Abstract
Background
In Iran, co-infections of Plasmodium vivax and Plasmodium falciparum are common and P. vivax infections are often exposed to sulphadoxine-pyrimethamine (SP). In the present study, the frequency distribution of mutations associated to SP resistance was investigated in pvdhfr and pvdhps genes from field isolates.
Methods
Clinical isolates of P. vivax were collected in two different malaria endemic regions in northern and south-eastern Iran, between 2001 and 2006. All 189 collected isolates were analysed for SNP/haplotypes at positions 13, 33, 57, 58, 61, 117 and 173 of the pvdhfr and 383 and 553 of pvdhps genes using nested PCR-RFLP methods
Results
All 189 examined isolates were found to carry wild-type amino acids at positions 13, 33, 61 and 173, while 57L and 58R and 117N mutations in pure form was detected among 1.1%, 17.5% and 26% examined samples, respectively, with no polymorphisms in different loci of dhps genes. Based on size polymorphism of pvdhfr genes at repeat region, among northern isolates, the frequency distribution for type A and B were 2.2% and 97.8% respectively. However, in southern samples the prevalence of type A, B and C were 7%, 89.5% and 7.7%, respectively. Mixed genotype infections (type B and C) were detected in only 4.2% (6/143) of southern, but in none of the northern isolates. The combination of pvdhfr and pvdhps haplotypes among all 189 samples demonstrated six distinct haplotypes. The two most prevalent haplotypes among all examined samples were I13P33F57S58T61S117I173/A383A553 (65.6%) and I13P33F57S58T61N117I173 (16.4%). Two other alleles with one point mutation I13P33F57R58T61S117I173/A383A553 and two mutations I13P33F57R58T61N117I173/A383A553 accounted for 7.4% and 9.5% of the total isolates.
Conclusion
The present molecular data provide important information for making decisions on population based drug use in Iran. In addition, since October 2005, with more availability of SP as first-line treatment, P. vivax isolates are more exposed to SP and the selection or spread of resistant pvdhfr and pvdhps alleles might increase in the near future in this region.
Similar content being viewed by others
Background
Plasmodium vivax remains the most widespread malaria parasite in areas outside of Africa [1] and chloroquine (CQ) remains the first-line treatment for vivax malaria in most endemic regions. In recent years, chloroquine-resistant P. vivax parasites have been reported in several locations, with high level resistance confirmed in parts of Thailand, Indonesia and New Guinea and in Central America [2–7]. In those countries, where sulfadoxine/pyrimethamine (SP) has been used extensively, high grade antifolate resistance has also emerged in P. vivax populations [8–13].
Unfortunately, resistance develops relatively quickly, when SP is widely used. Molecular and epidemiological studies of both Plasmodium falciparum and P. vivax have revealed that the dihyrofolate reductase (DHFR) and dihydropteroate synthase (DHFR) enzymes are the therapeutic targets of SP [10–17]. As a result, resistance to SP is determined by specific point mutations in the parasite dhfr and dhps genes. These mutations cause alterations of key amino acid residues in the active sites of these enzymes, which reduce the affinity of the enzyme for the drug [15, 18–25]. Therefore, detection of these mutations in wild isolates has proved valuable in the mapping and monitoring of resistance for guiding malaria control measures.
Vivax infections are not often treated with SP, but P. vivax isolates are exposed to SP because mixed infections are common in Asia and South America [26–29] and are often mis-diagnosed. As continuous in vitro culture of P. vivax remains unavailable, and it is difficult to monitor the susceptibility of P. vivax to anti-malarial drugs by in vitro tests [30, 31], the association between the various pvdhfr point mutations and resistance to pyrimethamine relies on epidemiological and clinical investigations. Different studies showed that in areas where there is a long history of extensive SP use, mutant alleles of pvdhfr gene are prevalent; however wild type pvdhfr has been found more commonly in areas with limited use of SP [10, 12, 13, 17]. So far, 20 non-synonymous mutations have been described in pvdhfr, [31] and different studies of P. vivax parasites in different malaria endemic areas, such as Thailand and India, showed that mutations at pvdhfr codons 57, 58, 61, 117 and 173, [13, 32] were found to be involved in clinical antifolate resistance [12, 31]. Five mutations have already been identified in pvdhps gene, at codons 382, 383, 512, 553, and 585.
Iran is located in the Middle East and, in this region, P. vivax is the most prevalent malaria parasite species, responsible for 80 to 90% of malaria cases. The disease is endemic in the south-east bordering with Pakistan and Afghanistan, but it re-emerged in the north of the country after a 20-year interruption of transmission [33]. In Iran, CQ has been used as the first line anti-malarial treatment and SP was used as second-line treatment [34] for uncomplicated P. falciparum infection. Furthermore, while in vitro studies have reported SP resistance since 1993 [35, 36], in vivo resistance of P. falciparum to SP is not yet common in these areas. With the spread of chloroquine resistance in P. falciparum, the Center for Diseases Management and Control (CDMC), decided in 2005 to revise its treatment policy and SP in combination with CQ has been recommended as the first-line anti-malarial treatment, with artemether-lumefantrine (Co-Artem®), as second-line [34]. Although SP may remain the treatment of choice for uncomplicated malaria, the high rates of treatment failures with CQ [34] and, therefore, inadequate efficacy of treatment with the SP/CQ combination, the CDMC decided in 2007 to revise its treatment policy again and SP/CQ was replaced with SP/artemisinin as the first-line recommendation for falciparum malaria. CQ remains the first choice for treatment of P. vivax mono-infections and resistance to either CQ or SP has not yet been recorded in Iran. In addition, in this area P. vivax co-exists with P. falciparum [26] but the correct diagnosis of mixed infections is not easy based on microscopic examination of blood films, and the clinical symptoms caused by the two species cannot be differentiated. As a result, P. vivax may often be treated with SP because of mixed infections and inaccurate diagnosis.
The pvdhfr and pvdhps genotype might be associated with treatment failure in individual vivax malaria patients, and while data on the genotypes of these two genes are available from Thailand, the Indian subcontinent and the Indonesian archipelago, such data are lacking in many regions, most notably Central and South America and the Middle East, with only a few isolates from those regions having been assessed for mutations in dhfr.
Methods
Study sites and P. vivax clinical isolates
In this study, 189 P. vivax clinical isolates were collected between March 2001 and March 2006, from P. vivax-infected patients, aged from one to > 60 years, living either in the tropical south-eastern region (Chabahar district in Sistan and Baluchistan) (n = 143), or in area of resurgent malaria (Pars Abad in Ardebil province) in the temperate northern endemic area (n = 46). The most prevalent parasite species in the temperate northern region is P. vivax, with Anopheles maculipennis and Anopheles sacharovi as the main vectors, and Anopheles superpictus and Anopheles hyrcanus as the secondary vectors. In the south-eastern region, transmission is year-round with two peaks, the first in May to August with P. vivax as the predominant species and the second peak from October to November when both P. falciparum and P. vivax infections are equally recorded. The main mosquito vectors in the south-eastern region are Anopheles stephensi, Anopheles culicifacies, Anopheles fluviatilis and Anopheles pulcherrimus.
All subjects had slide and PCR-proven infection by P. vivax. None of the subjects received SP therapy and infections were treated with chloroquine. In both study areas the patients have access to anti-malarial drugs through local health centers.
Informed consent was obtained from patients or parents of patients before inclusion in the study. The study was reviewed by, and received Ethical Clearance from Institut Pasteur of Iran. Blood samples were collected in tubes containing EDTA, stored at 4°C and then transported to the main laboratory in Tehran.
Parasite genomic DNA extraction
Parasite DNA was extracted from 250 μl infected whole blood by phenol/phenol-chloroform extraction and ethanol precipitation as described previously [37]. The DNA was dissolved in 30 μl TE buffer (10 mM Tris-HCL pH 8.0, 0.1 mM EDTA). For the detection of point mutations at residues 13, 33, 57, 58, 61, 117 and 173, the previously described PCR-RFLP protocols were used with some modifications [12, 13, 38].
Primary amplification of pvdhfr
In the first reaction, the entire P. vivax dhfr-ts gene was amplified by the primers VDT-OF: ATGGAGGACCTTTCAGATGTATTTGACATT and VDT-OR: GGCGGCCATCTCCATGGTTATTTTATCGTG [13]. The cycling conditions for the nest-1 reaction was as follows: 95°C for 5 min, 25 cycles of 64°C for 2 min, 72°C for 2 min, 94°C for 1 min followed by 64°C for 2 min and 72°C for 15 min.
PCR amplification of positions 13, 33, 58, and 61
One μl product of the first reaction was then used in second round of amplification using the following primers for positions 13, 33, 58, and 61:
VDFN13F: GACCTTTCAGATGTATTTGACATTTACGGC
VDFN13R: GGTACCTCTCCCTCTTCCACTTTAGCTTCT
The cycling conditions for the nest-2 reaction was as follows: 95°C for 5 min, 25 cycles of 66°C for 2 min, 72°C for 2 min, 94°C for 1 min followed by 66°C for 2 min and 72°C for 15 min.
RFLP
To detect mutation at position 13L, 10 μl of the PCR products were digested with 10 U HaeIII enzyme (Roche, Germany) for 1 h at 37°C in a total volume of 20 μl (232 bp = 32 bp + 200 bp). To detect mutations at residue P33L and S58R, 10 μl of the PCR products were digested with 10 U Cfr42I (SacII) (Fermentase, Vilnius, Lithuania) and AluI (Roche, Germany) for 4 h at 37°C in a total volume of 20 μl (232 bp = 94 bp + 138 bp for P33 and 232 bp = 25 bp + 207 bp for 58R, 232 bp = 25 bp+40 bp+167 bp for wild type S58), respectively. Mutation at residue 61 M was detected by digestion of 10 μl of the PCR products with 10 U Tsp45I (New England Biolab, Beverly, MA, USA) for 4 h at 37°C in a total volume of 20 μl (232 bp = 32 bp + 200 bp).
PCR amplification of positions 57 and 173
The product of first reaction was also amplified with oligonucleotide pair:
VDT-OF: ATGGAGGACCTTTCAGATGTATTTGACATT
VDFNR: TCACACGGGTAGGCGCCGTTGATCCTCGTG
to amplify positions 57 and 173. The cycling conditions for the nest-2 reaction was as follows: 95°C for 5 min, 25 cycles of 66°C for 2 min, 72°C for 2 min, 94°C for 1 min followed by 66°C for 2 min and 72°C for 15 min. It should be noted that because of presence of repeat regions that amplify the above-mentioned primers, there are size variations among the amplified PCR products, which affect the size of digested products. In the present study, the size of the digested PCR products, which is reported in the following RFLP section, is based on pvdhfr Accession no: X98123.
RFLP
To detect mutation at residue F57, 10 μl of the PCR products were digested with 10 U XmnI: (New England Biolab, Beverly, MA, USA) for 4 h at 37°C in a total volume of 20 μl (608 bp = 166 bp + 442 bp). Mutation at residue I173L was detected by digestion of 10 μl of the PCR products with 10 U Eco130I (StyI) (Fermentase, Vilnius, Lithuania) for 4 h at 37°C in a total volume of 20 μl (wild type I173: 608 bp = 136 bp + 472 bp and mutant type 173L: 608 bp = 73 bp + 97 bp + 438 bp).
PCR amplification of positions 57 and 117
One μl product of first reaction was also amplified with primers:
VDNF57:CATGGAAATGCAACTCCGTCGATATGATGT
VDF-NR:TCACACGGGTAGGCGCCGTTGATCCTCGTG
The cycling conditions for the this reaction was 95°C for 5 min, 25 cycles of 66°C for 2 min, 72°C for 2 min, 94°C for 1 min followed by 66°C for 2 min and 72°C for 15 min.
RFLP
To detect mutation at residue 57I, 10 μl of the PCR products were digested with 10 U BsrGI (New England Biolab, Beverly, MA, USA) for 4 h at 37°C in a total volume of 20 μl (472 bp = 28 bp + 444 bp). To detect mutation at residue S117N/T, 10 μl of the PCR products (472 bp) were digested with 10 U PvuII (New England Biolab, Beverly, MA, USA) (S117, 214 bp + 258 bp), for 4 h at 37°C and BsrI (New England Biolab, Beverly, MA, USA (117N, 219 bp + 253 bp), BstNI (New England Biolab, Beverly, MA, USA) (117T, 215 bp + 257 bp) for 1 h at 65°C in a total volume of 20 μl. All amplifications were carried out in a final volume of 25 μl, which included 1 μl of template from either genomic DNA or the primary reaction. The primers were used at a final concentration of 250 nM and the reaction mixture contained 10 mM Tris-HCL (pH 8.3), 50 mM KCl, 2 mM MgCl2, each of the four deoxynucleotide triphosphates at a concentration of 125 μM, and 0.2 U of Taq polymerase (Invitrogen, Carlsbad, CA). The DNA fragments obtained following PCR amplification or RFLP analysis were electrophoresed on 2.5% (Invitrogen, Carlsbad, CA) and 3% Metaphor (Invitrogen, Carlsbad, CA) agarose gels, respectively. All digested products of RFLP were subjected to electrophoresis on 3% MetaPhor agarose gels.
Analysis of pvdhfr gene at repeat region
This region was amplified using 1 μl of primary reaction with primers:
VDFN2F: CGGTGACGACCTACGTGGATGAGTCAAAGT
VDFN2R: TAGCGTCTTGGAAAGCACGACGTTGATTCT as described previously [12]. The cycling conditions for the this reaction was 95°C for 5 min, 25 cycles of 66°C for 2 min, 72°C for 2 min, 94°C for 1 min followed by 66°C for 2 min and 72°C for 15 min. The DNA fragments obtained following PCR amplification were analysed following electrophoresis on 3% Metaphor agarose gels.
Amplification of pvdhps
The pvdhps gene was amplified from genomic DNA by nested PCR. The primers used in the first and second round PCRs were described previously [38]. In the first reaction pvdhps was amplified with primers:
VDHPS-OF: ATTCCAGAGTATAAGCACAGCACATTTGAG
VDHPS-OR: CTAAGGTTGATGTATCCTTGTGAGCACATC
The second amplification was performed with the primers for detection of 383 mutation:
VDHPS-NF: AATGGCAAGTGATGGGGCGAGCGTGATTGA
VDHPS-NR: CAGTCTGCACTCCCCGATGGCCGCGCCACC
for detection of the 553 mutations the oligonuclotide primers
VDHPS-553OF: TTCTCTTTGATGTCGGCCTGGGGTTGGCCA
VDHPS-NR: CAGTCTGCACTCCCCGATGGCCGCGCCACC were used.
The cycling conditions for the nest-1 PCR reaction was as follows: 95°C for 5 min, 25 cycles of 58°C for 2 min, 72°C for 2 min, 94°C for 1 min followed by 58°C for 2 min and 72°C for 15 min and for nest-2 PCR was: 95°C for 5 min, 25 cycles of 50°C for 2 min, 72°C for 2 min, 94°C for 1 min followed by 50°C (for VDHPS-553OF and VDHPS-NR 50°C) for 2 min and 72°C for 15 min.
RFLP
To detect mutation at residue A383G, 10 μl of the PCR products were digested with 10 U MspI (HpaII) (Fermentase, Vilnius, Lithuania) (mutant 383G, 703 bp = 48 bp + 655 bp) and MscI (New England Biolab, Beverly, MA, USA) (wild A553, 170 bp = 27 bp + 143 bp) for 4 h at 37°C in a total volume of 20 μl.
Results
Distribution of mutations in pvdhfr and pvdhps
All 189 isolates from north and south were analysed for SNP/haplotypes at positions 13, 33, 57, 58, 61, 117 and 173 of the pvdhfr and 383 and 553 of pvdhps genes using PCR-RFLP methods [12, 13, 38]. In pvdhfr, polymorphisms at positions 57L, 58R and 117N have been found in 1.4%, 21.7% and 30% of southern isolates, respectively (Table 1). Among northern isolates mutations at 58R and 117N were found in 4.4% and 13% of the studied isolates, respectively (Table 1).
In total, all 189 examined isolates were found to carry wild-type amino acids at positions 13, 33, 61 and 173, while 57L and 58R and 117N mutations in pure form was detected among 1.1%, 17.5% and 26% examined samples, respectively (Table 1). In the case of pvdhps gene, polymorphisms in different loci of dhps (A383G and A553G) were investigated and no mutations were detected at all in the examined samples.
Size polymorphism of pvdhfr at repeat region
In this investigation all three types A, B and C [12] were found among southern isolates, but only types A and B were detected among northern isolates. The frequency distribution for type A and B were 2.2% (1/46) and 97.8% (45/46) among northern isolates, respectively. However, for southern samples the prevalence of the type A, B and C were 7%, 89.5% and 7.7%, respectively. Mixed genotype infections (type B and C) were detected in only 4.2% (6/143) of southern, but not in northern isolates.
Distribution of pvdhfr and pvdhps haplotypes in Iran
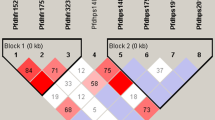
The combination of pvdhfr and pvdhps haplotypes among all 189 samples in this study demonstrated six distinct haplotypes (Figure 1). The two most prevalent haplotypes among all examined samples were I13P33F57S58T61S117I173/A383A553 (65.6%) and I13P33F57S58T61N117I173/A383A553 (16.4%). Two other alleles with one point mutation at position I13P33F57R58T61S117I173/A383A553 and two mutations at position 58R and 117N (I13P33F57R58T61N117I173/A383A553) accounted for 7.4% and 9.5% of the total isolates. This double mutant haplotype was the most frequently mutated haplotype observed among Iranian samples. Regarding these nine SNPs in pvdhfr and pvdhps genes, significant increasing in the prevalence of double mutated haplotypes (I13P33F57R58T61N117I173/A383A553) was observed in collected samples in year 2006 compare to 2001 (P > 0.005) (Table 2). In addition, the majority of isolates from north and also south were wild haplotype and I13P33L57R58T61S117I173/A383A553 (0.7%) mutant haplotype was only detected among southern but northern isolates (Table 1).
Frequency distribution of the combination pvdhfr/pvdhps haplotypes obtained from 189 isolates collected in Ardebil province in north and Sistan and Baluchistan province in southeastern of Iran. The haplotype I13P33F57S58T61S117I173/A383A553 was the most prevalent among northern (84.8%) and southern (59.4%) P. vivax isolates. All six haplotypes are indicated as A to F in the figure. Mutated amino acids are boldfaced. A) I13P33F57S58T61S117I173/A383A553. B) I13P33F57S58T61N117I173/A383A553. C) I13P33F57R58T61S117I173/A383A553. D) I13P33F57R58T61N117I173/A383A553. E) I13P33L57S58T61S117I173/A383A553. F) I13P33L57R58T61S117I173/A383A553.
Discussion
In regions where P. falciparum and P. vivax co-exist, it is crucial to identify effective treatment regimens that work against both parasite species. In malaria-endemic areas co-infection of P. vivax and P. falciparum is common and the long history of SP use has exposed P. vivax to this drug for decades. In the present study, the SP resistance-associated genes, pvdhfr and pvdhps, were analysed in samples collected (prior to introduction of SP as first-line anti-malarial), from re-emerged area in north and endemic region in south where both CQ and SP (in combination with primaquine) were used for treatment. In the south, although CQ still remains effective against P. vivax infection, the in vivo work in 2005 [39] showed that parasite clearance time increased compared to 2001 in Sistan and Baluchistan province, indicating that this could be an early sign of reduced susceptibility of the parasites to CQ. Therefore, effective alternative drug against P. vivax resistance to CQ might be needed. In this investigation, four and six distinct haplotypes of pvdhfr and only wildtype of pvdhps were detected among northern and southern isolates, respectively. The double mutant I13P33F57R58T61N117I173/A383A553 was the second frequent haplotype in our examined isolates. This is the first time that mutations at associated genes to SP resistance have been described in a large number of P. vivax isolates from the Middle East.
Mutations in pvdhfr, including 58R and 117N, have been implicated in in vivo pyrimethamine resistance and seem to arise first under drug pressure. The 58R was found in 17.5% of all examined isolates alone and in combination with 117N in 9.5%, despite the fact that SP had never been used as a first-line treatment for falciparum malaria before October 2005. The work carried out by Tahar and colleagues [40] showed that the 58R/117N mutant had a lower affinity for pyrimethamine and cycloguanil than did the wild type enzyme. In addition, Leartsakulpanich et al screened pyrimethamine resistance-associated genes and found that the 117N, 58R/117N, 58R and 173L mutant enzymes were more resistance to this drug than the wild type [41]. The similar work by Hastings and colleagues confirmed these results [11]. Regarding the clinical efficacy of SP against P. vivax, several workers concluded that the clinical response to SP depends on pvdhfr and pvdhps genotype [11, 12, 17]. They also showed that those patients who harboured triple and quadruple mutant parasites (57L/58R/61M/117T) compared with those who harboured wild type parasites were significantly associated more likely to SP treatment failure [11, 12, 17]. In addition, treatment failure was more frequently associated with multiple mutations in dhfr and dhps [38]. This correlation between two genes also reported for P. falciparum, as parasite carried wild type alleles of dhfr and dhps, the patient is likely to have an adequate clinical response to SP, but when the parasite carries mutant alleles of both genes, clinical effectiveness is compromised [42–48]. Although none of the P. vivax isolates have been tested in this study for their clinical response to SP, triple and quadruple mutant types, found in Thailand, India, and in Indonesia have previously been shown to be associated with a high risk of SP treatment failure [11, 12, 17].
The frequency distribution of pvdhfr mutant haplotypes was significantly higher in the endemic southern regions than re-emerged northern region (Pars-Abad, Ardebile). This might be caused by the level of disease endemicity in the south and longer usage of SP for treatment of P. falciparum infections. In other words, SP never used in north for treatment of malaria disease as any P. falciparum infections was not detected by microscopy method; therefore no SP pressure might be responsible for prevalence of mutant alleles of dhfr in this region. In fact, the limited diversity of pvdhfr mutant, particularly double mutants in northern compare to southern isolates may also be an indication for a founder effect linked to the introduction of malaria from Azerbaijan and Armenia to northern part of Iran. The presence of I13P33L57R58T61S117I173/A383A553 haplotype only among southern isolates may be related to human migration between Iran and Pakistan, resulting to introduction of such haplotypes from the Indian subcontinent, where it is prevalent [31].
In the present study, the most common haplotypes of pvdhfr were wild type and double mutant (58R and 117N); but triple and quadruple mutants were not detected among examined isolates. In contrast, molecular analysis of pvdhfr among Indian field isolates showed 14 haplotypes from wild type to quadruple mutant genotype, and wild type and double mutant were still the most common [31]. Since 2005, based on the national drug policy, the anti-malarial treatment in Iran has changed and the SP became the first choice drug for treatment. With more availability of SP, there is a risk of a changing pattern of resistance of both P. falciparum and P. vivax to SP, as this closely follows the intensity with which SP has been used [31]. In addition, other antifolates such as co-trimoxazole that are routinely used against urinary tract infections and chronic bronchitis in the region could add to the overall antifolate pressure in Iran.
Conclusion
The present molecular data provide important information for making decisions on population based drug use in Iran. In addition, previous study showed that in regions where the wild type or single mutated pvdhfr alleles are prevalent, SP could be a useful therapy [31] for the asexual erythrocytic stages of vivax in areas where CQ treatment failure has been reported.
References
Mendis K, Sina BJ, Marchesini P, Carter R: The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001, 64 (1–2 Suppl): 97-106. 1, 3
Baird JK, Basri H, Subianto B, Patchen LC, Hoffman SL: Resistance to chloroquine by Plasmodium vivax in Irian Jaya, Indonesia. Am J Trop Med Hyg. 1991, 4: 547-552.
Baird JK, Leksana B, Masbar S, Fryauff DJ, Sutanihardja MA, Suradi , Wignall FS, Hoffman SL: Diagnosis of resistance to chloroquine by Plasmodium vivax: timing of recurrence and whole blood chloroquine levels. Am J Trop Med Hyg. 1997, 56: 621-26.
Garg M, Gopinathan N, Bodhe P, Khirsagar NA: Vivax malaria resistant to chloroquine: case reports from Bombay. Trans R Soc Med Hyg. 1995, 89: 656-657. 10.1016/0035-9203(95)90432-8.
Kyaw MP, Kyaw MP, Myint O, Myint L, Thaw Z, Aye KH, Yin NN: Emergence of chloroquine-resistant Plasmodium vivax in Myanmar (Burma). Trans R Soc Trop Med Hyg. 1993, 87: 687-10.1016/0035-9203(93)90294-Z.
Murphy GS, Basri H, Purnomo , Andersen EM, Bangs MJ, Mount DL, Gorden J, Lal AA, Purwokusumo A, Harjosuwarno S, Sorensen K, Hoffman SL: Vivax malaria resistant to treatment and prophylaxis with chloroquine. Lancet. 1993, 341: 96-100. 10.1016/0140-6736(93)92568-E.
Rieckmann KH, Davis DR, Hutton DC: Plasmodium vivax resistance to chloroquine. Lancet. 1989, ii: 1183-1184. 10.1016/S0140-6736(89)91792-3.
Pukrittayakamee S, Chantra A, Simpson J, Vanijanonta S, Clemens R, Looareesuwan S, White NJ: Therapeutic responses to different antimalarial drugs in vivax malaria. Antimicrob Agents Chemother. 2000, 44: 1680-1685. 10.1128/AAC.44.6.1680-1685.2000.
Alam MT, Bora H, Bharti PK, Saifi MA, Das MK, Dev V, Kumar A, Singh N, Dash AP, Das B: Similar trends of pyrimethamine resistance-associated mutations in Plasmodium vivax and P. falciparum. Antimicrob Agents Chemother. 2007, 51: 857-863. 10.1128/AAC.01200-06.
de Pecoulas PE, Tahar R, Ouatas T, Mazabraud A, Basco LK: Sequence variations in the Plasmodium vivax dihydrofolate reductasethymidylate synthase gene and their relationship with pyrimethamine resistance. Mol Biochem Parasitol. 1998, 92: 265-273. 10.1016/S0166-6851(97)00247-8.
Hastings MD, Porter KM, Maguire JD, Susanti I, Kania W, Bangs MJ, Sibley CH, Baird JK: Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulphadoxinee plus pyrimethamine. J Infect Dis. 2004, 189: 744-750. 10.1086/381397.
Imwong M, Pukrittakayamee S, Looareesuwan S, Pasvol G, Poirreiz J, White NJ, Snounou G: Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulphadoxinee-pyrimethamine: geographical and clinical correlates. Antimicrob Agents Chemother. 2001, 45: 3122-3127. 10.1128/AAC.45.11.3122-3127.2001.
Imwong M, Pukrittayakamee S, Renia L, Letourneur F, Charlieu JP, Leartsakulpanich U, Looareesuwan S, White NJ, Snounou G: Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob Agents Chemother. 2003, 47: 1514-1521. 10.1128/AAC.47.5.1514-1521.2003.
Foote SJ, Galatis D, Cowman AF: Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci USA. 1990, 87: 3014-3017. 10.1073/pnas.87.8.3014.
Peterson DS, Milhous WK, Wellems TE: Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci USA. 1990, 87: 3018-3022. 10.1073/pnas.87.8.3018.
Peterson DS, Walliker D, Wellems TE: Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988, 85: 9114-9118. 10.1073/pnas.85.23.9114.
Tjitra E, Baker J, Suprianto S, Cheng Q, Anstey NM: Therapeutic efficacies of artesunate-sulphadoxinee-pyrimethamine and chloroquinesulphadoxinee-pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob Agents Chemother. 2002, 46: 3947-3953. 10.1128/AAC.46.12.3947-3953.2002.
Foote SJ, Cowman AF: The mode of action and the mechanism of resistance to antimalarial drugs. Acta Trop. 1994, 56: 157-171. 10.1016/0001-706X(94)90061-2.
Matthews DA, Alden RA, Bolin JT, Freer ST, Hamlin R, Xuong N, Kraut J, Poe M, Williams M, Hoogsteen K: Dihydrofolate reductase: X-ray structure of the binary complex with methotrexate. Science. 1977, 197: 452-455. 10.1126/science.17920.
Peterson DS, Milhous WK, Wellems TE: Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988, 85: 9114-9118. 10.1073/pnas.85.23.9114.
Sirawaraporn W, Prapunwattana P, Sirawaraporn R, Yuthavong Y, Santi DV: The dihydrofolate reductase domain of Plasmodium falciparum thymidylate synthase-dihydrofolate reductase: gene synthesis, expression, and anti-folate resistance mutants. J Biol Chem. 1993, 268: 21637-21644.
Snewin VA, England SM, Sims PFG, Hyde JE: Characterisation of the dihydrofolate reductase-thymidylate synthase gene from human malaria parasites highly resistant to pyrimethamine. Gene. 1989, 76: 41-52. 10.1016/0378-1119(89)90006-1.
Thaithong S, Chan SW, Songsomboon S, Wilairat P, Seesod N, Sueblinwong T, Goman M, Ridley R, Beale G: Pyrimethamine resistant mutations in Plasmodium falciparum. Mol Biochem Parasitol. 1992, 52: 149-158. 10.1016/0166-6851(92)90047-N.
Volz KW, Matthews DA, Alden RA, Freer ST, Hansch C, Kaufman BT, Kraut J: Crystal structure of avian dihydrofolate reductase containing phenyltriazine and NADPH. J Biol Chem. 1982, 257: 2528-2536.
Foote SJ, Cowman AF: The mode of action and the mechanism of resistance to antimalarial drugs. Acta Trop. 1994, 56: 157-171. 10.1016/0001-706X(94)90061-2.
Zakeri S, Najajabadi S, Zare A, Djadid N: Detection of malaria parasites by nested PCR in south-eastern, Iran: Evidence of highly mixed infections in Chabahar district. Malar J. 2002, 1: 2-10.1186/1475-2875-1-2.
Mayxay M, Pukrittayakamee S, Newton PN, White NJ: Mixed-species malaria infections in humans. Trends Parasitol. 2004, 20: 233-240. 10.1016/j.pt.2004.03.006.
Mehlotra RK, Lorry K, Kastens W, Miller SM, Alpers MP, Bockarie M, Kazura JW, Zimmerman PA: Random distribution of mixed species malaria infections in Papua New Guinea. Am J Trop Med Hyg. 2000, 62: 225-231.
Snounou G, White NJ: The co-existence of Plasmodium: sidelights from falciparum and vivax malaria in Thailand. Trends Parasitol. 2004, 20: 333-339. 10.1016/j.pt.2004.05.004.
Udomsangpetch R, Kaneko O, Chotivanich K, Sattabongkot J: Cultivation of Plasmodium vivax. Trends Parasitol. 2008, 24: 85-88. 10.1016/j.pt.2007.09.010.
Hawkins VN, Joshi H, Rungsihirunrat K, Na-Bangchang K, Sibley CH: Antifolates can have a role in the treatment of Plasmodium vivax. Trends Parasitol. 2007, 23 (5): 213-222. 10.1016/j.pt.2007.03.002.
Barnadas C, Tichit M, Bouchier C, Ratsimbasoa A, Randrianasolo L, Raherinjafy R, Jahevitra M, Picot S, Menard D: Plasmodium vivax dhfr and dhps mutations in isolates from Madagascar and therapeutic response to sulphadoxine-pyrimethamine. Malar J. 2008, 7: 35-10.1186/1475-2875-7-35.
Zakeri S, Mehrizi AA, Mamaghani S, Noorizadeh S, Snounou G, Djadid ND: Population structure analysis of Plasmodium vivax in areas of Iran with different malaria endemicity. Am J Trop Med Hyg. 2006, 74 (3): 394-400.
Zakeri S, Afsharpad M, Kazemzadeh T, Mehdizadeh K, Shabani A, Djadid ND: Association of pfcrt but not pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum. Am J Trop Med Hyg. 2008, 78: 633-640.
Edrissian GH, Afshar A, Sayedzadeh A, Mohsseni GH, Satvat MT: Assessment of the response in vivo and in vitro of Plasmodium falciparum to sulphadoxine-pyrimethamine in the malarious areas of Iran. J Trop Med Hyge. 1993, 96: 237-240.
Heidari A, Dittrich S, Jelink T, Kheirandish A, Banihashemi K, Keshavarz H: Genotypes and in vivo resistance of Plasmodium falciparum isolates in an endemic region of Iran. Parasitol Res. 2007, 100: 589-592. 10.1007/s00436-006-0291-z.
Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN: High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993, 61: 315-320. 10.1016/0166-6851(93)90077-B.
Imwong M, Pukrittayakamee S, Cheng Q, Moore C, Looareesuwan S, Snounou G, White NJ, Day NPJ: Limited Polymorphism in the Dihydropteroate Synthetase Gene (dhps) of Plasmodium vivax Isolates from Thailand. Antimicrob Agents Chemother. 2005, 49: 4393-4395. 10.1128/AAC.49.10.4393-4395.2005.
Nateghpour M, Sayedzadeh SA, Edrissian GhH, Raeisi A, Jahantigh A, Motevalli-Haghi A, Mohseni Gh, Rahimi A: Evaluation of sensitivity of Plasmodium vivax to chloroquine. Iranian J Publ Health. 2007, 36: 60-63.
Tahar R, de Pécoulas PE, Basco LK, Chiadmi M, Mazabraud A: Kinetic properties of dihydrofolate reductase from wild-type and mutant Plasmodium vivax expressed in Escherichia coli. Mol Biochem Parasitol. 2001, 113: 241-249. 10.1016/S0166-6851(01)00230-4.
Leartsakulpanich U, Imwong M, Pukrittayakamee S, White NJ, Snounou G, Sirawaraporn W, Yuthavong Y: Molecular characterization of dihydrofolate reductase in relation to antifolate resistance in Plasmodium vivax. Mol Biochem Parasitol. 2002, 119: 63-73. 10.1016/S0166-6851(01)00402-9.
Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco JG, Doumbo OK: Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulphadoxinee use and resistance. J Infect Dis. 1997, 176: 1590-1596.
Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RAG, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV: Molecular markers for failure of sulphadoxinee-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002, 185: 380-388. 10.1086/338566.
Nzila AM, Mberu EK, Sulo J, Dayo H, Winstanley PA, Sibley CH, Watkins WM: Towards an understanding of the mechanism of pyrimethamine-sulphadoxinee resistance in Plasmodium falciparum: genotyping of dihydrofolate reductase and dihydropteroate synthase of Kenyan parasites. Antimicrob Agents Chemother. 2000, 44: 991-996. 10.1128/AAC.44.4.991-996.2000.
Mutabingwa T, Nzila A, Mberu E, Nduati E, Winstanley P, Hills E, Watkins W: Chlorproguanil-dapsone for treatment of drug-resistant falciparum malaria in Tanzania. Lancet. 2001, 358: 1218-1223. 10.1016/S0140-6736(01)06344-9.
Kyabayinze D, Cattamanchi A, Kamya MR, Rosenthal PJ, Dorsey G: Validation of a simplified method for using molecular markers to predict sulphadoxinee-pyrimethamine treatment failure in African children with falciparum malaria. Am J Trop Med Hyg. 2003, 69: 247-252.
Staedke SG, Sendagire H, Lamola S, Kamya MR, Dorsey G, Rosenthal PJ: Relationship between age, molecular markers, and response to sulphadoxine-pyrimethamine treatment in Kampala, Uganda. Trop Med Int Health. 2004, 9: 624-629. 10.1111/j.1365-3156.2004.01239.x.
Nzila A: The past, present and future of antifolates in the treatment of Plasmodium falciparum infection. J Antimicrob Chemother. 2006, 57: 1043-1054. 10.1093/jac/dkl104.
Acknowledgements
We acknowledge with deep respect to Prof. H. Malek Afzali. (the former Deputy for Research, Ministry of Health, I.R. Iran) for his invaluable support; and also co-operation of the Center for Diseases Management and Control (CDMC), particularly Dr. M.M. Gouya and Dr. A. Raeisi. We are grateful for the hospitality and generous collaboration of Zahedan University of Medical Sciences, staff in Public Health Department, Sistan and Baluchistan province, Chabahar district, and also Pars-Abad, Ardebil for their assistance in collecting blood samples from the field. We are indebted to the patients and their families in Sistan and Baluchistan and Ardebil provinces for their willingness to parcipitate in this study. This study was partially supported by grants from Deputy for Research, Ministry of Health, and Institut Pasteur of Iran.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SZ designed and supervised the study, analysed the data and wrote the manuscript. SRM contributed in the laboratory work and MA contributed in the laboratory work and helped with analysis of the data. NDD helped with the preliminary analysis of the data and also critical reading of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zakeri, S., Motmaen, S.R., Afsharpad, M. et al. Molecular characterization of antifolates resistance-associated genes, (dhfr and dhps) in Plasmodium vivax isolates from the Middle East. Malar J 8, 20 (2009). https://doi.org/10.1186/1475-2875-8-20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-8-20