Abstract
Background
Multidrug resistant Pseudomonas aeruginosa harbours integrons and other mobile genetic elements such as plasmids and transposons, which easily disseminate antibiotic resistance genes among clinical strains of P. aeruginosa.
Methodology
Plasmid extraction of 54 clinical isolates of P. aeruginosa was carried out by alkaline lysis method; and plasmid size estimation was done by using E. coli V517 standard plasmid marker. Fifty-four clinical strains of P. aeruginosa were isolated from 5 hospitals in 3 Southwestern states of Nigeria between March and September 2010. Plasmid extraction of isolates was carried out by alkaline lysis method; and plasmid size estimation was done by using E. coli V517 standard plasmid marker. PCR amplification for the 3 classes of resistance integrons, and gene cassette characterization were carried out using specific primers and by sequencing of PCR products. Conjugal mating of the integron positive P. aeruginosa strains with E. coli DH5α was performed to demonstrate transferability of integrons and gene cassettes.
Result
Agarose gel electrophoresis of plasmid DNA revealed that all the 54 P. aeruginosa harboured 1–4 plasmids with sizes ranging from 2.2 – >58 kb. Class 1 integron was identified in 31 (57%) strains; but none of them carried class 2 and class 3 integrons. High prevalence of aadA gene conferring resistance to streptomycin/spectinomycin was detected in the strains positive for class 1 integron. Sequencing of the 1.6 kb and 1.2 kb amplified band of gene cassettes revealed the presence of aadA6-orfD and aadA13 respectively.
Conclusion
This study demonstrates the presence of plasmids and integrons harbouring resistance gene cassettes, which may collectively constitute an efficient system for dissemination of resistance genes in P. aeruginosa. Disturbingly, the rapid and unabated spread of class 1 integron-associated multidrug resistant P. aeruginosa in Southwest Nigeria may greatly hamper successful treatment of infections caused by such strains. This necessitates the establishment of functional antimicrobial resistance surveillance programmes in Nigeria.
Similar content being viewed by others
Introduction
The worldwide threat of nosocomial multidrug resistant Pseudomonas aeruginosa is a growing concern among hospitalized patients. Infections caused by P. aeruginosa are severe and mostly associated with high mortality and morbidity rates. P. aeruginosa frequently develops resistance during therapy; hence, it becomes difficult to treat [1]. P. aeruginosa is naturally resistant to many structurally unrelated antibiotics because of its dominant array of chromosomal and plasmid-mediated antibiotic resistance factors [1]; as well as its dynamic propensity to acquire newer resistance genes from bacteria from other genera such as Acinetobacter baumannii, Klebsiella pneumoniae and Salmonella spp [2].
Mobile genetic elements such as plasmids, transposons and integrons are means of acquiring resistance mechanisms contributing to P. aeruginosa multidrug resistance [2]. These mobile genetic elements play an important role in the dissemination of resistance genes among bacteria. Several studies have related antimicrobial resistance of P. aeruginosa to the presence of one or more of these genetic elements [3–6]. Integrons are genetic elements that capture and integrate gene cassettes by site-specific recombination and convert them to functional genes. The essential components of an integron include the integrase gene (intI), the gene for the adjacent recombination site (attI) and the promoter (Pc), which promotes the expression of any suitably integrated gene [7]. Genes carried by integrons usually encodes multiple resistance mechanisms such as resistance to beta-lactams, aminoglycosides and other antimicrobial agents [8, 9].
There are three main well characterized classes of antibiotic resistance integrons designated class 1, 2 and 3 [10, 11]. The class 1 integrons remain the most common integrons found in members of the family Enterobacteriaceae such as Enterobacter spp, K. pneumoniae, E. coli and Proteus spp., as well as other clinically important Gram-negative bacteria such as P. aeruginosa and Acinetobacter baumannii [6, 12]. Detection of class 2 and class 3 integrons among these nosocomial pathogens is not widely reported [12]. Though, a recent study from Southern China found class 2 integron carrying Tn7-like dfrA1-sat1-aadA1 conferring resistance to trimethoprim, streptothricin and streptomycin/spectinomycin respectively in clinical strains of P. aeruginosa [13].
In Nigeria, bacterial resistance to cheap and readily available antimicrobials has necessitated the wide use of broad-spectrum antibiotics [14]. Consequently, multidrug-resistant P. aeruginosa, as well as other resistant nosocomial pathogens have been associated with serious infections due to indiscriminate use of these broad-spectrum antibiotics [15]. Integrons, plasmids and other mobile genetic elements have been implicated in the evolution and dissemination of multidrug resistant bacteria [11, 12]. However, reports of multidrug resistance in P. aeruginosa associated with mobile genetic elements are scanty in our region. The aim of this study was to investigate integron-associated resistance and characterize the respective gene cassettes among multidrug resistant P. aeruginosa from Nigeria.
Materials and method
Bacterial isolates
Fifty-four non-duplicate multidrug resistant P. aeruginosa strains isolated during a seven-month period (March - September 2010) from University College Hospital Ibadan (n = 20), Catholic Hospital Oluyoro Ibadan (n = 12), Catholic Hospital Eleta Ibadan (n = 7), Federal Medical Centre Akure (n = 5), Federal Medical Centre Abeokuta (n = 10) were included in this study. The isolates were obtained from different clinical samples (wound swab, urine, pus, ear swab, blood and vagina swab) and were verified using standard biochemical methods as described previously [16]. All isolates were collected under approved ethical standards.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined by the disk diffusion method on Mueller-Hinton agar (Oxoid UK), according to the Clinical and Laboratory Standards Institute guidelines [17]. Antimicrobial susceptibility testing was performed for 13 antimicrobial agents: ceftriaxone (CRO, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg), carbenicillin (CAR, 100 μg), piperacillin (PRL, 100 μg), levofloxacin (LEV, 5 μg), ciprofloxacin (CIP, 5 μg), gentamicin (GEN, 10 μg), amikacin (AMK, 30 μg), streptomycin (STR, 10 μg), tetracycline (TET, 30 μg), imipenem (IPM, 10 μg) and ticarcillin/clavulanic acid (75/10 μg). P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used for quality control of the susceptibility testing.
Plasmid and genomic DNA extraction
Plasmid extraction was carried out by alkaline lysis method [18], with E.coli V517 used for plasmid size estimation as described previously [19]. Plasmid DNA bands were visualized and photographed by using Gel Documentation system with UV transillumination. Genomic DNA extraction was carried out as described previously [20] with modifications. Briefly, the P. aeruginosa isolates were inoculated into 2 ml of Trypticase Soy broth (Difco, Detroit, MI) and incubated overnight at 37°C. The bacterial cells were harvested by centrifugation at 8,000 × g for 5 min and the supernatant was completely removed using sterile Pasteur pipette. The pellet was resuspended in 500 μl of Tris EDTA (TE) buffer. The cells were lysed by boiling for 10 min in a water bath, cooled on ice, and centrifuged at 14,000 × g for 5 min to remove any cell debris before it was stored at -20°C. Aliquots of 2 μl of the template DNA were used for polymerase chain reaction (PCR).
Integron detection
Class 1, 2 and 3 integrons were initially detected by PCR using degenerate primer set hep35 and hep36 (Table 1), which hybridizes to conserve regions of integron-encoded integrase genes intI1, intI2, and intI3[21]. The class of integron was determined by analyzing integrase PCR products through enzyme digestion using HinfI as described previously [22]. Specific primers for class 2 and 3 integrons were also used in separate PCR assays [23] [Table 1].
Characterization of class 1 integron cassette arrays and DNA sequencing
Class 1 integron cassette regions were amplified using hep58 and hep59 primers as described previously [21] using the following modified conditions; initial denaturation of 94°C for 2 min, 35 cycles of 94°C for 30 s, 55°C for 45 s, extension at 72°C for 45 s and final extension at 72°C for 7 min. Amplified gene cassette of the same fragment sizes, were further sequenced to determine the cassette array present. The amplicons were purified and sequenced at the sequencing facility of Chromous Biotech, Bangalore, India.
Sequence analysis and nucleotide sequence accession number
Homology searches were performed with BLAST (http://www.ncbi.nlm.nih.gov). Sequences were assembled with BioEdit, and multiple sequences were aligned with Clustal W (http://www.ebi.ac.uk/). The sequences obtained from the gene cassette analysis have been deposited in GenBank/EMBL/DDBJ under accession numbers JX195555 for aadA6-ofrD of P. aeruginosa strain ODM-24 and JX195556 for aadA13 of strain ODM-08.
Conjugation assay
Conjugation was carried out using previously described protocol [24] with modification. Briefly, donor P. aeruginosa and recipient nalidixic acid-resistant E. coli DH5α cells were grown to logarithmic phase in Luria broth (LB), equal volume (500 μl) of each strains were mixed thoroughly in a tube and centrifuged at 5000 rpm for 3 min. The pellets were inoculated on Mueller Hinton agar plates containing no antibiotics and incubated at 37°C for 24 h. Transconjugants were selected on nutrient agar plates containing 30 μg nalidixic acid plus one of the antibiotics to which the donor strain was resistant as described previously [24].
Results
Susceptibility testing
Susceptibility testing of the 31 integron positive P. aeruginosa isolates showed that all the isolates were resistant to ≥ 5 of the 13 antimicrobial agents tested in this study. Resistance was mostly observed for tetracycline (100%), amoxicillin/clavulanate (100%), followed by streptomycin (90.3%), ceftriaxone (87.1%), kanamycin (83.8%), carbenicillin (80.6%), cefotaxime (77.4%) and to a lesser degree amikacin (25.5%), ceftazidime (22.5%), and imipenem (9.6%) (Table 2).
Plasmid profile
Agarose gel electrophoresis of the plasmid DNA revealed that all the 54 P. aeruginosa strains harboured 1–4 plasmids with sizes ranging from 2.2 – >58 kb as compared with the corresponding E. coli V517 standard marker (Table 3). Eleven distinct plasmid profiles labelled A-K were identified. Thirty-seven (69%) P. aeruginosa strains carried a single large plasmid (58 kb or >58 kb) while 17 (32%) isolates harboured 2 – 4 variety of plasmids that differed in sizes (Table 3).
Intgenes
PCR assay with primers hep35 and hep36 revealed that 31 (57%) of the 54 P. aeruginosa strains were integrase positive (Figure 1). Analysis of the integrase PCR product by restriction fragment length polymorphism (RFLP) by HinfI also yielded a single fragment of 491 bp among the 31 strains, confirming class 1integron as described previously [21, 22]. None of the isolates were positive for class 2 and 3 integrons specific PCR assays.
Gene cassette array
Using the hep58 and hep59 primers for gene cassette characterization, two different fragments sizes of approximately 1.6 kb and 1.2 kb were obtained for all the 31 intI1-positive isolates (Figure 2). Twenty-three (74%) isolates yielded a single fragment of approximately 1.6 kb while the remaining 8 isolates gave a single fragment of 1.2 kb. Sequence data [accession number JX195555] obtained from sequencing the 1.6 kb gene cassette fragment from strain ODM-24 gave 100% homology with aadA6, conferring resistance to streptomycin and spectinomycin and orfD of unknown function [25, 26] whereas the 1.2 kb fragment of strain ODM-08 was 100% identical to aadA13, which also confer resistance to streptomycin and spectinomycin [accession number JX195556].
Transfer of antibiotic resistance and genetic location of integrons
All the 31 class 1integron positive P. aeruginosa strains were tested for conjugal transfer of resistance plasmids and gene cassettes. Conjugation experiment was successful in 10 (32%) donor strains but none of the E. coli DHα transconjugants plasmids was positive for class 1 integron and gene cassette by PCR.
Discussion
To our knowledge, this is the first study to report carriage of class 1 integrons and associated gene cassettes in P. aeruginosa isolates from Nigeria. The 57.4% incidence rate of class 1 integrons observed in this study is higher than previously reported rates of 41.5% from Brazil [3], 45.8% from China [13], and 56.3% from Iran [27]. The prevalence of class 1 integrons in clinical isolates of P. aeruginosa from Southwest Nigeria is of great concern because these genetic elements are highly stable among resistant pathogens; and also capable of easy spread and capture of other multidrug resistance gene cassettes leading to increase in resistance to broad-spectrum antibiotics [28]. Consistent with other previous studies that documented low detection of class 2 and class 3 integrons in clinical P. aeruginosa[13, 29], none of the isolates in this study harboured class 2 or class 3 integron.
In Africa, class 1 integrons have been reported previously in clinical isolates of P. aeruginosa. Labuschagne et al. [30] reported blaGES-5 and blaGES-5-like genes as part of the variable region of class 1 integrons, occurring in three clinical P. aeruginosa isolates from South Africa. Another study reported class 1 integron containing blaVIM-2, aacA7 and aacA4, as well as aadB and arr6, a novel rifampin resistance gene among 35 clonally related P. aeruginosa isolated from a hospital in Tunisia [31]. This suggests that class 1 integrons are strongly associated with multiple drug resistance and are frequently detected among clinical isolates of P. aeruginosa [32].
Clinically significant antimicrobial resistance was found among isolates that were positive for class 1 integrons, with notable high resistance rates to cefotaxime (77.4%), carbenicillin (80.6%), ceftriaxone (87.1%), streptomycin (90.3%) and tetracycline (100%). This is comparable with previously reported high rates of resistance to ceftriaxone (88.7%) and cefotaxime (90.1%) [5], tetracycline (100%) and gentamicin (78.6%) [29], among integron positive clinical P. aeruginosa from China. The high resistance rates observed in this study against beta-lactam antibiotics (ceftriaxone, cefotaxime, carbenicillin and piperacillin) is disturbing because resistance to this class of antibiotics in Gram-negative bacteria including P. aeruginosa is usually mediated by the production of extended-spectrum β-lactamase enzymes that are mostly plasmid encoded [33].
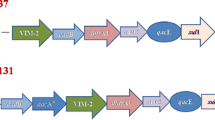
The gene cassettes mostly detected in all the class 1 integrons belong to aadA family conferring resistance to streptomycin and spectinomycin [25]. The aadA6-orfD gene cassette array derived from sequencing of the 1.6 kb gene cassette fragment from isolate ODM-24 showed complete homology with the aadA6-ofrD of class 1 integron reported from previous studies [25, 34]. Naas et al. [25] first reported gene cassette aadA6 as novel gene cassette in P. aeruginosa; and it has been subsequently reported to be highly conserved among the class 1 integrons gene cassettes of the Enterobacteriaceae most especially E. coli [35, 36]. The complete sequence of aadA13 obtained from sequencing ODM-08 was identical to previously reported aadA13 sequences [accession numbers DQ779002, DQ779001]. Interestingly, there are few reports of aadA13 in clinical strains of P. aeruginosa, unlike other aadA family, aadA13 gene cassette are novel and rarely reported in P. aeruginosa. In a recent report, Yuan et al.[37] reported aadA13 cassette, which was present in a new array of aac(6′)-II-aadA13-cmlA8-oxa-10 gene cassette from P. aeruginosa.
P. aeruginosa is well known for harbouring multiple copies of plasmids, some of which are conjugative with resistance genes that are responsible for multiple drug resistance. P. aeruginosa strains from this study harboured 1 to 4 plasmids with sizes ranging from 2.2 kb to >58 kb. This findings contradict previous studies, [38, 39] which reported low molecular weight plasmids (<2 kb) in clinical isolates of P. aeruginosa from Nigeria. The relationship between plasmid profiles and multiple drug resistance patterns observed in this study suggests that plasmids may have played a significant role in the multidrug resistance of P. aeruginosa strains from this study because multiple antibiotic resistance genes as well as virulence genes have often been found clustered together on a single plasmid [40].
Though, conjugation experiment was successful in 10 (32%) P. aeruginosa parental strains, PCR amplification with specific primers revealed the absence of integrons and gene cassettes among the transconjugants; indicating the non-transference of the integrons and associated gene cassettes along with the conjugative plasmids. This is probably suggestive of chromosomal location of the integrons and gene cassettes as previously observed in previous studies that non-plasmid lateral exchange of resistance regions may be common in P. aeruginosa. [32] In addition, our result also concurs with a recent paper on the dispersal of resistance regions from chromosomally located class 1 integrons possibly serving as the major genetic element of global dissemination in P. aeruginosa[41].
In summary, our data demonstrate the presence of multiple drug resistant P. aeruginosa harbouring antibiotic resistant plasmids, class 1 integrons and gene cassettes that can be easily dispersed among other bacteria, resulting in the rapid spread of antibiotic resistance genes. Disturbingly, the widespread dissemination of the class 1 integron and associated gene cassettes in P. aeruginosa and other clinically important pathogens would gravely complicate treatments of infections in our hospitals if not properly monitored. Hence, functional surveillance of antimicrobial resistance and appropriate, effective measures geared towards curbing indiscriminate and unregulated use of antibiotics are urgently needed to prevent outbreaks of multidrug resistant bacteria in Southwest Nigeria hospitals.
References
Ali A, Solh E, Alhajhusain A: Update on the treatment of Pseudomonas aeruginosa pneumonia. J Antimicrob Chemother. 2009, 64: 229-238. 10.1093/jac/dkp201.
Bonomo RA, Szabo D: Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis. 2006, 43: 49-56. 10.1086/504804.
Fonseca EL, Vieira VV, Cipriano R, Vicente AC: Class 1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol Med Microbiol. 2005, 44: 303-309. 10.1016/j.femsim.2005.01.004.
Yan JJ, Hsueh PR, Lu JJ, Chang FY, Ko WC, Wu JJ: Characterization of acquired beta-lactamases and their genetic support in multidrug-resistant Pseudomonas aeruginosa isolates in Taiwan: the prevalence of unusual integrons. J Antimicrob Chemother. 2006, 58: 530-536. 10.1093/jac/dkl266.
Chen J, Su Z, Liu Y, Wang S, Dai X, Li Y, Peng S, Shao Q, Zhang H, Wen P, Yu J, Huang X, Xu H: Identification and characterization of class 1 integrons among Pseudomonas aeruginosa isolates from patients in Zhenjiang, China. Int J Infect Dis. 2009, 13: 717-721. 10.1016/j.ijid.2008.11.014.
Ruiz-Martínez L, López-Jiménez L, Fusté E, Vinuesa T, Martínez JP, Viñas M: Class 1 integrons in environmental and clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents. 2011, 38: 398-402. 10.1016/j.ijantimicag.2011.06.016.
Carattoli A: Importance of integrons in the diffusion of resistance. Vet Res. 2001, 32: 243-259. 10.1051/vetres:2001122.
Elbourne LD, Hall M: Gene cassette encoding a 3-N -aminoglycoside acetyltransferase in a chromosomal integron. Antimicrob Agents Chemother. 2006, 50: 2270-2271. 10.1128/AAC.01450-05.
Jeong JH, Shin KS, Lee JW, Park EJ, Son SY: Analysis of a novel class 1 integron containing metallo-beta-lactamase gene VIM-2 in Pseudomonas aeruginosa. J Microbiol. 2009, 47: 753-759. 10.1007/s12275-008-0272-2.
Mazel D, Dychinco B, Webb VA, Davies J: A distinctive class of integron in the Vibrio cholerae genome. Science. 1998, 280: 605-608. 10.1126/science.280.5363.605.
Mazel D: Integrons and the origin of antibiotic resistance gene cassettes. ASM News. 2004, 70: 11-
Fluit AC, Schmitz FJ: Resistance integrons and super-integrons. Clin Microbiol Infect. 2004, 10: 272-288. 10.1111/j.1198-743X.2004.00858.x.
Xu Z, Li L, Shirtliff ME, Alam MJ, Yamasaki S, Shi L: Occurrence and characteristics of class 1 and 2 integrons in pseudomonas aeruginosa isolates from patients in Southern China. J Clin Microbiol. 2009, 47: 230-234. 10.1128/JCM.02027-08.
Lamikanra A, Crowe JL, Lijek RS, Odetoyin BW, Wain J, Aboderin AO, Okeke IN: Rapid evolution of fluoroquinolone-resistant Escherichia coli in Nigeria is temporally associated with fluoroquinolone use. BMC Infect Dis. 2011, 11: 312-2334. 10.1186/1471-2334-11-312.
Okonko IO, Soleye FA, Amusan TA, Ogun AA, Ogunnusi TA, Ejembi J: Incidence of multi-drug resistance (MDR) organisms in Abeokuta, southwestern Nigeria. Global J Pharmacol. 2009, 3: 69-80.
Murray PR, Baron EJ, Jorgensen JH, Pfaller M, Yolken R: Manual of Clinical Microbiology. 2003, Washington DC: ASM Press, 8
CLSI: Performance standards for antimicrobial susceptibility testing; 20th informational supplement. M100-S20. 2010, Wayne, Pa: Clinical and Laboratory Standards Institute
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning; a laboratory manual. 1989, Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 2
Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM: A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmids molecules. Plasmid. 1978, 1: 417-420. 10.1016/0147-619X(78)90056-2.
Queipo-Ortuñ MI, Colmenero JD: Macias M, Bravo MJ, Morata P: Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in Real-Time PCR for serum samples from patients with Brucellosis. Clin Vaccine Immunol. 2008, 15: 293-296. 10.1128/CVI.00270-07.
White PA, McIver CJ, Deng YM, Rawlinson WD: Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol Lett. 2000, 182: 265-269. 10.1111/j.1574-6968.2000.tb08906.x.
White PA, Mclver CJ, Rawlinson W: Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother. 2001, 45: 2658-2661. 10.1128/AAC.45.9.2658-2661.2001.
Ploy MC, Denis F, Courvalin P, Lambert T: Molecular characterization of integrons in Acinetobacter baumanii: description of an hybrid class 2 integron. Antimicrob Agents Chemother. 2000, 44: 2684-2688. 10.1128/AAC.44.10.2684-2688.2000.
Shohayeb M, Sonbol F: Conjugal transfer of antibiotics resistance of Shigella isolated in Tanta. Alex J Pharm Sci. 1994, 8: 177-180.
Naas T, Poirel L, Nordmann P: Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim Biophys Acta. 1999, 1489: 445-451. 10.1016/S0167-4781(99)00202-X.
Davies J: Microbes have the last word. EMBO Rep. 2007, 8: 616-621. 10.1038/sj.embor.7401022.
Yousefi S, Nahaei MR, Farajnia S, Ghojazadeh M, Akhi MT, Sharifi Y, Milani M, Ghotaslou R: Class 1 integron and imipenem resistance in clinical isolates of Pseudomonas aeruginosa: prevalence and antibiotic susceptibility. Iran J Microbiol. 2010, 2: 113-119.
Tenover FC: Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006, 119: 3-10.
Gu B, Tong M, Zhao W, Liu G, Ning M, Pan S, Zhao W: Prevalence and characterization of class I integrons among pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbio. 2007, 45: 241-243. 10.1128/JCM.01318-06.
Labuschagne CJ, Weldhagen GF, Ehlers MM, Dove MG: Emergence of class 1 integron-associated GES-5 and GES-5-like extended-spectrum beta-lactamases in clinical isolates of Pseudomonas aeruginosa in South Africa. Int J Antimicrob Agents. 2008, 31: 527-530. 10.1016/j.ijantimicag.2008.01.020.
Hammami S, Gautier V, Ghozzi R, Da Costa A, Ben-Redjeb S, Arlet G: Diversity in VIM-2-encoding class 1 integrons and occasional blaSHV2a carriage in isolates of a persistent, multidrug-resistant Pseudomonas aeruginosa clone from Tunis. Clin Microbiol Infect. 2010, 16: 189-193. 10.1111/j.1469-0691.2009.03023.x.
Martinez E, Marquez C, Ingold A, Merlino J, Djordjevic SP, Stokes HW, Chowdhurya PR: Diverse mobilized class 1 integrons Are common in the chromosomes of pathogenic Pseudomonas aeruginosa clinical isolates. Antimicrob Agent Chemother. 2012, 56: 2169-2172. 10.1128/AAC.06048-11.
Bush K, Jacoby GA: Updated functional classification of β-Lactamases. Antimicrob Agents Chemother. 2010, 54: 969-976. 10.1128/AAC.01009-09.
Sekiguchi JI, Asagi T, Akiyama MT, Atsushi K, Mizuguchiy , Araake M, et al: Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community ospitals in Japan. J Clin Microbiol. 2007, 45: 979-989. 10.1128/JCM.01772-06.
Zhao S, White DG, Ge B, Ayers S, Friedman S, English L, et al: Identification and characterization of integron-mediated antibiotics resistance among the shiga toxin-producing Escherichia coli isolates. Appl Environ Microbiol. 2001, 67: 1558-1564. 10.1128/AEM.67.4.1558-1564.2001.
Roe MT, Vega E, Pillai SD: Antimicrobial resistance markers of class 1 and class 2 integron-bearing Escherichia coli from irrigation water and sediments. Emerg Infect Dis. 2003, 9: 822-826. 10.3201/eid0907.020529.
Yuan W, Hui L, Jun L, Hang ZH: Detection of Pseudomonas aeruginosa carried a new array of gene cassettes within class 1 integron isolated from a teaching hospital in Nanjing. J Microbol. 2008, 46: 687-691. 10.1007/s12275-008-0021-6.
Yah SC, Eghafona NO, Enabulele IO: Prevalence of plasmids mediated pseudomonas aeruginosa resistant genes from burn wound patients at the university of Benin teaching hospital Benin City, Nigeria. J Biomed Sc. 2006, 5: 61-68.
Olayinka AT, Onile BA, Olayinka BO: Prevalence of multi-drug resistant (MDR) Pseudomonas aeruginosa isolates in surgical units of Ahmadu Bello University Teaching Hospital, Zaria, Nigeria: an indication for effective control measures. Afr Med. 2004, 3: 13-16.
Villa L, Carattoli A: Integrons and transposons on the Salmonella enterica serovar typhimurium virulence plasmid. Antimicrob Agents Chemother. 2005, 49: 1194-1197. 10.1128/AAC.49.3.1194-1197.2005.
Stokes HW, Martinez E, Chowdhury PR, Djordjevic S: Class 1 integron-associated spread of resistance regions in Pseudomonas aeruginosa: plasmid or chromosomal platforms?. J Antimicrob Chemother. 2012, 67: 1799-1800. 10.1093/jac/dks116.
Acknowledgements
This work was supported by TWAS/CSIR 2010 Fellowship Award to BTO.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare no competing interests.
Authors’ contributions
BTO and BAA planned this study. BTO performed the experiments under the guidance of RC. BTO wrote the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Odumosu, B.T., Adeniyi, B.A. & Chandra, R. Analysis of integrons and associated gene cassettes in clinical isolates of multidrug resistant Pseudomonas aeruginosafrom Southwest Nigeria. Ann Clin Microbiol Antimicrob 12, 29 (2013). https://doi.org/10.1186/1476-0711-12-29
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1476-0711-12-29