Abstract
Background
Anopheles cruzii (Diptera: Culicidae) has long been known as a vector of human and simian malaria parasites in southern and south-eastern Brazil. Previous studies have provided evidence that An. cruzii is a species complex, but the status of the different populations and the number of sibling species remains unclear. A recent analysis of the genetic differentiation of the timeless gene among An. cruzii populations from south and south-east Brazil has suggested that the population from Itatiaia, Rio de Janeiro State (south-east Brazil), is in a process of incipient speciation.
Methods
A ~180 bp fragment of cpr, a gene encoding the NADPH-cytochrome P450 reductase, an enzyme involved in metabolic insecticide resistance and odorant clearance in insects, was used in this study as a molecular marker to analyse the divergence between five An. cruzii populations from south and south-east Brazil.
Results
Analysis of the genetic differentiation in the cpr gene revealed very high F ST values and fixed differences between Itatiaia and the other four populations studied (Florianópolis, Cananéia, Juquitiba and Santa Teresa). In addition, the data also provided preliminary evidence that seems to indicate the occurrence of two sympatric sibling species in Itatiaia.
Conclusions
Population genetics analysis of An. cruzii samples from different localities using a fragment of the cpr gene suggests that the Itatiaia sample represents at least one new sibling species in this complex.
Similar content being viewed by others
Background
Anopheles cruzii has long been known as a vector of human and simian malaria parasites in southern and south-eastern Brazil [1, 2]. This species, which belongs to the subgenus Kerteszia, is found from the coast of Rio Grande do Sul State in southern Brazil to Sergipe State in north-eastern Brazil [3, 4], all along the Brazilian Atlantic forest. This forest provides an excellent environment for An. cruzii, since it is an ecosystem abundant in bromeliads, the larval habitat for this anopheline [2, 5, 6].
The possibility that An. cruzii could represent more than a single species was first supported by morphological differences observed among populations from the states of Santa Catarina and Rio de Janeiro [3]. Later it was revealed that An. cruzii is polymorphic for chromosome rearrangements [7, 8]. Differences in inversion frequencies and X chromosome banding patterns from populations in south-eastern and southern Brazil have suggested a process of incipient speciation [9, 10]. Malafronte et al[11] compared sequences of ITS2 (Internal Spacer Region 2) from several An. cruzii populations from south and south-east Brazil and found differences between sequences from different localities, although they considered premature to conclude based on their results that there are distinct sibling species in the areas investigated. Similar results were observed by Calado et al[12], using PCR-RAPD and PCR-RFLP of the ITS2 region.
Finally, isoenzyme analysis indicated two genetically isolated groups, one from Bahia State (north-eastern Brazil), and the other from south-eastern and southern Brazil (Rio de Janeiro, São Paulo and Santa Catarina States) [13]. Supporting the isoenzyme results, analysis of the molecular polymorphism and genetic differentiation of the timeless gene among Brazilian populations of An. cruzii also indicated two cryptic species, one occurring in the north-east (Bahia State) and another in south and south-east Brazil (Espírito Santo, Rio de Janeiro, São Paulo and Santa Catarina States). In addition, the timeless gene sequences also suggested that populations from the south and south-east regions might also constitute different incipient species within this complex in Brazil [14]. Since previous X chromosome analyses suggested the existence of sibling species in these Brazilian regions [9, 10], it would be interesting to analyse the same populations with an X-linked molecular marker to see whether a higher level of differentiation is found.
The gene encoding the NADPH-cytochrome P450 reductase (CPR) has been cloned from several insect species [15–17]. In Anopheles gambiae, this protein is encoded by a single copy gene located on the X chromosome [17, 18]. Previous studies have shown that the cytochrome P450 gene family, which is involved in metabolic insecticide resistance, requires CPR to function [19, 20]. Additionally, knockdown of CPR expression increases An. gambiae sensitivity to the insecticide permethrin [21]. Another putative function associated with CPR in insects is odorant clearance, since the cpr gene is highly expressed in the antennae of the fruit fly Drosophila melanogaster and more specifically at the base of olfactory sensilla in the moth Mamestra brassicae[16, 22]. Olfactory cues are important environmental stimuli affecting mosquito behaviour, playing significant roles in the location of food sources, mates and oviposition sites [23]. Therefore, genes involved in the regulation of antennal response to pheromones can be potentially important in maintaining sexual isolation between closely related species, and in this case, the CPR would be an interesting molecular marker for population studies of the An. cruzii complex.
In this study, an analysis of intraspecific variability and genetic divergence among five Brazilian populations of An. cruzii was carried out using a fragment of the cpr gene.
Methods
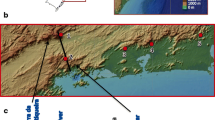
The mosquitoes used in this study were females captured at different localities along the Brazilian Atlantic forest and identified on the basis of their morphology according to Consoli and Lourenço-de-Oliveira [4]. A total of 56 individuals were used for the molecular analysis: 14 from Florianópolis, Santa Catarina State (SC) (27°31'S/48°30'W), 12 from Cananéia, São Paulo State (SP) (25°01'S/47°55'W), 12 from Juquitiba, São Paulo State (SP) (23°57'S/47°03'W), 11 from Itatiaia, Rio de Janeiro State (RJ) (22°27'S/44°36'W) and 7 from Santa Teresa, Espírito Santo State (ES) (19°56'S/40°35'W) (Figure 1).
For the isolation of a fragment of the An. cruzii cpr gene, initially a pair of degenerated primers was designed based on conserved regions of the CPR proteins from Drosophila melanogaster, Drosophila pseudoobscura, Musca domestica, Aedes aegypti and An. gambiae (Table 1 and Additional file 1). These primers, named here 5'Cpr01deg and 3'Cpr01deg, were used in PCR with An. cruzii genomic DNA extracted according to Jowett [24]. PCR was carried out with an Eppendorf Mastercycler® thermocycler using the following conditions: 15 cycles at 94°C for 60 s, 50°C (decreasing 1°C/cycle) for 90 s and 72°C for 60 s, followed by 20 cycles of 94°C for 60 s, 50°C for 90 s and 72°C for 60 s. PCR products were then purified and cloned using Zero Blunt TOPO PCR cloning kit (Invitrogen).
Sequencing of positive clones was carried out in an ABI Prism 3730 DNA sequencer (PDTIS-FIOCRUZ DNA sequencing facility) using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems). The identity of the cloned fragments was confirmed by BlastX analysis using the GenBank [25]. Based on these initial sequences, two new specific primers named 5'cpr01ancruzii and 3'cpr01ancruzii (Table 1 and Additional file 1) were designed to amplify a ~180 bp fragment of the An. cruzii cpr gene from individual mosquitoes of the different localities listed above. This short fragment includes an intron of variable size (see below) and two small segments of the flanking exons (Additional file 1). PCR amplification using the specific primers was carried out for 35 cycles at 94°C for 30 s, 50°C for 60 s and 72°C for 90 s using the proofreading Pfu DNA polymerase (Biotools). PCR fragments were cloned using either Zero Blunt TOPO PCR cloning kit (Invitrogen) or pMOS Blue vector blunt-ended cloning kit (GE Healthcare) and at least eight clones of each mosquito were sequenced.
Sequences were edited and in most cases consensus sequences representing the two alleles were generated. In a number of individuals only one haplotype was observed among the eight sequences and in these cases the mosquitoes were classified as homozygotes. The probability of incorrectly classifying a heterozygote as a homozygote individual with this procedure is less than 1%. Eight homozygotes were found in Florianópolis, four in Cananéia, seven in Juquitiba, three in Itatiaia and two in Santa Teresa. The sequences from homozygote mosquitoes were duplicated prior to analysis. However, the analysis was also carried out without duplicating the homozygote sequences with similar results.
The cpr sequences were aligned using ClustalX software [26] and the population genetics analysis was carried out using DNASP4.0 [27], PROSEQv 2.91 [28] and Arlequin 3.0 [29] softwares. The haplotype network was estimated using TCS1.21 [30].
Results
Polymorphism and divergence among An. cruzii populations
A total of 112 sequences were obtained (28 from Florianópolis, 24 from Cananéia, 24 from Juquitiba, 22 from Itatiaia and 14 from Santa Teresa). The sequences were submitted to the GenBank (accession numbers: GU072619 - GU072730). An alignment of the variable sites in shown in Figure 2 (an alignment of the whole sequences is presented in Additional file 2). The small segments of the flanking exons in this cpr fragment are totally conserved among the five populations analysed. Therefore, all base substitutions occurred in the intron which also shows a number of indels, including three polymorphic dinucleotide repetitions (Figure 2 and Additional file 2). Table 2 shows the number of copies of each dinucleotide repeat in all An. cruzii populations. In Itatiaia only one repeat of CG dinucleotide was found, while in the other four populations there are two or three repeats. Similar pattern was observed for the CT repeat which shows four copies in Itatiaia while there are six to nine in the other populations (Table 2).
Alignment of the variable sites in the cpr fragment of An. cruzii. Alignment of the variable positions in the DNA sequences from the cpr gene fragment from all populations of An. cruzii analysed. The sequences of homozygote individuals were grouped and are represented as a/b. Dots represent the identity of the first nucleotide sequence and asterisks represent the identity of all sequences. Flo: individuals from Florianópolis; Can: Cananéia; Juq: Juquitiba; Ita: Itatiaia; San: Santa Teresa.
Table 3 shows the pair-wise estimates of population differentiation between the An. cruzii populations. Because this cpr fragment contains a number of indels, the F ST values were calculated in two different ways. In the first one (FST (1)) the gaps were treated as single mutations in the analysis performed with the PROSEQv 2.91 software. In the second one (FST(2)), the Arlequin 3.0 software was used to calculate the differentiation values considering the three types of dinucleotide repeats as microsatellite loci. In all cases the F ST values were significant and the two types of estimates showed similar results in most cases. Very high F ST values (ranging from ~0.6 to 0.8) were found between Itatiaia and the other populations. Albeit significant, the pair-wise F ST values in the comparisons among the samples of Florianópolis, Cananéia and Santa Teresa are usually under 0.2, while those between Juquitiba and the other three populations are moderately high.
Table 3 also shows the average number of nucleotide substitutions per site (D xy ), the number of net nucleotide substitutions per site between populations (D a ) and the distribution of the four mutually exclusive categories of segregating sites observed in each comparison: the number of exclusive polymorphisms for each population (S 1 and S 2 ), the number of shared polymorphisms (S s ) and the number of fixed differences (S f ). As in the case of the F ST , the highest D xy and D a values are those involving the Itatiaia population. In addition, this sample shows few shared polymorphisms and it is the only one presenting fixed differences in comparisons with the other populations.
Genealogy of the An. cruzii cpr sequences
A network of genealogical relationships of An. cruzii haplotypes was estimated using the method of Templeton et al[31] available in the TCS programme (Figure 3). Gaps were treated as a 5th state. A network was also estimated ignoring the gaps, but in this case much of the divergence among the sequences was lost and the network was not informative. The haplotype network shows that the Itatiaia population is clearly separated in an isolated group. A less clear separation was found among the sequences of the other populations.
Haplotype network of cpr sequences. Each colour represents one population of An. cruzii. Each circle represents a different haplotype with size proportional to its relative frequency. Haplotype numbers are given in Roman and the number of sequences of each haplotype is given in brackets. The small white circles represent missing intermediates and the lines connecting the haplotypes represent one mutational step between two observed haplotypes. Each individual of Itatiaia population is discriminated next to the respective haplotype.
Divergence between Itatiaia A and Itatiaia B
Inspection of the cpr sequences presented in Figure 2 and Additional file 2 suggests that the Itatiaia sample might include two different sets of individuals. Based on the number of uninterrupted AG repeats between positions 32 to 49 (Figure 2) the Itatiaia population was divided in two groups: the first one, called henceforth Itatiaia A, has more than three AG repeats (04 to 06 repeats) and the second, called henceforth Itatiaia B, has exactly three AG repeats (Table 2). According to this classification the individuals Ita2, Ita3, Ita4, Ita8, Ita10 and Ita11 belong to Itatiaia A (genotype "4-6/4-6"), the mosquitoes Ita5, Ita6, Ita7 and Ita9 belong to Itatiaia B (genotype "3/3") and individual Ita12 is the only "hybrid" between the two groups (genotype "3/4-6"). Therefore, the Itatiaia sample is not in Hardy-Weinberg equilibrium (X2 = 7.24; d.f. = 1; P < 0.01) suggesting the possibility that two sympatric sibling species might exist in this locality. The separation between the two groups is also evident in Figure 3. Besides, the F ST value (considering gaps as single mutations) between Itatiaia A and B is quite large (0.6678) and highly significant (P < 0.001) despite the small sample sizes.
Finally, to test the hypothesis that the Itatiaia population might include two different sympatric sibling species, the recently published timeless data [14] from the same sample were reanalysed. As for the cpr data, the timeless sequences were divided into Itatiaia A (Ita2, Ita3, Ita4, Ita8, Ita10 and Ita11) and Itatiaia B (Ita5, Ita6, Ita7 and Ita9). The timeless gene also suggests that the sequences might belong to two different sibling species with a F ST value (0.3418) that is highly significant (P < 0.001) and the occurrence of two fixed differences. There is also a clear separation between the Itatiaia A and B timeless sequences in a haplotype network (Additional file 3).
Discussion
The X chromosome seems to be enriched in genes that cause reproductive isolation between species in the genus Drosophila[32]. In addition, many sibling species including the An. gambiae complex are outcomes of recent speciation processes associated with paracentric inversions involving this chromosome [33]. The X chromosome banding patterns and inversion frequencies studies of Brazilian south and south-east An. cruzii populations, showed three X chromosomal forms (A, B and C), suggesting a process of incipient speciation [9, 10]. The authors observed that the majority of mosquitoes from Juquitiba population had form A, while form B predominated in Cananéia [9, 10]. In the current study, although there are no fixed differences in the cpr gene between Juquitiba and Cananéia, a moderately high F ST value was observed. In An. gambiae, cpr is located on the X chromosome. Therefore, if this molecular marker has a similar chromosomal location in An. cruzii, it might be associated with the chromosomal forms described by Ramirez & Dessen [9, 10].
Comparisons of the F ST values observed with cpr and timeless in all pair-wise comparisons involving the five populations analysed in the current study and in Rona et al[14] show only a partial consistence but this is expected. Wang-Sattler et al[34] demonstrated that the phylogenetic relationships in the An. gambiae complex could vary widely between different genomic regions, thus indicating the mosaic nature of the genome of these species [34].
As mentioned above the cpr gene in An. gambiae is X-linked, while timeless is autosomal. Assuming these two markers have similar locations in An. cruzii, cpr is expected to be under more efficient selection than the timeless, since in species with X/Y sex determination, as An. cruzii, rare recessive mutations are fully expressed in the heterogametic sex, which could lead to 'faster-X evolution' if a large proportion of mutations are fixed by positive selection [35]. If positive selection is more efficient on the X chromosome, one expects it to harbour less variability than the autosomes [36]. The X chromosome is indeed less variable than the autosomes in non-African populations of Drosophila simulans[37]. Comparing timeless and cpr, the first is more polymorphic than the latter, but the latter shows higher differentiation among the southern populations of An. cruzii.
Since An. cruzii is polymorphic for chromosomal inversions and Ramirez & Dessem [9, 10] found evidence for sibling species carrying different X chromosomal forms, another hypothesis that might explain the differences between the two markers is the suppressed-recombination model of speciation proposed by Coluzzi [33, 38].
Analysis of the molecular polymorphism and genetic differentiation of the timeless gene among Brazilian populations of An. cruzii suggested that the population from Itatiaia (Rio de Janeiro State) is in a process of differentiation and incipient speciation [14]. High F ST values between Itatiaia (Rio de Janeiro State) and the other populations from south and south-east Brazil was reported here. In addition, comparison of Itatiaia with other populations revealed some fixed differences and only a few shared polymorphisms. Moreover, the haplotype network shows that Itatiaia is clearly separated in an isolated group (Figure 3). These results, therefore, suggest that this population represents a different species in the An. cruzii complex.
Preliminary evidence was also presented here that raised the possibility of the existence of two different sympatric incipient species in Itatiaia. This is based on the analysis of the genetic differentiation of the cpr gene and a reanalysis of the recently published timeless data [14]. Although a putative heterozygote was found in cpr analysis considering the AG repeats and shared polymorphisms were observed in timeless, high F ST values were detected between Itatiaia A and Itatiaia B in these two molecular markers, as well as fixed differences which seem to indicate that these two groups might represent different incipient species. Inspection of the neighbour-joining tree presented in the timeless study [14] reveals that the individuals classified here as Itatiaia A are clearly isolated in a separated branch. However, the individuals classified as Itatiaia B are mixed with the other individuals from south and south-east populations. In that study, only one putative heterozygote was found (Ita01) carrying alleles of the two Itatiaia groups. Unfortunately this DNA sample was lost and therefore it was not possible to analyse the cpr gene of this individual mosquito. The sample sizes available for the two Itatiaia groups are quite small and further work is clearly needed to determined beyond any doubt that two sympatric incipient sibling species exist in this locality but the results presented here seems to indicate that might be the case.
Analysis of a number of other molecular markers will allow a more precise estimate of the Itatiaia population differentiation and might provide a more complete representation of the divergence history of this species complex.
Conclusions
Evidence was presented here suggesting the existence of at least one new sibling species within the Anopheles (Kerteszia) cruzii complex in Itatiaia, south-east Brazil, a finding that supports a previous timeless gene study. In addition, according to cpr and timeless gene analyses, the Itatiaia sample might be in fact composed by two sympatric incipient species, named here Itatiaia A and Itatiaia B.
References
Deane LM, Ferreira-Neto JA, Deane SP, Silveira IP: Anopheles (Kerteszia) cruzii, a natural vector of the monkey malaria parasites, Plasmodium simium and Plasmodium brasilianum. Trans R Soc Trop Med Hyg. 1970, 64 (647):
Rachou RG: Anofelinos do Brasil: Comportamento das espécies vetoras de malária. Rev Bras Malariol Doencas Trop. 1958, 10: 145-181.
Zavortink TJ: A review of the subgenus Kerteszia of Anopheles. Cont Am Entomol Inst. 1973, 9: 1-54.
Consoli RAGB, Lourenço-de-Oliveira R: Principais mosquitos de importância sanitária no Brasil. 1994, Rio de Janeiro: Ed. Fiocruz
Pittendrigh CS: The ectopic specialization of Anopheles homunculus, and its relation to competition with An. bellator. Evolution. 1949, 4: 64-78. 10.2307/2405534.
Veloso HP, De Moura JV, Klein RM: Ecological limitation of Anopheles of the Subgenus Kerteszia in the coastal region of Southern Brazil. Mem Inst Oswaldo Cruz. 1956, 54: 517-548.
Ramirez CC, Dessen EM: Cytogenetics analysis of a natural population of Anopheles cruzii. Rev Bras Genet. 1994, 17: 41-46.
Ramirez CC, Dessen EM, Otto PA: Inversion polymorphism in a natural population of Anopheles cruzii. Caryologia. 1994, 47: 121-130.
Ramirez CC, Dessen EM: Chromosomal evidence for sibling species of the malaria vector Anopheles cruzii. Genome. 2000, 43: 143-151. 10.1139/gen-43-1-143.
Ramirez CC, Dessen EM: Chromosome differentiated populations of Anopheles cruzii: evidence for a third sibling species. Genetica. 2000, 108: 73-80. 10.1023/A:1004020904877.
Malafronte Rdos S, Marrelli MT, Ramirez CC, Nassar MN, Marinotti O: Intraspecific variation of second internal transcribed spacer of nuclear ribosomal DNA among populations of Anopheles (Kerteszia) cruzii (Diptera: Culicidae). J Med Entomol. 2007, 44: 538-542. 10.1603/0022-2585(2007)44[538:IVOSIT]2.0.CO;2.
Calado DC, Navarro-Silva MA, Sallum MAM: PCR-RAPD and PCR-RFLP polymorphism detected in Anopheles cruzii (Diptera, Culicidae). Rev Bras Entomol. 2006, 50: 423-430. 10.1590/S0085-56262006000300014.
Carvalho-Pinto CJ, Lourenço-de-Oliveira R: Isoenzymatic analysis of four Anopheles (Kerteszia) cruzii (Díptera: Culicidae) populations of Brazil. Mem Inst Oswaldo Cruz. 2004, 99: 471-475. 10.1590/S0074-02762004000500002.
Rona LD, Carvalho-Pinto CJ, Gentile C, Grisard EC, Peixoto AA: Assessing the molecular divergence between Anopheles (Kerteszia) cruzii populations from Brazil using the timeless gene: Further evidence of a species complex. Malar J. 2009, 8: 60-10.1186/1475-2875-8-60.
Koener JF, Carino FA, Feyereisen R: The cDNA and deduced protein sequence of house fly NADPH-cytochrome P450 reductase. Insect Biochem Mol Biol. 1993, 23: 439-447. 10.1016/0965-1748(93)90051-S.
Hovemann BT, Sehlmeyer F, Malz J: Drosophila melanogaster NADPH-cytochrome P450 oxidoreductase: pronounced expression in antennae may be related to odorant clearance. Gene. 1997, 189: 213-219. 10.1016/S0378-1119(96)00851-7.
Nikou D, Ranson H, Hemingway J: An adult-specific CYP6 P450 gene is overexpressed in a pyrethroid-resistant strain of the malaria vector, Anopheles gambiae. Gene. 2003, 318: 91-102. 10.1016/S0378-1119(03)00763-7.
Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL: The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002, 298: 129-149. 10.1126/science.1076181.
Smith GC, Tew DG, Wolf CR: Dissection of NADPH-cytochrome P450 oxidoreductase into distinct functional domains. Proc Natl Acad Sci. 1994, 91: 8710-8714. 10.1073/pnas.91.18.8710.
Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, Wolf CR: Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem. 2003, 278: 13480-13486. 10.1074/jbc.M212087200.
Lycett GJ, McLaughlin LA, Ranson H, Hemingway J, Kafatos FC, Loukeris TG, Paine MJ: Anopheles gambiae P450 reductase is highly expressed in oenocytes and in vivo knockdown increases permethrin susceptibility. Insect Mol Biol. 2006, 15: 321-327. 10.1111/j.1365-2583.2006.00647.x.
Maibeche-Coisne M, Merlin C, François MC, Porcheron P, Jacquin-Joly E: P450 and P450 reductase cDNAs from the moth Mamestra brassicae: cloning and expression patterns in male antennae. Gene. 2005, 346: 195-203. 10.1016/j.gene.2004.11.010.
Takken W, Knols BG: Odor-mediated behavior of Afrotropical malaria mosquitoes. 1999, 44: 131-157.
Jowett T: Preparation of nucleic acids. Drosophila, A Practical Approach. 1998, IRL press, Oxford: Roberts DB, 347-371.
GenBank database. [http://www.ncbi.nlm.nih.gov/BLAST/]
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25: 4876-4882. 10.1093/nar/25.24.4876.
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R: DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003, 19: 2496-2497. 10.1093/bioinformatics/btg359.
Filatov DA, Charlesworth D: DNA polimorphism, haplotype structure and balancing selection in the Leavenworthia PgiC locus. Genetics. 1999, 153: 1423-1434.
Excoffier L, Laval G, Schneider S: Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005, 1: 47-50.
Clement M, Posada D, Crandall KA: TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000, 9: 1657-1659. 10.1046/j.1365-294x.2000.01020.x.
Templeton AR, Crandall KA, Sing CF: A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA-sequence data. III. Cladogram estimation. Genetics. 1992, 132: 619-633.
Tao Y, Chen S, Hartl DL, Laurie CC: Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics. 2003, 164: 1383-1397.
Ayala FJ, Coluzzi M: Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc Natl Acad Sci. 2005, 102: 6535-6542. 10.1073/pnas.0501847102.
Wang-Sattler R, Blandin S, Ning Y, Blass C, Dolo G, Touré YT, della Torre A, Lanzaro GC, Steinmetz LM, Kafatos FC, Zheng L: Mosaic genome architecture of the Anopheles gambiae species complex. PLoS One. 2007, 2 (e1249):
Vicoso B, Charlesworth B: Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006, 7: 645-653. 10.1038/nrg1914.
Betancourt AJ, Kim Y, Orr HA: A pseudohitchhiking model of X vs. autosomal diversity. Genetics. 2004, 168: 2261-2269. 10.1534/genetics.104.030999.
Schofl G, Schlotterer C: Patterns of microsatellite variability among X chromosomes and autosomes indicate a high frequency of beneficial mutations in non-African D. simulans. Mol Biol Evol. 2004, 21: 1384-1390. 10.1093/molbev/msh132.
Coluzzi M: Spatial distribution of chromosomal inversions and speciation in Anopheline mosquitoes. Mechanisms of Speciation. 1982, New York: Liss AR, 143-153.
Nei M, Kumar S: Molecular Evolution and Phylogenetics. 2000, New York: Oxford University Press
Acknowledgements
The authors would like to thank Dr Rosely Malafronte (Instituto de Medicina Tropical de São Paulo), Dr Monique Motta (FIOCRUZ - Rio de Janeiro) and Claudiney dos Santos (Unidade de Medicina Tropical - Universidade Federal do Espirito Santo) for providing most of the samples used in this work. The authors are also indebted to Robson da Silva for technical assistance, to Dr André Pitaluga for helping prepare Figure 1, and to PDTIS-FIOCRUZ for use of its DNA sequencing facility. This work was supported by grants from the Howard Hughes Medical Institute, FIOCRUZ, Faperj and CNPq.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LDPR participated in data generation and analysis, and drafted the manuscript. She also helped capture mosquitoes in Florianópolis. CJCP carried out the capture and morphological identification of mosquitoes collected in Florianópolis. AAP is the principal investigator, participated in its design and coordination, and helped to write the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12936_2009_1114_MOESM1_ESM.DOC
Additional file 1: CPR protein multiple alignment and primer positions. The putative fragment of An. cruzii CPR deduced protein is aligned with D. melanogaster, D. pseudoobscura, M. domestica, An. gambiae and Ae. aegypti homologues. Arrows point to the approximated positions of the primers used in this study. The inverted triangle represents the position of the intron. (DOC 23 KB)
12936_2009_1114_MOESM2_ESM.DOC
Additional file 2: Alignment of the An. cruzii cpr sequences. Alignment of the DNA sequences from the cpr gene fragment from all populations of An. cruzii analysed. The translated amino acid sequence is shown above the alignment and the intron is highlighted in grey. Dots represent identity and dashed represent gaps. The asterisks in the bottom line represent identity of all sequences. Flo: individuals from Florianópolis; Can: Cananéia; Juq: Juquitiba; Ita: Itatiaia; San: Santa Teresa. (DOC 52 KB)
12936_2009_1114_MOESM3_ESM.PPT
Additional file 3: Haplotype network using timeless nucleotide sequences of the Itatiaia population. Each circle represents a different haplotype with size proportional to its relative frequency. Haplotype numbers are given in Roman and the number of sequences of each haplotype is given in brackets. The small white circles represent missing intermediates and the lines connecting the haplotypes represent one mutational step between two observed haplotypes. Each individual of Itatiaia population is discriminated next to respective haplotype. (PPT 119 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rona, L.D., Carvalho-Pinto, C.J. & Peixoto, A.A. Molecular evidence for the occurrence of a new sibling species within the Anopheles (Kerteszia) cruzii complex in south-east Brazil. Malar J 9, 33 (2010). https://doi.org/10.1186/1475-2875-9-33
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-9-33