (as well as other health related fields such as disaster risk management or conservation), for policy and planning. The most useful one would be to improve information on population attributes of interest. The disease impact in terms of morbidity, mortality, and speed of spread varies substantially with demographic profiles, so that identifying the most exposed or affected populations becomes a key aspect of planning and targeting interventions. It is not feasible for global population databases to generate on-demand maps for each variable of interest, nevertheless the potential to leverage current freely available population databases appears large, as was recently discussed in Tatem et al. [137]. These authors proposed a strategy for building an open-access database of spatial demographic data that is tailored to epidemiological applications.
Over the next few years, improvements in population distribution modelling methods and infectious disease distribution mapping will allow further refinements in PAR estimation and intervention targeting. Continued efforts to resolve the remaining challenges in accurate spatial population datasets construction will be required to obtain the full benefits from these potentially powerful methodologies.
References
Ostfeld RS, Glass GE, Keesing F: Spatial epidemiology: an emerging (or re-emerging) discipline. Trends Ecol Evol. 2005, 20: 328-336. 10.1016/j.tree.2005.03.009.
Elliott P, Wartenberg D: Spatial Epidemiology: Current Approaches and Future Challenges. Environ Health Perspect. 2004, 112: 998-1006. 10.1289/ehp.6735.
Lambin EF, Tran A, Vanwambeke SO, Linard C, Soti V: Pathogenic landscapes: Interactions between land, people, disease vectors, and their animal hosts. Int J Health Geogr. 2010, 9: 54-10.1186/1476-072X-9-54.
Hagenaars TJ, Donnelly CA, Ferguson NM: Spatial heterogeneity and the persistence of infectious diseases. J Theor Biol. 2004, 229: 349-359. 10.1016/j.jtbi.2004.04.002.
Grenfell B, Harwood J: (Meta) population dynamics of infectious diseases. Trends Ecol Evol. 1997, 12: 395-399. 10.1016/S0169-5347(97)01174-9.
Hess G, Randolph S, Arneberg P, Chemini C, Furlanello C, Harwood J, Roberts M, Swinton J: Spatial aspects of disease dynamics. The Ecology of Wildlife Diseases. 2002, Oxford Univ. Press, 102-118.
Ngwa GA, Shu WS: A mathematical model for endemic malaria with variable human and mosquito populations. Math Comput Model. 2000, 32: 747-763. 10.1016/S0895-7177(00)00169-2.
Anderson RM, May RM, Boily MC, Garnett GP, Rowley JT: The spread of HIV-1 in Africa: sexual contact patterns and the predicted demographic impact of AIDS. Nature. 1991, 352: 581-589. 10.1038/352581a0.
Hethcote HW: An age-structured model for pertussis transmission. Math Biosci. 1997, 145: 89-136. 10.1016/S0025-5564(97)00014-X.
World Health Organization: The global burden of disease: 2004 update. 2008, WHO Press. Geneva Switzerland
Murray CJ, Lopez AD: The global burden of disease: a comprehensive assessment of mortality and disability from diseases, injuries, and risk factors in 1990 and projected to 2020. 1996, Cambridge, Massachusetts: Harvard University Press
Murray CJ, Lopez AD: Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet. 1997, 349: 1269-1276. 10.1016/S0140-6736(96)07493-4.
Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C: Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002, 2: 25-32. 10.1016/S1473-3099(01)00170-0.
Black RE, Morris SS, Bryce J: Where and why are 10 million children dying every year?. Lancet. 2003, 361: 2226-2234. 10.1016/S0140-6736(03)13779-8.
Kosek M, Bern C, Guerrant RL: The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003, 81: 197-204.
De Silva NR, Brooker S, Hotez PJ, Montresor A, Engels D, Savioli L: Soil-transmitted helminth infections: updating the global picture. Trends Parasitol. 2003, 19: 547-551. 10.1016/j.pt.2003.10.002.
Riley S: Large-scale spatial-transmission models of infectious disease. Science. 2007, 316: 1298-10.1126/science.1134695.
Hay SI, Randolph SE, Rogers DJ: Remote sensing and geographical information systems in epidemiology. 2000, Oxford, UK: Academic Press, 47-
Beck LR, Lobitz BM, Wood BL: Remote sensing and human health: new sensors and new opportunities. Emerg Infect Dis. 2000, 6: 217-10.3201/eid0603.000301.
Graham AJ, Atkinson PM, Danson FM: Spatial analysis for epidemiology. Acta Trop. 2004, 91: 219-225. 10.1016/j.actatropica.2004.05.001.
Herbreteau V, Salem G, Souris M, Hugot JP, Gonzalez JP: Thirty years of use and improvement of remote sensing, applied to epidemiology: From early promises to lasting frustration. Health Place. 2007, 13: 400-403. 10.1016/j.healthplace.2006.03.003.
Huh OK, Malone JB: New tools: potential medical applications of data from new and old environmental satellites. Acta Trop. 2001, 79: 35-47. 10.1016/S0001-706X(01)00101-2.
Rogers DJ, Randolph SE: Studying the global distribution of infectious diseases using GIS and RS. Nat Rev Microbiol. 2003, 1: 231-236. 10.1038/nrmicro776.
Pfeiffer DU, Robinson TP, Stevenson M, Stevens KB, Rogers DJ, Clements ACA: Spatial analysis in epidemiology. 2008, Oxford; New York: Oxford University Press
Robinson TP: Spatial statistics and geographical information systems in epidemiology and public health. Adv Parasitol. 2000, 47: 81-128.
Bharti N, Tatem AJ, Ferrari MJ, Grais RF, Djibo A, Grenfell BT: Explaining Seasonal Fluctuations of Measles in Niger Using Nighttime Lights Imagery. Science. 2011, 334: 1424-1427. 10.1126/science.1210554.
Carneiro I, Roca-Feltrer A, Griffin JT, Smith L, Tanner M, Schellenberg JA, Greenwood B, Schellenberg D: Age-Patterns of Malaria Vary with Severity, Transmission Intensity and Seasonality in Sub-Saharan Africa: A Systematic Review and Pooled Analysis. PLoS One. 2010, 5: e8988-10.1371/journal.pone.0008988.
Okiro EA, Al-Taiar A, Reyburn H, Idro R, Berkley JA, Snow RW: Age patterns of severe paediatric malaria and their relationship to Plasmodium falciparum transmission intensity. Malar J. 2009, 8: 4-10.1186/1475-2875-8-4.
Teixeira MG, Costa MC, Coelho G, Barreto ML: Recent shift in age pattern of dengue hemorrhagic fever, Brazil. Emerg Infect Dis. 2008, 14: 1663-10.3201/eid1410.071164.
Brooker S, Donnelly CA, Guyatt HL: Estimating the number of helminthic infections in the Republic of Cameroon from data on infection prevalence in schoolchildren. Bull World Health Organ. 2000, 78: 1456-1465.
Petney TN: Environmental, cultural and social changes and their influence on parasite infections. Int J Parasitol. 2001, 31: 919-932. 10.1016/S0020-7519(01)00196-5.
Sumilo D, Asokliene L, Avsic-Zupanc T, Bormane A, Vasilenko V, Lucenko I, Golovljova I, Randolph SE: Behavioural responses to perceived risk of tick-borne encephalitis: Vaccination and avoidance in the Baltics and Slovenia. Vaccine. 2008, 26: 2580-2588. 10.1016/j.vaccine.2008.03.029.
Šumilo D, Bormane A, Asokliene L, Vasilenko V, Golovljova I, Avsic-Zupanc T, Hubalek Z, Randolph SE: Socio-economic factors in the differential upsurge of tick-borne encephalitis in Central and Eastern Europe. Rev Med Virol. 2008, 18: 81-95. 10.1002/rmv.566.
Tatem AJ, Campiz N, Gething PW, Snow RW, Linard C: The effects of spatial population dataset choice on population at risk of disease estimates. Popul Health Metr. 2011, 9: 4-10.1186/1478-7954-9-4.
Hay SI, Noor AM, Nelson A, Tatem AJ: The accuracy of human population maps for public health application. Trop Med Int Health. 2005, 10: 1073-10.1111/j.1365-3156.2005.01487.x.
Tatem AJ, Guerra CA, Kabaria CW, Noor AM, Hay SI: Human population, urban settlement patterns and their impact on Plasmodium falciparum malaria endemicity. Malar J. 2008, 7: 218-10.1186/1475-2875-7-218.
Deichmann U: A review of spatial population database design and modeling. 1996, University of California, Santa Barbara: National Center for Geographic Information and Analysis
Jones HR: Population geography. 1990, New York: Guilford Press
Salvatore M, Pozzi F, Ataman E, Huddleston B, Bloise M: Mapping global urban and rural population distributions. 2005, Rome: Food and Agriculture Organization of the United Nations
Wright JK: A method of mapping densities of population: With Cape Cod as an example. Geogr Rev. 1936, 26: 103-110. 10.2307/209467.
Balk D, Yetman G: The Global Distribution of Population: Evaluating the gains in resolution refinement. 2004, New York: Center for International Earth Science Information Network (CIESIN)
Balk DL, Deichmann U, Yetman G, Pozzi F, Hay SI, Nelson A: Determining global population distribution: methods, applications and data. Adv Parasitol. 2006, 62: 119-156.
Balk D, Pozzi F, Yetman G, Deichmann U, Nelson A: The distribution of people and the dimension of place: Methodologies to improve the global estimation of urban extents. Proceedings of the Urban Remote Sensing Conference. 2005, Tempe, Arizona: International Society for Photogrammetry and Remote Sensing
Mennis J: Generating surface models of population using dasymetric mapping. Prof Geogr. 2003, 55: 31-42.
Deichmann U, Balk D, Yetman G: Transforming population data for interdisciplinary usages: From census to grid. 2001, Washington DC: Center for International Earth Science Information Network
Tobler WR: Smooth pycnophylactic interpolation for geographical regions. J Am Stat Assoc. 1979, 74: 519-530. 10.2307/2286968.
Tobler W, Deichmann U, Gottsegen J, Maloy K: The global demography project. 1995, Santa Barbara: National Center for Geographic Information and Analysis, Department of Geography, University of California
Tobler W, Deichmann U, Gottsegen J, Maloy K: World population in a grid of spherical quadrilaterals. Int J Popul Geogr. 1997, 3: 203-225. 10.1002/(SICI)1099-1220(199709)3:3<203::AID-IJPG68>3.0.CO;2-C.
Mennis J: Dasymetric Mapping for Estimating Population in Small Areas. Geography Compass. 2009, 3: 727-745. 10.1111/j.1749-8198.2009.00220.x.
Linard C, Gilbert M, Snow RW, Noor AM, Tatem AJ: Population Distribution, Settlement Patterns and Accessibility across Africa in 2010. PLoS One. 2012, 7: e31743-10.1371/journal.pone.0031743.
AfriPop project.http://www.afripop.orghttp://www.afripop.org
Asia Population Database Documentation.http://na.unep.net/siouxfalls/globalpop/asia/http://na.unep.net/siouxfalls/globalpop/asia/
Hyman G, Lema G, Nelson A, Deichmann U: Latin American and Caribbean Population Database Documentation. 2004, Mexico: International Center for Tropical Agriculture
African population database documentation.http://na.unep.net/siouxfalls/globalpop/africa/Africa_index.htmlhttp://na.unep.net/siouxfalls/globalpop/africa/Africa_index.html
Deichmann U, Eklundh L: Global digital datasets for land degradation studies: A GIS approach. 1991, Nairobi, Kenya: United Nations Environment Programme, Global Resource Information Database, Case Study No. 4
Dobson JE, Bright EA, Coleman PR, Durfee RC, Worley BA: LandScan: a global population database for estimating populations at risk. Photogramm Eng Remote Sensing. 2000, 66: 849-857.
Bhaduri B, Bright E, Coleman P, Urban M: LandScan USA: a high-resolution geospatial and temporal modeling approach for population distribution and dynamics. Geo Journal. 2007, 69: 103-117.
Bhaduri B, Bright E, Coleman P, Dobson J: LandScan: Locating people is what matters. Geoinformatics. 2002, 5: 34-37.
Mba CJ: Assessing the reliability of the 1986 and 1996 Lesotho census data. J Soc Dev Afr. 2003, 18: 111-128.
Mba CJ: Challenges of population census enumeration in Africa: an illustration with the age-sex data of the Gambia. Inst Afr Stud Res Rev. 2004, 20: 9-
Tatem AJ, Noor AM, von Hagen C, Di Gregorio A, Hay SI: High resolution population maps for low income nations: combining land cover and census in East Africa. PLoS One. 2007, 2: e1298-10.1371/journal.pone.0001298.
Linard C, Gilbert M, Tatem AJ: Assessing the use of global land cover data for guiding large area population distribution modelling. Geo Journal. 2010, 76: 525-538.
United Nations Population Division: World Population Prospects: The 2010 Revision. 2010, New York: United Nations
Lutz W, Samir KC: Dimensions of global population projections: what do we know about future population trends and structures?. Philos Trans R Soc Lond B Biol Sci. 2010, 365: 2779-2791. 10.1098/rstb.2010.0133.
Mennis J, Hultgren T: Intelligent dasymetric mapping and its application to areal interpolation. Cartogr Geogr Inf Sci. 2006, 33: 179-194. 10.1559/152304006779077309.
Gregory IN: The accuracy of areal interpolation techniques: standardising 19th and 20th century census data to allow long-term comparisons. Comput Environ Urban Syst. 2002, 26: 293-314. 10.1016/S0198-9715(01)00013-8.
Sabesan A, Abercrombie K, Ganguly AR, Bhaduri B, Bright EA, Coleman PR: Metrics for the comparative analysis of geospatial datasets with applications to high-resolution grid-based population data. Geo Journal. 2007, 69: 81-91.
Linard C, Alegana VA, Noor AM, Snow RW, Tatem AJ: A high resolution spatial population database of Somalia for disease risk mapping. Int J Health Geogr. 2010, 9: 45-10.1186/1476-072X-9-45.
Tatem AJ, Noor AM, Hay SI: Assessing the accuracy of satellite derived global and national urban maps in Kenya. Remote Sens Environ. 2005, 96: 87-97. 10.1016/j.rse.2005.02.001.
Herold M, Mayaux P, Woodcock CE, Baccini A, Schmullius C: Some challenges in global land cover mapping: An assessment of agreement and accuracy in existing 1 km datasets. Remote Sens Environ. 2008, 112: 2538-2556. 10.1016/j.rse.2007.11.013.
Potere D, Schneider A, Angel S, Civco D: Mapping urban areas on a global scale: which of the eight maps now available is more accurate?. Int J Remote Sens. 2009, 30: 6531-6558. 10.1080/01431160903121134.
Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P: Global trends in emerging infectious diseases. Nature. 2008, 451: 990-993. 10.1038/nature06536.
Est'ıvariz CF, Watkins MA, Handoko D, Rusipah R, Deshpande J, Rana BJ, Irawan E, Widhiastuti D, Pallansch MA, Thapa A: A large vaccine-derived poliovirus outbreak on Madura Island-Indonesia, 2005. J Infect Dis. 2008, 197: 347-354. 10.1086/525049.
Fischer EAJ, Pahan D, Chowdhury SK, Richardus JH: The spatial distribution of leprosy cases during 15 years of a leprosy control program in Bangladesh: An observational study. BMC Infect Dis. 2008, 8: 126-10.1186/1471-2334-8-126.
Kalipeni E, Zulu LC: HIV and AIDS in Africa: a geographic analysis at multiple spatial scales. Geo Journal. 2010, DOI: 10.1007/s10708-010-9358-6
Reid RS, Kruska RL, Deichmann U, Thornton PK, Leak SG: Human population growth and the extinction of the tsetse fly. Agric Ecosyst Environ. 2000, 77: 227-236. 10.1016/S0167-8809(99)00103-6.
Hay SI, Guerra CA, Tatem AJ, Atkinson PM, Snow RW: Urbanization, malaria transmission and disease burden in Africa. Nat Rev Microbiol. 2005, 3: 81-90. 10.1038/nrmicro1069.
Pfeiffer DU, Minh PQ, Martin V, Epprecht M, Otte MJ: An analysis of the spatial and temporal patterns of highly pathogenic avian influenza occurrence in Vietnam using national surveillance data. Vet J. 2007, 174: 302-309. 10.1016/j.tvjl.2007.05.010.
Rogers DJ, Wilson AJ, Hay SI, Graham AJ: The global distribution of yellow fever and dengue. Adv Parasitol. 2006, 62: 181-
Kelly-Hope LA, McKenzie FE: The multiplicity of malaria transmission: a review of entomological inoculation rate measurements and methods across sub-Saharan Africa. Malar J. 2009, 8: 19-10.1186/1475-2875-8-19.
Gemperli A, Sogoba N, Fondjo E, Mabaso M, Bagayoko M, Briet OJT, Anderegg D, Liebe J, Smith T, Vounatsou P: Mapping malaria transmission in West and Central Africa. Trop Med Int Health. 2006, 11: 1032-1046. 10.1111/j.1365-3156.2006.01640.x.
Henning J, Pfeiffer DU, Vu LT: Risk factors and characteristics of H5N1 Highly Pathogenic Avian Influenza (HPAI) post-vaccination outbreaks. Vet Res. 2009, 40: 15-10.1051/vetres:2008053.
Wint GR, Robinson TP, Bourn DM, Durr PA, Hay SI, Randolph SE, Rogers DJ: Mapping bovine tuberculosis in Great Britain using environmental data. Trends Microbiol. 2002, 10: 441-444. 10.1016/S0966-842X(02)02444-7.
Gilbert M, Xiao X, Pfeiffer DU, Epprecht M, Boles S, Czarnecki C, Chaitaweesub P, Kalpravidh W, Minh PQ, Otte MJ: Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proc Natl Acad Sci USA. 2008, 105: 4769-10.1073/pnas.0710581105.
Napier M: Application of GIS and modeling of dengue risk areas in the Hawaiian islands. 2003, Hawaii: Pacific Disaster Centre
Johansson MA, Dominici F, Glass GE: Local and Global Effects of Climate on Dengue Transmission in Puerto Rico. PLoS Negl Trop Dis. 2009, 3 (2): e382-10.1371/journal.pntd.0000382.
Moffett A, Shackelford N, Sarkar S: Malaria in Africa: vector species' niche models and relative risk maps. PLoS One. 2007, 2: e824-10.1371/journal.pone.0000824.
Schur N, Hürlimann E, Garba A, Traoré MS, Ndir O, Ratard RC, Tchuem Tchuenté L-A, Kristensen TK, Utzinger J, Vounatsou P: Geostatistical Model-Based Estimates of Schistosomiasis Prevalence among Individuals Aged ≤ 20 Years in West Africa. PLoS Negl Trop Dis. 2011, 5: e1194-10.1371/journal.pntd.0001194.
Kiszewski A, Mellinger A, Spielman A, Malaney P, Sachs SE, Sachs J: A global index representing the stability of malaria transmission. Am J Trop Med Hyg. 2004, 70: 486-498.
You L, Diao X: Assessing the Potential Impact of Avian Influenza on Poultry in West Africa: A Spatial Equilibrium Analysis. Journal of Agricultural Economics. 2007, 58: 348-367. 10.1111/j.1477-9552.2007.00099.x.
Ferguson NM, Cummings DA, Cauchemez S, Fraser C, Riley S, Meeyai A, Iamsirithaworn S, Burke DS: Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005, 437: 209-214. 10.1038/nature04017.
Rakowski F, Gruziel M, Bieniasz-Krzywiec Ł, Radomski JP: Influenza epidemic spread simulation for Poland -- a large scale, individual based model study. Physica A. 2010, 389: 3149-3165. 10.1016/j.physa.2010.04.029.
Rao DM, Chernyakhovsky A, Rao V: Modeling and analysis of global epidemiology of avian influenza. Environ Model Software. 2009, 24: 124-134. 10.1016/j.envsoft.2008.06.011.
Balcan D, Colizza V, Gonçalves B, Hu H, Ramasco JJ, Vespignani A: Multiscale mobility networks and the spatial spreading of infectious diseases. Proc Natl Acad Sci USA. 2009, 106: 51-10.1073/pnas.0901953106.
Ferguson NM, Cummings DAT, Fraser C, Cajka JC, Cooley PC, Burke DS: Strategies for mitigating an influenza pandemic. Nature. 2006, 442: 448-452. 10.1038/nature04795.
Riley S, Ferguson NM: Smallpox transmission and control: spatial dynamics in Great Britain. Proc Natl Acad Sci USA. 2006, 103: 12637-10.1073/pnas.0510873103.
Merler S, Ajelli M: The role of population heterogeneity and human mobility in the spread of pandemic influenza. Proc Biol Sci. 2009, 277: 557-565.
Balcan D, Hu H, Goncalves B, Bajardi P, Poletto C, Ramasco JJ, Paolotti D, Perra N, Tizzoni M, Broeck WV: Seasonal transmission potential and activity peaks of the new influenza A (H 1 N 1): a Monte Carlo likelihood analysis based on human mobility. BMC Med. 2009, 7: 45-10.1186/1741-7015-7-45.
Bajardi P, Poletto C, Balcan D, Hu H, Goncalves B, Ramasco J, Paolotti D, Perra N, Tizzoni M, Van den Broeck W, Colizza V, Vespignani A: Modeling vaccination campaigns and the Fall/Winter 2009 activity of the new A(H1N1) influenza in the Northern Hemisphere. Emerg Health Threats J. 2009, 2: e11-
Tatem AJ, Smith DL: International population movements and regional Plasmodium falciparum malaria elimination strategies. Proc Natl Acad Sci USA. 2010, 107: 12222-12227. 10.1073/pnas.1002971107.
Brooker S, Clements AC: Spatial heterogeneity of parasite co-infection: Determinants and geostatistical prediction at regional scales. Int J Parasitol. 2009, 39: 591-597. 10.1016/j.ijpara.2008.10.014.
Keeling MJ: Dynamics of the 2001 UK Foot and Mouth Epidemic: Stochastic Dispersal in a Heterogeneous Landscape. Science. 2001, 294: 813-817. 10.1126/science.1065973.
Smith DL, Lucey B, Waller LA, Childs JE, Real LA: Predicting the spatial dynamics of rabies epidemics on heterogeneous landscapes. Proc Natl Acad Sci USA. 2002, 99: 3668-
Simarro PP, Cecchi G, Paone M, Franco JR, Diarra A, Ruiz JA, Fèvre EM, Courtin F, Mattioli RC, Jannin JG: The Atlas of human African trypanosomiasis: a contribution to global mapping of neglected tropical diseases. Int J Health Geogr. 2010, 9: 57-10.1186/1476-072X-9-57.
Snow RW, Craig MH, Deichmann U, Le Sueur D: A preliminary continental risk map for malaria mortality among African children. Parasitol Today. 1999, 15: 99-104. 10.1016/S0169-4758(99)01395-2.
Snow RW, Craig M, Deichmann U, Marsh K: Estimating mortality, morbidity and disability due to malaria among Africa's non-pregnant population. Bull World Health Organ. 1999, 77: 624-
Cox J, Hay SI, Abeku TA, Checchi F, Snow RW: The uncertain burden of Plasmodium falciparum epidemics in Africa. Trends Parasitol. 2007, 23: 142-148. 10.1016/j.pt.2007.02.002.
Riedel N, Vounatsou P, Miller J, Gosoniu L, Chizema-Kawesha E, Mukonka V, Steketee R: Geographical patterns and predictors of malaria risk in Zambia: Bayesian geostatistical modelling of the 2006 Zambia national malaria indicator survey (ZMIS). Malar J. 2010, 9: 37-10.1186/1475-2875-9-37.
Guerra CA, Snow RW, Hay SI: Defining the global spatial limits of malaria transmission in 2005. Adv Parasitol. 2006, 62: 157-179.
Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI: The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005, 434: 214-217. 10.1038/nature03342.
Hay SI, Okiro EA, Gething PW, Patil AP, Tatem AJ, Guerra CA, Snow RW: Estimating the Global Clinical Burden of Plasmodium falciparum Malaria in 2007. PLoS Med. 2010, 7: e1000290-10.1371/journal.pmed.1000290.
Guerra CA, Gikandi PW, Tatem AJ, Noor AM, Smith DL, Hay SI, Snow RW: The limits and intensity of Plasmodium falciparum transmission: implications for malaria control and elimination worldwide. PLoS Med. 2008, 5: e38-10.1371/journal.pmed.0050038.
Hay SI, Guerra CA, Gething PW, Patil AP, Tatem AJ, Noor AM, Kabaria CW, Manh BH, Elyazar IRF, Brooker S, Smith DL, Moyeed RA, Snow RW: A World Malaria Map: Plasmodium falciparum Endemicity in 2007. PLoS Med. 2009, 6: e48-10.1371/journal.pmed.1000048.
Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, Temperley WH, Kabaria CW, Tatem AJ, Manh BH, Elyazar IRF, Baird JK, Snow RW, Hay SI: The International Limits and Population at Risk of Plasmodium vivax Transmission in 2009. PLoS Negl Trop Dis. 2010, 4: e774-10.1371/journal.pntd.0000774.
Brooker S, Miguel EA, Waswa P, Namunyu R, Moulin S, Guyatt H, Bundy DAP: The potential of rapid screening methods for Schistosoma mansoni in western Kenya. Ann Trop Med Parasitol. 2001, 95: 343-351. 10.1080/00034980120063437.
Brooker S, Beasley M, Ndinaromtan M, Madjiouroum EM, Baboguel M, Djenguinabe E, Hay SI, Bundy DA: Use of remote sensing and a geographical information system in a national helminth control programme in Chad. Bull World Health Organ. 2002, 80: 783-789.
Brooker S, Hotez PJ, Bundy DA, Raso G: Hookworm-related anaemia among pregnant women: a systematic review. PLoS Negl Trop Dis. 2008, 2: e291-10.1371/journal.pntd.0000291.
Lindsay SW, Thomas CJ: Mapping and estimating the population at risk from lymphatic filariasis in Africa. Trans R Soc Trop Med Hyg. 2000, 94: 37-45. 10.1016/S0035-9203(00)90431-0.
Simarro PP, Cecchi G, Franco JR, Paone M, Fèvre EM, Diarra A, Postigo JAR, Mattioli RC, Jannin JG: Risk for Human African Trypanosomiasis, Central Africa, 2000-2009. Emerg Infect Dis. 2011, 17: 2322-2324. 10.3201/eid1712.110921.
Hales S, de Wet N, Maindonald J, Woodward A: Potential effect of population and climate changes on global distribution of dengue fever: an empirical model. Lancet. 2002, 360: 830-834. 10.1016/S0140-6736(02)09964-6.
Brooker S, Clements AC, Hotez PJ, Hay SI, Tatem AJ, Bundy DA, Snow RW: The co-distribution of Plasmodium falciparum and hookworm among African schoolchildren. Malar J. 2006, 5: 99-10.1186/1475-2875-5-99.
Brooker S, Akhwale W, Pullan R, Estambale B, Clarke SE, Snow RW, Hotez PJ: Epidemiology of plasmodium-helminth co-infection in Africa: populations at risk, potential impact on anemia, and prospects for combining control. Am J Trop Med Hyg. 2007, 77: 88-
Dellicour S, Tatem AJ, Guerra CA, Snow RW, ter Kuile FO: Quantifying the Number of Pregnancies at Risk of Malaria in 2007: A Demographic Study. PLoS Med. 2010, 7: e1000221-10.1371/journal.pmed.1000221.
Gething PW, Kirui VC, Alegana VA, Okiro EA, Noor AM, Snow RW: Estimating the Number of Paediatric Fevers Associated with Malaria Infection Presenting to Africa's Public Health Sector in 2007. PLoS Med. 2010, 7: e1000301-10.1371/journal.pmed.1000301.
Noor AM, Mutheu JJ, Tatem AJ, Hay SI, Snow RW: Insecticide-treated net coverage in Africa: mapping progress in 2000-07. Lancet. 2008, 373: 58-67.
Teklehaimanot A, McCord GC, Sachs JD: Scaling up malaria control in Africa: an economic and epidemiological assessment. Am J Trop Med Hyg. 2007, 77: 138-
Tatem AJ, Smith DL, Gething PW, Kabaria CW, Snow RW, Hay SI: Ranking of elimination feasibility between malaria-endemic countries. Lancet. 2010, 376: 1579-1591. 10.1016/S0140-6736(10)61301-3.
Snow RW, Guerra CA, Mutheu JJ, Hay SI: International Funding for Malaria Control in Relation to Populations at Risk of Stable Plasmodium falciparum Transmission. PLoS Med. 2008, 5: e142-10.1371/journal.pmed.0050142.
Abeku TA, Hay SI, Ochola S, Langi P, Beard B, de Vlas SJ, Cox J: Malaria epidemic early warning and detection in African highlands. Trends Parasitol. 2004, 20: 400-405. 10.1016/j.pt.2004.07.005.
Hay SI, Rogers DJ, Shanks GD, Myers MF, Snow RW: Malaria early warning in Kenya. Trends Parasitol. 2001, 17: 95-99. 10.1016/S1471-4922(00)01763-3.
Hay SI, Tatem AJ, Guerra CA, Snow RW: Infectious Diseases: preparing for the future - T8.2: Population at malaria risk in Africa: 2005, 2015 and 2030. 2006, Centre for Geographic Medicine KEMRI/Wellcome Trust Collaborative Programme, Kenya; University of Oxford, UK
Peterson AT: Shifting suitability for malaria vectors across Africa with warming climates. BMC Infect Dis. 2009, 9: 59-10.1186/1471-2334-9-59.
Rogers DJ, Randolph SE: The global spread of malaria in a future, warmer world. Science. 2000, 289: 1763-
Van Lieshout M, Kovats RS, Livermore MTJ, Martens P: Climate change and malaria: analysis of the SRES climate and socio-economic scenarios. Glob Environ Change. 2004, 14: 87-99. 10.1016/j.gloenvcha.2003.10.009.
Utzinger J, Keiser J: Urbanization and tropical health then and now. Ann Trop Med Parasitol. 2006, 100: 517-533. 10.1179/136485906X97372.
Tatem AJ, Adamo S, Bharti N, Burgert CR, Castro M, Dorelien A, Fink G, Linard C, Mendelsohn J, Montana L, Montgomery MR, Nelson A, Noor AM, Pindolia D, Yetman G, Balk D: Mapping populations at risk: Improving spatial demographic data for infectious disease modeling and health metric derivation. Popul Health Metr.
Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW: The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004, 4: 327-336. 10.1016/S1473-3099(04)01043-6.
Gething PW, Patil AP, Hay SI: Quantifying Aggregated Uncertainty in Plasmodium falciparum Malaria Prevalence and Populations at Risk via Efficient Space-Time Geostatistical Joint Simulation. PLoS Comput Biol. 2010, 6: e1000724-10.1371/journal.pcbi.1000724.
Brooker S, Clements AC, Bundy DA: Global epidemiology, ecology and control of soil-transmitted helminth infections. Adv Parasitol. 2006, 62: 221-261.
Noma M, Nwoke BEB, Nutall I, Tambala PA, Enyong P, Namsenmo A, Remme J, Amazigo UV, Kale OO, Seketeli A: Rapid epidemiological mapping of onchocerciasis (REMO): its application by the African Programme for Onchocerciasis Control (APOC). Ann Trop Med Parasitol. 2002, 96: 29-39. 10.1179/000349802125000637.
Pullan RL, Gething PW, Smith JL, Mwandawiro CS, Sturrock HJW, Gitonga CW, Hay SI, Brooker S: Spatial Modelling of Soil-Transmitted Helminth Infections in Kenya: A Disease Control Planning Tool. PLoS Negl Trop Dis. 2011, 5: e958-10.1371/journal.pntd.0000958.
Clements AC, Firth S, Dembelé R, Garba A, Touré S, Sacko M, Landouré A, Bosqué-Oliva E, Barnett AG, Brooker S, Fenwick A: Use of Bayesian geostatistical prediction to estimate local variations in Schistosoma haematobium infection in western Africa. Bull World Health Organ. 2009, 87: 921-929. 10.2471/BLT.08.058933.
Gilbert M, Mitchell A, Bourn D, Mawdsley J, Clifton-Hadley R, Wint W: Cattle movements and bovine tuberculosis in Great Britain. Nature. 2005, 435: 491-496. 10.1038/nature03548.
Beasley M, Brooker S, Ndinaromtan M, Madjiouroum EM, Baboguel M, Djenguinabe E, Bundy DA: First nationwide survey of the health of schoolchildren in Chad. Trop Med Int Health. 2002, 7: 625-10.1046/j.1365-3156.2002.00900.x.
Acknowledgements
CL is supported by the Fonds National de la Recherche Scientifique (F.R.S./FNRS). AJT is supported by grants from the Bill and Melinda Gates Foundation (#49446, and #OPP1032350). AJT also acknowledges funding support from the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health. This work forms part of the AfriPop and AsiaPop Projects http://www.afripop.org, http://www.asiapop.org, principally funded by the Fondation Philippe Wiener - Maurice Anspach, and the Malaria Atlas Project (MAP, http://www.map.ox.ac.uk), principally funded by the Wellcome Trust, U.K. The F.R.S./FNRS, the Fondation Philippe Wiener - Maurice Anspach, the Bill and Melinda Gates Foundation and the Wellcome Trust have no intellectual or editorial input into the content of this Review.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CL conducted the core literature review and drafted the manuscript. AJT helped to draft the manuscript and revised it critically. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Linard, C., Tatem, A.J. Large-scale spatial population databases in infectious disease research. Int J Health Geogr 11, 7 (2012). https://doi.org/10.1186/1476-072X-11-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-072X-11-7