Abstract
Background
Plasmodium vivax circumsporozoite variants have been identified in several geographical areas. The real implication of the genetic variation in this region of the P. vivax genome has been questioned for a long time. Although previous studies have observed significant association between VK210 and the Duffy blood group, we present here that evidences of this variation are limited to the CSP central portion.
Methods
The phylogenetic analyses were accomplished starting from the amplification of conserved domains of 18 SSU RNAr and Cyt B. The antibodies responses against the CSP peptides, MSP-1, AMA-1 and DBP were detected by ELISA, in plasma samples of individuals infected with two P. vivax CS genotypes: VK210 and P. vivax-like.
Results
These analyses of the two markers demonstrate high similarity among the P. vivax CS genotypes and surprisingly showed diversity equal to zero between VK210 and P. vivax-like, positioning these CS genotypes in the same clade. A high frequency IgG antibody against the N- and C-terminal regions of the P. vivax CSP was found as compared to the immune response to the R- and V- repetitive regions (p = 0.0005, Fisher's Exact test). This difference was more pronounced when the P. vivax-like variant was present in the infection (p = 0.003, Fisher's Exact test). A high frequency of antibody response against MSP-1 and AMA-1 peptides was observed for all P. vivax CS genotypes in comparison to the same frequency for DBP.
Conclusions
This results target that the differences among the P. vivax CS variants are restrict to the central repeated region of the protein, mostly nucleotide variation with important serological consequences.
Similar content being viewed by others
Background
The circumsporozoite surface protein (CSP) is the most abundant polypeptide present in the sporozoite covering. This protein is involved in the motility and invasion of the sporozoite during its entrance in the hepatocyte [1, 2].
Some years ago, CSP was studied as the main goal for anti-malarial vaccine development; however the existence of variations in the repetitive sequence of its central portion has been hindering these studies. Plasmodium vivax CSP sequences analyses revealed that parasites have repeats belonging to one of two types of nonapeptide repeat units, GDRA(A/D)GQPA or ANGA(G/D)(N/D)QPG, named VK210 or VK247 respectively [3, 4]. In 1993, a new human malaria parasite from a P. vivax-infected person was identified by Qari et al [5], who named it P. vivax-like. The CSP sequence of P. vivax-like has an 11-mer repeat sequence, APGANQ(E/G)GGAA, and is different to the two previously described variants [5, 6].
All P. vivax CS genotypes have a worldwide distribution and have been identified for several authors [7–17]. In Brazil, the occurrence of the three genotypes in pure and mixed infections was described [11, 17]. Seroreactivity tests have identified the presence of three variant genotypes in samples from the State of São Paulo [10, 16] and in indigenous populations [8, 9] and other communities of the Amazon region [13]. Studies have also reported differences in the infectivity of anophelines to the variant genotypes, indicating that Anopheles darlingi and Anopheles pseudopunctipennis were more susceptible to the infection by VK210 [18, 19]. These findings could be a consequence of differences in the emergence of this genotype in specific geographical regions or suggest that the VK210 genotype is the best-adapted variant in the world [11].
The successful of the vaccine against malaria can be related to the immunological intervention in the development of the parasite in the human host or mosquito vector. To improve the health and quality of more than one billion people around the world, several efforts have been addressed for the identification and antigenic characterization of different P. vivax antigens, among these the preerythrocytic antigens such as circumsporozoite protein (CSP) [20], the blood-stage proteins as merozoite surface protein 1 (MSP-1) [21, 22], apical membrane antigen 1 (AMA-1) [22, 23], and the Duffy binding protein (DBP), an merozoite antigen that interacts with the Duffy blood group in the host cells surface [22, 24]. Currently, several authors have considered the CSP of P. vivax as the major target for the development of recombinant malaria vaccines, since the synthetic peptides starting from this protein induce a high and specific humoral response as the induced by natural exposure of humans to malaria [25–31]. Moreover, starting from the description of the P. vivax CS genotypes, VK210, VK247 and P. vivax-like, several studies proposed the existence of differences among those that seem to go besides variations in the repetitive portion of the protein, as geographical distribution, transmission intensity, vectorial competence, immune and treatment responses and drug resistance [11, 18, 19, 32–34].
Many studies are being conducted to better understand the age and origin of the P. vivax as a human parasite [35, 36]. Low microsatellite and tandem repeat variability indicate that P. vivax infected humans recently (10,000 years ago) [37]. Indeed, a different study based on polymorphisms of two nuclear and one mitochondrial gene places this parasite origin between 45,000 and 81,000 years ago [35]. In addition, P. vivax seems to be related to the clade of parasites found in Asian cercopithecines, indicating its origin in Asia via a host-switch from parasites found in non-human primates, such as macaques [35, 36, 38]. Little it is known about the characterization of the P. vivax variants, since analyses of the non-repetitive portion of CS gene showed that these genotypes belong to a same clade, including several types of primate Plasmodium species [35, 39]. Nevertheless, the important question remains whether the P. vivax CS repeated region is a limited, mostly simple base variation [40] or if these variants represent the existence of a new species or subspecies of Plasmodium causing human malaria, with major biological consequences [6].
Here, this work contributes to the understanding of the implication of the central repetitive region variation of the CS in the P. vivax genome by phylogenetic tools and to the evaluation of the humoral immune response against different parasite antigens.
Methods
Subjects
After given written informed consent, peripheral blood samples were drawn from malaria patients living in four Brazilian Amazon endemic areas (Macapá, Amapá State; Novo Repartimento, Pará State; Porto Velho, Rondônia State and Plácido de Castro, Acre State). All individuals enrolled in this study complied with the following criteria: they sought medical assistance for clinical malaria symptoms, were over 18 years old and had a positive malaria diagnosis by thick blood film for P. vivax. The genomic DNA was extracted by the phenol-chloroform method [41] or using a commercially available kit (Easy-DNA™, Invitrogen, USA), and a semi-nested PCR was performed using P. vivax-specific small-subunit (SSU) rDNA primers to confirm the Plasmodium diagnosis [42]. The P. vivax CS genotypes were determined as described by Alves et al [43].
Molecular analyses
For the phylogenetic reconstruction, a subset of the P. vivax field samples was used and data for non-human Plasmodium spp. samples were obtained from GenBank. Natural hosts type, geographic origins and GenBank accession numbers of the out groups are described in additional file 1: Hosts type, geographic origins and GenBank accession numbers of the out groups. All amplification reactions were performed in a thermocycler (DNA MasterCycler; Eppendorf, USA). The PCR-amplified products were purified by using GFX (GE Healthcare, United Kingdom) and EXOSAP (USB, USA) PCR purification kits, according to the manufacturer's protocol. DNA sequencing was performed using the Big Dye™ Terminator V3.1 Cycle Sequencing kit on ABI 3100 Genetic Analyzer (Applied Biosystems, USA).
Amplification of the molecular markers
18 small sub unit ribosomal RNA (SSU rRNA) gene analyses
The amplification of a target area between variable regions 7 and 8 of the 18 SSU rRNA gene from P. vivax was designed as described by Santos-Ciminera et al [44]. PCR was performed using the primer pairs VAR1 (5'- CTT GGA TGG TGA TGC ATG GCC - 3') and VAR2 (5'- ATC TTT CAA TCG GTA GGA GCG AC - 3'). The reaction mixture contained buffer 10 mM Tris-HCl with pH 8.3, 50 mM KCl, 200 μM of each of the four dNTPs, 10 μM of each oligonucleotide primer, 1 μg DNA template and 0.5 U of ampli-Taq DNA polymerase (Invitrogen, USA) to a final volume of 25 μL. All amplification cycles included to an initial cycle of 95°C for 15 min, followed by 30 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min, then a final extension at 72°C for 10 min.
Cytochrome B gene analyses
The cytochrome B (Cyt B) sequences were amplified by PCR using sets of primers: PC1 (5'- GCTACAGGTGCATCTCTTGTATTC - 3') and PC2 (5' - CACTTACAGTATATCCTCCACATAACCA - 3'). A reaction mixture of buffer 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 200 μM of each of the four dNTPs, 10 μM of each oligonucleotide primer, 1 μg DNA template and 0.5 U of ampli-Taq DNA polymerase (Invitrogen, USA). The amplification conditions were as follows: first, 1 min at 94°C, followed by 30 cycles with 0.5 min of denaturation at 94°C, annealing at 40°C for 0.5 min and elongation at 72°C for 1.5 min. After 30 cycles, a final elongation step at 72°C for 3 min was carried out. The agarose gels were stained with ethidium bromide and analysed with a Gel Doc 2000 illuminator (Bio-Rad Laboratories, USA).
Sequence alignment and phylogenetic analyses
The sequences were edited and aligned with the programs MEGA (version 4.1) and BioEdit Sequence Alignment Editor by the CLUSTAL W tool. Phylogenetic analyses were performed with neighbor-joining (NJ), using the program MEGA (version 4.1), with p distance which takes into account the possibility of high bias in the transition/transversion and in G+C content, derived of the position of the first, second and third codon [45]. The reliability of the NJ trees is assessed by the bootstrap method with 500 replications [46].
Assessment of the serological response against P. vivax CS genotypes in the current infections
IgG antibodies against four CSP peptides (N-terminal [N] and C-terminal [C], repetitive region corresponding to the VK210 [R] and repetitive region corresponding to the VK247 [V]) [47], MSP-1 N- terminal fragment (rPv 200L) [48], recombinant peptide of the AMA-1 [49] and of the DBP [50] were detected by ELISA (enzyme-linked immunosorbent assay), in plasma samples in infected individuals with P. vivax CS genotypes.
Statistical analysis
The serological data were performed using R version 2.8.1 statistical software The R Foundation for Statistical Computing, Vienna, Austria [51]. Differences among the frequencies of responders were analysed using Pearson's chi-square or, alternatively, the Fisher's exact test. Differences were considered significant when p-value ≤ 0.05.
Results
Phylogeny of P. vivax CS genotypes, VK210 and P. vivax-like
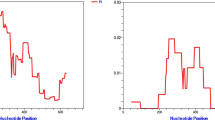
The analyses of the two markers show high similarity among the P. vivax CS genotypes, with nucleotide diversity equal to zero (p = 0.224, t Student's test), positioning the genotypes VK210 and P. vivax-like in the same clade (Figure 1 and Figure 2). The genetic distances between CS genotypes from the Plasmodium species analysed are described in Additional file 2 and Additional file 3: Genetic distances between 18 SSU rRNA and Cyt B genes from Plasmodium spp. The blood samples infected with VK247 genotypes were not included in this study, because of the reduced numbers of VK247 samples (n = 4) and low quality of the material.
Antibody response against the CSP, MSP-1, AMA-1 and DBP peptides
A higher frequency IgG antibody against the N- and C-terminal regions of the P. vivax CSP was found as compared to the immune response to the R- and V- repetitive regions (p = 0.0005, Fisher's exact test). Antibody responses against the peptides of the CSP repetitive central region [R] and [V] compared displayed a lower frequency against the [V] peptide, which corresponds to the central region of the VK247 variant, in individuals with VK210 genotype (p < 0.005, Fisher's exact test). When P. vivax-like genotype was present in the infection a lower antibody response against [R] and [V] peptides was observed (p = 0.003, Fisher's exact test). None other significant association was found with de CS genotypes in the infection (Table 1).
A high frequency of antibody response against MSP-1 and AMA-1 peptides was observed for all P. vivax CS genotypes in comparison to the same frequency for DBP. A high frequency of antibody response against MSP-1 (rPv 200L) and AMA-1 peptides was observed for all the P. vivax CS genotypes (Table 2) in comparison to same frequency for DBP (p = 0.003, Fisher's exact test). However, significant differences were not observed among the immune responses of individuals infected with the P. vivax CS genotypes for none of the analysed peptides.
Discussion
Starting from the description of the P. vivax CS genotypes, VK210, VK247 and P. vivax-like, several studies proposed the existence of differences among those that seem to go besides variations in the repetitive portion of the protein, as geographical distribution, transmission intensity, vectorial competence, immune and treatment responses and drug resistance [11, 18, 19, 32–34]. The real implication of the genetic variation in this region of the P. vivax genome has been questioned for a long time. Although previous studies of our group have observed significant association between VK210 and the Duffy blood group [17], this work presents here evidences of this variation is limited to the CSP central portion.
Studies based on molecular marker analysis represent an important tool for the phylogenetic characterization of malaria parasites. Similarities between P. vivax-like and Plasmodium simiovale have been reported in phylogenetic studies with conserved domains of the CS gene and, some authors suggested that this variant genotype could be a subspecies or a new species [6]. However, previous phylogenetic studies were designed with the CS gene as the only molecular marker in a way that prevents an explanation on the evolutionary relationship among the three CS genotypes as well as its relativity to other primate parasites that possess molecular similarities with P. vivax [35]. In this study, the results were obtained through the phylogenetic analysis of the 18 SSU rRNA and Cyt B Plasmodium spp. recognized markers and surprisingly showed diversity equal to zero between both P. vivax CS genotypes, VK210 and P. vivax-like. This analyses positioned VK210 and P. vivax-like as members of the same clade, in accordance with previous data [35]. Although the absence of VK247 genetic sequences, a limitation of the present study, the results point to the fact that P. vivax CS genotypes merely represent markers of intra-specific genetic variations.
Supporting the above mentioned hypothesis, the evaluation of the serological response profile against the different parasite peptides corroborates the idea that this variation is restricted to central portion of CSP, once significant associations were not observed between the presence of certain genotype and frequency of the antibodies responses against the three analysed merozoite peptides, MSP1 (Pv 200L), AMA-1, DBP and against the CSP conserved fractions in the sporozoite, N-terminal and C-terminal. Besides, when the antibody responses against the peptides corresponding to the CSP repetitive central region were evaluated, significant associations were detected against the peptides [R] and [V], which corresponds to the protein sequences of VK210 and VK247 genotypes, respectively. In individuals infected by the VK210 genotype, a lower antibody response against [V] was observed whereas in those infected by the P. vivax-like genotype observed an even lower antibody response against these two fragments ([R] and [V]) were observed. Once VK210 represents the classic P. vivax CS variant form and also the most prevalent in all Brazilian endemic areas [17], these results were expected. Moreover, the repeated region of the P. vivax-like CS is the most genetically distinct compared to the other variants [5, 6].
The report that variations in the central repetitive portion of CSP do not provide significant differences in antibody responses against P. vivax merozoite and sporozoite conserved regions peptides represents key information in the future design of vaccine assays. On the other hand, studies based in CSP should consider the influences of this variation in the modulation of the epidemiology and to consider the use of chimerical constructs including the sequences of the different CS genotypes in order to obtain a vaccine indeed protecting.
Conclusion
These results target that the differences among the P. vivax CS variants are restrict to the central repeated region of the protein, mostly nucleotide variation with important serological consequences. This variation can represent intra-specific biological signatures that must be considered for P. vivax CSP malaria vaccine trial.
References
Kappe SH, Buscaglia CA, Nussenzweig V: Plasmodium sporozoite molecular cell biology. Annu Rev Cell Dev Biol. 2004, 20: 29-59. 10.1146/annurev.cellbio.20.011603.150935.
Mota MM, Rodriguez A: Migration through host cells: the first steps of Plasmodium sporozoites in the mammalian host. Cell Microbiol. 2004, 20: 29-59.
Arnot DE, Barnwell JW, Tam JP, Nussenzweig V, Nussenzweig RS: Circumsporozoite protein of Plasmodium vivax: gene cloning and characterization of the immonodominant epitope. Science. 1985, 230: 815-817. 10.1126/science.2414847.
Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, Prasittisuk C: Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989, 245: 973-976. 10.1126/science.2672336.
Qari SH, Shi YP, Goldman IF, Udhayakumar V, Alpers MP, Collins WE, Lal AA: Identification of Plasmodium vivax-like human malaria parasite. The Lancet. 1993, 341: 780-783. 10.1016/0140-6736(93)90559-Y.
Qari SH, Shi YP, Póvoa MM Alpers MP, Deloron P, Murphy GS, Harjosuwarno S, Lal AA: Global occurrence of Plasmodium vivax-like human malaria parasite. J Infect Dis. 1993, 168: 1485-1489.
Kain KC, Brown AE, Webster HK, Witz RA, Keystone JS, Rodriguez MH, Kinahan J, Rowland M, Lanar DE: Circumsporozoite genotyping of global isolates of Plasmodium vivax from dried blood specimens. J Clin Microbiol. 1992, 30: 1863-1866.
Arruda ME, Aragaki C, Gagliardi F, Halle RW: A seroprevalence and descriptive epidemiological study of malaria among Indian tribes of the Amazon basin of Brazil. Ann Trop Med Parasitol. 1996, 90: 135-143.
Arruda ME, De Souza RC, Veiga ME, Ferreira AF, Zimmerman RH: Prevalence of Plasmodium vivax variants VK247 and P. vivax-like human malaria: a retrospective study in Indian populations of the Amazon region of Brazil. Trans R Soc Trop Med Hyg. 1998, 92: 628-10.1016/S0035-9203(98)90788-X.
Curado II, Duarte AMRC, Lal AA, Nussenzweig RS, Oliveira S, Kloetzel JK: Serological investigation of human Plasmodium vivax-like malaria in several localities in the State of São Paulo, Brazil. Mem Inst Oswaldo Cruz. 1995, 90 (Suppl 1): 284-
Machado RLD, Póvoa MM: Distribution of Plasmodium vivax variants (VK210, VK247 and P. vivax-like) in three endemic areas of Amazonian Brazil and their correlation with chloroquine-treatment. Trans R Soc Trop Med Hyg. 2000, 94: 377-381. 10.1016/S0035-9203(00)90110-X.
Gonzalez JM, Hurtado S, Arévalo-Herrera M, Herrera S: Variants of the Plasmodium vivax circumsporozoite protein (VK210 and VK247) in Colombian isolates. Mem Inst Oswaldo Cruz. 2001, 96: 709-712.
Oliveira-Ferreira J, Pratt-Riccio LR, Arruda M, Santos F, Ribeiro CT, Goldberg AC, Banic DM: HLA class II and antibody responses to circumsporozoite protein repeats of P. vivax (VK210, VK247 and P. vivax-like) in individuals naturally exposed to malaria. Acta Trop. 2004, 92: 63-69. 10.1016/j.actatropica.2004.02.011.
Imwong M, Pukrittayakamee S, Gruner AC, Renia L, Letourneur F, Looaresuwan S, White NJ, Snounou G: Plasmodium vivax: polymerase chain reaction amplification artifacts limit the suitability of pvgamI as genetic marker. Exp Parasitol. 2005, 99: 175-179. 10.1006/expr.2001.4646.
Zakeri S, Abouie-Mehrizi A, Djadid ND, Snounou G: Circumsporozoite protein gene diversity among temperate and tropical Plasmodium vivax isolates from Iran. Trop Med Int Health. 2006, 11: 729-737. 10.1111/j.1365-3156.2006.01613.x.
Curado I, Malafronte RS, Duarte AMRC, Kirchgatter K, Branquinho MS, Galati EAB: Malaria epidemiology in low-endemicity areas of the Atlantic Forest in the Vale do Ribeira, São Paulo, Brazil. Acta Trop. 2006, 100: 54-62. 10.1016/j.actatropica.2006.09.010.
Storti-Melo LM, Souza-Neiras WC, Cassiano GC, Joazeiro ACP, Fontes CJ, Bonini-Domingos CR, D'Almeida Couto AAR, Povoa MM, Mattos LC, Cavasini CE, Rossit ARB, Machado RLD: Plasmodium vivax circumsporozoite variants and Duffy blood group genotypes in the Brazilian Amazon region. Trans R Soc Trop Med Hyg. 2009, 103: 672-678. 10.1016/j.trstmh.2008.07.018.
Da Silva ANM, Santos CCB, Lacerda RN, Machado RLD, Póvoa MM: Susceptibility of Anopheles aquasalis and An.darlingi to Plasmodium vivax VK210 and VK247. Mem Inst Oswaldo Cruz. 2006, 101: 547-550.
Gonzalez-Ceron L, Rodríguez MH, Entel JC, Villarreal C, Kain KC, Hernandez JE: Differential susceptibilities of Anopheles albimanus and Anopheles pseudopunctipennis to infections with coindigenous Plasmodium vivax variants VK210 and VK247 in Southern Mexico. Infect Immun. 1999, 67: 410-412.
Vilai CS, Looareesuwan S, Good MF: Analysis of circumsporozoite protein-specific immune responses following recent infection with Plasmodium vivax. Am J Trop Med Hyg. 2004, 71: 29-39.
Soares IS, Levitus G, Souza JM, Del Portillo HA, Rodrigues MM: Acquired immune responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1 in individuals exposed to malaria. Infect Immun. 1997, 65: 1606-1614.
Barbedo MB, Ricci R, Jimenez MCS, Cunha MG, Yazdani SS, Chitnis CE, Rodrigues MM, Soares IS: Comparative recognition by human IgG antibodies of recombinant proteins representing three asexual erythrocytic stage vaccine candidates of Plasmodium vivax. Mem Inst Oswaldo Cruz. 2007, 102: 335-339. 10.1590/S0074-02762007005000040.
Múfalo BC, Gentil F, Bargieri DY, Costa FT, Rodrigues MM, Soares IS: Plasmodium vivax apical membrane antigen-1: comparative recognition of different domains by antibodies induced during natural human infection. Microbes Infect. 2008, 10: 1266-1273. 10.1016/j.micinf.2008.07.023.
Arevalo-Herrera M, Castellanos A, Yazdani SS, Shakri AR, Chitnis CE, Dominik R, Herrera S: Immunogenicity and protective efficacy of recombinant vaccine based on the receptor-binding domain of the Plasmodium vivax Duffy binding protein in Aotus monkeys. Am J Trop Med Hyg. 2005, 73: 25-31.
Herrera S, Bonelo A, Perlaza BL, Fernandez OL, Victoria L, Lenis AM, Soto L, Hurtado H, Acuna LM, Velez JD, Palacios R, Chen-Mok M, Corradin G, Arevalo-Herrera M: Safety and elicitation of humoral and cellular responses in Colombian malaria-naive volunteers by a Plasmodium vivax circumsporozoite protein-derived synthetic vaccine. Am J Trop Med Hyg. 2005, 73: 3-9.
Arevalo-Herrera M, Herrera S: Plasmodium vivax malaria vaccine development. Mol Immunol. 2001, 38: 443-455. 10.1016/S0161-5890(01)00080-3.
Rodrigues MH, Rodrigues KM, Oliveira TR, Cômodo AN, Rodrigues MM, Kocken CH, Thomas AW, Soares IS: Antibody response of naturally infected individuals to recombinant Plasmodium vivax apical membrane antigen-1. Int J Parasitol. 2005, 35: 185-192. 10.1016/j.ijpara.2004.11.003.
Herrera S, Corradin G, Arevalo-Herrera M: An update on the search for a Plasmodium vivax vaccine. Trends Parasitol. 2007, 23: 122-128. 10.1016/j.pt.2007.01.008.
Beeson JG, Crabb BS: Towards a vaccine against Plasmodium vivax malaria. Plos Med. 2007, 4: 1862-1864. 10.1371/journal.pmed.0040350.
Penny MA, Maire N, Studer A, Schapira A, Smith TA: What should vaccine developers ask? Simulation of the effectiveness of malaria vaccines. Plos One. 2008, 3: e3193-10.1371/journal.pone.0003193.
King CL, Michon P, Shakri AR, Marcotty A, Stanisic D, Zimmerman PA, Cole-Tobian JL, Mueller I, Chitnis CE: Naturally acquired Duffy-binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci USA. 2008, 105: 8363-8368. 10.1073/pnas.0800371105.
Branquinho MS, Marrelli MT, Curado I, Natal D, Barata JM, Tubaki R, Carreri-Bruno GC, de Menezes RT, Kloetzel JK: Infection of Anopheles (Kerteszia) cruzii by Plasmodium vivax and Plasmodium vivax variant VK247 in the municipalities of Sao Vicente and Juquitiba, Sao Paulo. Rev Panam Salud Publica. 1997, 2: 189-193. 10.1590/S1020-49891997000900004.
Marrelli MT, Branquinho MS, Hoffman EH, Taipe-Lagos CB, Natal D, Kloetzel JK: Correlation between positive serology for Plasmodium vivax-like/Plasmodium simiovale malaria parasites in the human and anopheline populations in the state of Acre, Brazil. Trans R Soc Trop Med Hyg. 1998, 92: 149-151. 10.1016/S0035-9203(98)90723-4.
Machado RLD, Figueriredo-Filho AF, Calvosa VSP, Nascimento JM, Póvoa MM: Correlation between Plasmodium vivax variants in Belém, Pará State, Brazil and symptoms and clearence of parasitemia. J Infec Dis. 2003, 7: 175-177.
Escalante AA, Barrio E, Ayala FJ: Evolutionary origin of human and primate malarias: Evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995, 12: 616-626.
Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, Su X-Z: Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005, 22: 1686-1693. 10.1093/molbev/msi160.
Leclerc MC, Durand P, Gauthier C, Patot S, Billotte N, Menegon M, Severini C, Ayala FJ, Renaud F: Meager genetic variability of the human malaria agent Plasmodium vivax. Proc Natl Acad Sci USA. 2004, 101: 14455-14460. 10.1073/pnas.0405186101.
Carlton JM, Escalante AA, Neafsey D, Volkman SK: Comparative evolutionary genomics of human malaria parasites. Trends Parasitol. 2008, 24 (12): 545-550. 10.1016/j.pt.2008.09.003.
Snewin VA, Longacre S, David PH: Plasmodium vivax: older and wiser?. Res Immunol. 1991, 142: 631-636. 10.1016/0923-2494(91)90140-E.
Rongnoparut P, Supsamran N, Sattabongkot J, Suwanabun N, Rosenberg R: Phenotype and genotype diversity in the circumsporozoite proteins of Plasmodium vivax in Thailand. Mol Biochem Parasitol. 1995, 74: 201-210. 10.1016/0166-6851(95)02504-9.
Pena SDJ, Macedo AM, Gontijo NF, Medeiros AM, Ribeiro JC: DNA bioprints: simple non-isotopic DNA fingerprints with biotinnylated probes. Eletrophoresis. 1991, 12: 14-52. 10.1002/elps.1150120209.
Kimura M, Kneko O, Liu Q, Zhou M, Kawamoto F, Wataya Y, Otani S, Yamaguchi Y, Tanake K: Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol Intern. 1997, 46: 91-95. 10.1016/S1383-5769(97)00013-5.
Alves RT, Póvoa MM, Goldman IF, Cavasini CE, Rossit AR, Machado RLD: A new polymerase chain reaction/restriction fragment lenght polymorphism protocol for Plasmodium vivax circumsporozoite protein genotype (VK210, VK247, and P. vivax-like) determination. Diag Microbiol Infec Dis. 2007, 59: 415-419. 10.1016/j.diagmicrobio.2007.06.019.
Santos-Ciminera PD, Alecrim MGC, Roberts DR, Quinnan GV: Molecular epidemiology of Plasmodium vivax in the State of Amazonas, Brazil. Acta Trop. 2007, 102: 38-46. 10.1016/j.actatropica.2007.02.013.
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetics trees. Mol Biol Evol. 1987, 17: 1542-1557.
Kumar S, Tamura K, Nei M: Molecular evolutionary genetics analysis, version 1.0. Pennsylvania State University, University Park.
Valderrama-Aguirre A, Quintero G, Gomez A, Castellanos A, Pérez Y, Méndez F, Arévalo-Herrera M, Herrera S: Antigenicity, Immunogenicity and protective efficacy of Plasmodium vivax MSP1 PV 200L: a potencial malaria vaccine subunit. Am J Trop Med Hyg. 2005, 73: 16-24.
Herrera S, Bonelo A, Perlaza BL, Valencia AZ, Cifuentes C, Hurtado S, Quintero G, López JA, Corradin G, Arevalo-Herrera M: Use of long synthetic peptides to study the antigenicity and immunogenicity of the Plasmodium vivax circumsporozoite protein. Int J Parasitol. 2004, 34: 1535-46. 10.1016/j.ijpara.2004.10.009.
Rodrigues MH, Rodrigues KM, Oliveira TR, Comodo AN, Rodrigues MM, Kocken CH, Thomas AW, Soares IS: Antibody response of naturally infected individuals to recombinant Plasmodium vivax apical membrane antigen-1. Int J Parasitol. 2005, 35: 185-192. 10.1016/j.ijpara.2004.11.003.
Cerávolo IP, Bruna-Romero O, Braga EM, Fontes CJ, Brito CF, Souza JM, Krettli AU, Adams JH, Carvalho LH: Anti-Plasmodium vivax duffy binding protein antibodies measure exposure to malaria in the Brazilian Amazon. Am J Trop Med Hyg. 2005, 72: 75-681.
The R Foundation for Statistical Computing. [http://www.r-project.org]
Acknowledgements
To all individuals enrolled in this study. We thank the following people for assistance in obtaining samples: Dr. Carlos Eugênio Cavasini, Dr. Aline Barroso, Dr. Maria Cristina Figueredo and Dr. Mauro Tada. To Professor Dr. Luiz Hildebrando Pereira da Silva for facilities at CEPEM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
WCSN carried out the molecular genetic studies, participated in the sequence alignment, phylogenetics analyses and drafted the manuscript. LMSM carried out the immunoassays and participated in the design of the study and performed of the statistical analysis part. ISS, LHC and MGC designed serological experiments and provided reagents. GCC, VSCAC, ISS, LHC, SH, MAH, ARBR critically revised the manuscript. CMAC participated in the sequence alignment and phylogenetics analyses. RLDM conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Wanessa C Souza-Neiras, Luciane M Storti-Melo contributed equally to this work.
Electronic supplementary material
12936_2009_1259_MOESM1_ESM.DOC
Additional file 1: Hosts type, geographic origins and GenBank accession numbers of the out groups. GenBank accession numbers. (DOC 46 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Souza-Neiras, W.C., Storti-Melo, L.M., Cassiano, G.C. et al. Plasmodium vivax circumsporozoite genotypes: a limited variation or new subspecies with major biological consequences?. Malar J 9, 178 (2010). https://doi.org/10.1186/1475-2875-9-178
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-9-178