Abstract
Background
Molecular markers of insecticide resistance can provide sensitive indicators of resistance development in malaria vector populations. Monitoring of insecticide resistance in vector populations is an important component of current malaria control programmes. Knockdown resistance (kdr) confers resistance to the pyrethroid class of insecticides with cross-resistance to DDT through single nucleotide polymorphisms (SNPs) in the voltage-gated sodium channel gene.
Methods
To enable detection of kdr mutations at low frequency a method was developed that uses polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA)-based technology, allowing rapid, reliable and cost-effective testing of large numbers of individual mosquitoes. This was used to assay mosquitoes from sites in lower Moshi, Tanzania.
Results
Sequence-specific oligonucleotide probes (SSOP) were used for simultaneous detection of both East and West African kdr mutations with high specificity and sensitivity. Application of the SSOP-ELISA method to 1,620 field-collected Anopheles arabiensis from Tanzania identified the West African leucine-phenylalanine kdr mutation in two heterozygous individuals, indicating the potential for resistance development that requires close monitoring.
Conclusion
The presence of the West African kdr mutation at low frequency in this East African population of An. arabiensis has implications for the spread of the kdr gene across the African continent.
Similar content being viewed by others
Introduction
With efforts to scale-up the coverage of insecticide-treated nets (ITN) in Africa [1] there is increasing concern regarding the potential impact of insecticide resistance on malaria control [2, 3]. The knockdown resistance (kdr) mechanism confers resistance to pyrethroid insecticides and DDT through point mutations in the voltage-gated sodium channel gene [4, 5]. Resulting single amino acid changes in the domain II region of the sodium channel reduce the sensitivity of the insect nervous system to these compounds. Two different kdr mutations have been identified in resistant Anopheles gambiae s.l. populations from East and West Africa. The West African kdr is characterized by a leucine to phenylalanine substitution at position 104 of the voltage-gated sodium channel sequence (L104F) [4], while the East African kdr involves a serine substitution at the same position (L104S) [6]. Recent reports of both kdr mutations in An. gambiae from Uganda [7]indicate, however, that this geographic distinction may be too simplified. While the kdr allele has predominantly been found in An. gambiae s.s. [8–10], several reports have also identified the presence of kdr in the sibling species Anopheles arabiensis [7, 11, 12].
Models of antimalarial drug resistance spread have demonstrated the potential for a rapid increase in the frequency of resistance, rising quickly from undetectable levels to levels that result in control failure [13]. Models of insecticide resistance show an equally rapid rise when the frequency reaches levels as low as 0.1% [14]. Since the kdr allele is incompletely recessive, conventional bioassay methods that measure phenotypic resistance cannot reliably detect the heterozygous proportion of the population [8]. A more sensitive approach is direct genotyping, which can identify heterozygotes and, thus, facilitate early detection of resistance development.
However, current methods for kdr genotyping have limitations for testing large numbers of individuals, which is often needed in routine monitoring programmes. Multiplex polymerase chain reaction (PCR) methods [4, 6] are time consuming and the visualization of the PCR product by gel electrophoresis uses toxic reagents. The PCR-dot blot method [15] is also relatively time consuming, and scoring of the product is subjective if done by eye, or requires costly equipment for automated scoring. The HOLA technique [16] allows more simple and reliable detection of kdr mutations, but requires two thermal cycling steps, necessitating additional time and cost. The FRET/MCA technique requires costly real-time PCR equipment [7], which is not affordable by most resource poor laboratories.
A rapid, high-throughput method for kdr screening was developed that is appropriate for use in laboratories in malaria endemic countries. This method combines a PCR assay with visualization of the product using sequence-specific oligonucleotide probes (SSOP) in an enzyme-linked immunosorbent assay (ELISA) format. It is based on the SSOP-ELISA method of Alifrangis et al. [17] for detecting drug resistance mutations in Plasmodium falciparum, with some modifications. The is rapid, allowing analysis of more than 150 samples in a single day, convenient, using a 96-well plate format for easy transfer of reagents and samples with multichannel pipettes, and cost-effective, amounting to roughly 1 USD per sample. Following optimization, the method was evaluated by analysis of field-collected material from northern Tanzania, where insecticide resistance monitoring and ITN evaluation activities are ongoing.
Methods
Anopheles gambiae reference strains
Three laboratory reference strains of An. gambiae s.s. were used to verify the specificity and sensitivity of the assay, and these were used as controls in subsequent experiments. R70 from Tanzania possesses the wildtype susceptible (104L) genotype; VKPR, homozygous for the L104F West African kdr allele; and RSP, homozygous for the L104S East African kdr allele. In addition, artificial heterozygotes were created by combining DNA extracted from individuals of susceptible and resistant strains, L(R70)/F(VKPR) for the West African kdr and L(R70)/S(RSP) for the East African kdr.
Field-collected samples
Field studies were conducted in villages in the lower Moshi area on the slopes beneath Mt Kilimanjaro in northeast Tanzania.
Mabogini
Female An. arabiensis were collected by indoor-resting catch from houses in Mabogini village (3°22' S, 37°19' E) during June/July 2004, and exposed to 0.75% permethrin in WHO susceptibility tests [18]. Survivors and dead individuals from these tests (n = 822) were processed for circumsporozoite protein (CSP) ELISA for detection of P. falciparum sporozoites, i.e. head and thorax of individual mosquitoes homogenized in grinding buffer [19], then stored in 96-well plates in grinding buffer at -20°C. Experimental hut trials conducted in the same area to evaluate permethrin-treated nets and sheets provided survivors and dead mosquitoes (n = 156), which were stored on silica gel at 4°C.
Msitu wa Tembo
Female An. arabiensis (n = 642) from light trap collections carried out in 2004 in the village of Msitu wa Tembo, Tanzania (3°33' S, 37°17' E) were processed for CSP ELISA and stored in 96-well plates at -20°C.
Extraction of DNA from mosquitoes
Individual mosquitoes, dried on silica gel or processed for CSP ELISA, were homogenized in 50 μl STE buffer (1 mM EDTA, 10 mM Tris-HCl, 50 mM NaCl), incubated at 95°C for 12 minutes then centrifuged at 13,000 rpm for four minutes at room temperature. Aliquots of the supernatant containing suspended DNA were transferred into fresh tubes and stored at -20°C until use. DNA extraction was carried out in individual tubes or in 96-well PCR plates using a clean pipette tip for homogenization of each sample. DNA extracted from individual, dried mosquitoes using the method described by Collins et al. [20] was used for comparison. Ten microlitres of the DNA extract from two individuals were pooled in each well, and this pooled DNA was used in the polymerase chain reaction.
Polymerase chain reaction
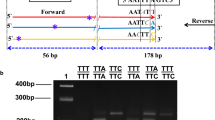
Forward and reverse primers developed by Kolaczinski et al. [21] were used to amplify a 216 bp fragment of the voltage-gated sodium channel gene, with a biotin modification of the reverse primer at the 5' end (MWG Biotech, Riskov, Denmark). Each 20 μL PCR reaction consisted of 0.25 mM each dNTP, 0.1 μM each primer, one unit HotStarTaq polymerase (Qiagen, Albertslund, Denmark) in buffer containing 1.5 mM MgCl2 (Qiagen) and 2 μL extracted DNA. Reaction conditions were 94°C for 15 minutes followed by 35 cycles of 94°C for one minute, 55°C for one minute and 72°C for one minute, with a final extension at 72°C for four minutes; the PCR product was kept at 4°C until use. Amplifications were performed in 96-well PCR plates and the reaction mixture was overlaid with one drop of mineral oil. PCR products from controls and several samples from each plate were confirmed by electrophoresis on a 1.5% agarose gel. Species identifiation of members of the An. gambiae complex was performed on individual mosquitoes according to Scott et al. [22].
SSOP-ELISA for kdr detection
The ELISA plates (Maxisorp; Nunc, Roskilde, Denmark) were coated with streptavidin in phosphate buffered saline (PBS) (1 g/mL), covered, and left overnight at 4°C. Prior to use, the plates were washed three times in washing buffer (1 × PBS containing 0.05% Tween 20). The PCR products were diluted 1:1 in water in a 96-well PCR plate, denatured at 95°C for five minutes, and immediately thereafter cooled to 4°C until use. The 3'-end digoxigenin-conjugated SSOPs (MWG Biotech, Riskov, Denmark) (shown in Table 1) were diluted to a 4 nM concentration in tetramethyl ammonium chloride (TMAC; Sigma Aldrich, Dorset, UK) solution (3 M TMAC, 50 mM Tris, pH 8.0, 0.1% sodium dodecyl sulfate, 2 mM EDTA, pH 8.0), heated to 53°C, and 100 μL was then added to each well of the ELISA plate. Two microlitres of the diluted PCR products was subsequently added. Replicate ELISA plates were made to enable simultaneous probing with SSOPs targeting all three kdr genotypes. The plates were incubated in a hybridization oven (AH Diagnostics, Aarhus, Denmark) at 53°C on a shaking device for one hour and washed three times in washing buffer. This was followed by two rounds of washing and incubation (14 minutes per round for 104L and 104S probes, 15 minutes per round for the 104F probe) in TMAC solution at 68°C. To remove TMAC, the plates were then washed three times in washing buffer, and peroxidase-conjugated anti-digoxigenin antibody in dilution buffer (1:1,000) (Roche Diagnostics, Mannheim, Germany) was added to each well. After incubation for one hour at room temperature, the plates were washed three times in washing buffer and 100 μL of room temperature TMB substrate (Sigma Aldrich, Dorset, UK) was added to the plates. The reaction was stopped after five minutes by adding 0.5 M H2SO4 and the optical density (OD) at 450 nm was measured in an ELISA reader. A flow diagram of the methodology is presented in Figure 1.
Scoring of ELISA data
For each probe, the non-complementary control strains served as negative controls (e.g. for the 104L probe, wells that contained R70 served as positive controls while wells on the same plate that contained VKPR or RSP served as negative controls). Some between-experiment variation in the OD values of positive and negative controls was apparent, possibly due to marginal differences in the probe binding strength and the washing force during high stringency washes. While this rarely compromised specificity, no fixed threshold could be specified for SNP tests and for each experiment a threshold of positivity was set for each SSOP corresponding to twice the maximum negative control OD value. Samples with OD values exceeding this threshold for each SSOP were considered positive for the corresponding SNP, or combination of SNPs in the case of heterozygotes.
Evaluation of SSOP-ELISA compared to multiplex PCR
A set of 12 samples, consisting of six individual controls, five mixtures of controls and one field-collected sample were used in a double blind trial to test the consistency of the SSOP-ELISA method in relation to the multiplex PCR methods described by Martinez-Torres et al. [4] and Ranson et al. [6]. The susceptible Dondotha strain of An. arabiensis was used in addition to R70 as a wildtype control.
Sequencing
Kdr primers with additional 5' EcoR1 and 3' Not1 site were used to generate PCR products, which were cloned into pAcGP67 (BD biosciences). Plasmids were prepared using MiniPrep spin columns (Omega Biotech). Sequencing was done on an ABI Prism 377 (Perkin-Elmer) using the Big Dye terminator reaction mix (Perkin-Elmer) and ABI Prism proof-reading and translation software.
Results
Specificity and sensitivity of SSOP-ELISA
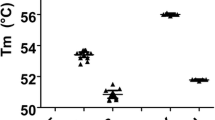
To evaluate the specificity and sensitivity of the ELISA, the three reference strains of An. gambiae s.s., R70, VKPR and RSP known to carry the different kdr genotypes, as well as the artificial heterozygote mixtures of these reference strains, were tested. DNA extracted from samples by STE or the Collins method worked equally well, and further evaluation was carried out using DNA extracted by the STE method. Figure 2 shows that the assay correctly identifies the SNPs of the different strains using SSOP for 104L, 104F and 104S for single individuals. The difference in OD value between the threshold of positivity and the positive reactions, ΔOD, was always greater than 1.3. For mixtures of two individuals representing artificial heterozygotes (Figure 3), the OD obtained with kdr-specific probes (104F or 104S) was slightly lower than the OD obtained with corresponding probes for single individuals; however, the ΔODs for the different SNPs were at least 0.9. The OD using the wildtype probe (104L) was lower in both mixtures compared to single individuals, with a ΔOD of 0.9 for the L(R70)/F(VKPR) mixture and 0.8 for the L(R70)/S(RSP) mixture; however, the signal strength was sufficient for visual detection. Figure 4 shows the visual results of the SSOP-ELISA test for control strains, individually, in combination as artificial heterozygotes, and with an additional dilution, in three columns of an ELISA plate representing the different SSOP.
In two dilution series with pools containing four or five mosquitoes possessing the resistant and susceptible genotypes, the SSOP-ELISA could reliably detect one resistant individual in a pool containing three other susceptible individuals (1F:3L or 1S:3L), with the ΔOD in relation to the negative control of 0.9 for 104F and 0.7 for 104S. However, when the pool size was increased to five individuals, sensitivity to detect one resistant mosquito in a pool containing four other susceptible individuals (1F:4L or 1S:4L) was reduced, and the ΔOD for the minority genotype was <0.5.
Evaluation of SSOP-ELISA versus multiplex PCR
Results of the double blind trial are presented in Table 2. All six individual controls were successfully genotyped by multiplex PCR and correctly identified by SSOP-ELISA. For five artificial heterozygotes, multiplex PCR failed to amplify the 137 bp susceptible band in both L/S mixtures, and failed to amplify the 195 bp resistant band in one of two L/F mixtures, while the F/S mixture was successfully genotyped. This was in contrast to the SSOP-ELISA which detected both SNPs in all mixtures with success. The field sample of unknown genotype was identified as 104L by multiplex PCR and SSOP-ELISA.
Analysis of field-collected samples
To ensure detection of heterozygous individuals in field-collected specimens, pools of two individuals only were used representing a total of four alleles. The individuals from positive pools were tested separately for confirmation. All 978 individuals from susceptibility tests and experimental huts in Mabogini tested homozygous for the wildtype susceptible (104L) genotype. However, the SSOP-ELISA detected two out of 642 individuals from Msitu wa Tembo village that were heterozygous for the L104F kdr genotype (allele frequency = 0.16%). These L104F kdr genotypes were confirmed by DNA sequencing. It was not possible to correlate the kdr genotype with insecticide susceptibility status since these mosquitoes had been collected by light trap catch for the purpose of sporozoite rate measurement.
Discussion
A SSOP-ELISA method is described that allows simple, high-throughput detection of kdr SNPs in An. gambiae s.l. This method can be established and operated in malaria endemic countries to assist local or national insecticide resistance monitoring programmes. Multiplex PCR methods are often unreliable for detection of heterozygotes [7, 16] and other methods have apparent limitations in terms of time and cost. The SSOP-ELISA method has an advantage of using equipment that is readily available in research laboratories in malaria endemic countries. In addition its increased sensitivity can facilitate detection of the kdr alleles when still present at low frequency, as demonstrated by detection of two heterozygotes in Tanzania where the kdr mutation has not previously been recorded.
While the leucine-phenylalanine kdr mutation is widespread in West Africa, it has not, until recently, been found further east of the Central African Republic [12, 23]. In Uganda, this mutation was found in combination with the leucine-serine kdr mutation in An. gambiae s.s. [7]. Interestingly, the presence of the leucine-phenylalanine kdr allele in two An. arabiensis from northern Tanzania represents the first account of this mutation in East African populations of this sibling species. More widespread monitoring of the distribution of both kdr mutations in the An. gambiae complex across Africa may therefore reveal less definite geographic restrictions than previously thought. The kdr genotype of An. arabiensis from Msitu wa Tembo village could not be linked to the resistance phenotype, yet this finding raises potential implications for malaria control. While high frequencies of the same kdr mutation in An. gambiae s.s. in Côte d'Ivoire have not impacted on the control achieved with insecticide-treated nets [24, 25], kdr may interact with other genes to eventually have a serious impact on malaria control. The small-scale farming village where kdr was detected in the present study is situated nearby the irrigated rice-growing Mabogini area of Tanzania, where survivors of bioassays did not posess the kdr genotype [26] suggesting that an additional resistance mechanism, possibly metabolic, may be present in the local vector population.
Present insecticide use in the area where the two kdr heterozygotes were collected is negligible, with only small-scale, subsistence agricultural production. However, the river that serves as a focus for vector breeding in this village [27] flows downstream from large-scale agricultural areas where pyrethroids and organochlorines have previously been used (Kulkarni et al. in preparation). Early insecticide use may have selected for resistance in local vector populations, and the mutation may have since existed at low undetectable frequencies. Resistance monitoring using bioassays has not detected significant reductions in insecticide susceptibility in this area (Kulkarni et al. in preparation). However, there is clearly the potential for resistance development in populations where the kdr mutation exists at low levels [14] as was demonstrated in western Kenya, where the kdr L104S allele pre-dated ITN use and increased significantly in frequency after introduction of ITN [12]. Investigation of local heterogeneity in the frequency of kdr in Tanzania and neighbouring countries in relation to history of insecticide use may reveal areas that contributed to early selection of resistance and environmental conditions that might be favouring growth of resistance.
Continued monitoring of vector populations is essential and will benefit from screening for genetic markers of resistance. The occurrence of the typically West African leucine-phenylalanine kdr mutation in Tanzania emphasizes the need to test for both kdr mutations regardless of geographic location. Use of the cost-effective, high-throughput SSOP-ELISA method to detect kdr alleles may contribute to resistance monitoring efforts in many regions of Africa, where little is known on the insecticide resistance status of malaria vector populations.
References
RBM: The Abuja declaration and the plan of action. In: The African Summit on Roll Back Malaria, Abuja, Nigeria. 2003, Geneva , Roll Back Malaria Partnership Secretariat
Curtis CF, Miller JE, Hodjati MH, Kolaczinski JH, Kasumba I: Can anything be done to maintain the effectiveness of pyrethroid-impregnated bednets against malaria vectors?. Philosophical Transactions of the Royal Society of London B. 1998, 353: 1769-1775. 10.1098/rstb.1998.0329.
Sina BJ, Aultman K: Resisting resistance. Trends in Parasitology. 2001, 17 (7): 305-306. 10.1016/S1471-4922(01)02007-4.
Martinez-Torres D, Chandre F, Williamson MS, Darriet F, Berge JB, Devonshire AL, Guillet P, Pasteur N, Pauron D: Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Molecular Biology. 1998, 7 (2): 179-184. 10.1046/j.1365-2583.1998.72062.x.
Williamson MS, Martinez-Torres D, Hick CA, Devonshire AL: Identification of mutations in the housefly para-type sodium channel gene associated with knockdown resistance (kdr) to pyrethroid insecticides. Mol Gen Genet. 1996, 252: 51-60. 10.1007/BF02173204.
Ranson H, Jensen B, Vulule JM, Wang X, Hemingway J, Collins FH: Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Molecular Biology. 2000, 9 (5): 491-497. 10.1046/j.1365-2583.2000.00209.x.
Verhaeghen K, Van Bortel W, Roelants P, Backeljau T, Coosemans M: Detection of the East and West African kdr mutation in Anopheles gambiae and Anopheles arabiensis from Uganda using a new asssay based on FRET/Melt Curve analysis. Malaria Journal. 2006, 5: 16-10.1186/1475-2875-5-16.
Chandre F, Darriet F, Duchon S, Finot L, Manguin S, Carnevale P, Guillet P: Modifications of pyrethroid effects associated with kdr mutation in Anopheles gambiae. Medical and Veterinary Entomology. 2000, 14: 81-88. 10.1046/j.1365-2915.2000.00212.x.
Etang J, Manga L, Chandre F, Guillet P, Fondjo E, Mimpfoundi R, Toto JC, Fontenille D: Insecticide susceptibility status of Anopheles gambiae s.l. (Diptera: Culicidae) in the Republic of Cameroon. Journal of Medical Entomology. 2003, 40 (4): 491-497.
Awolola TS, Brooke BD, Koekemoer LL, Coetzee M: Absence of the kdr mutation in the molecular 'M' form suggests different pyrethroid resistance mechanisms in the malaria vector mosquito Anopheles gambiae s.s. Tropical Medicine and International Health. 2003, 8 (5): 420-422. 10.1046/j.1365-3156.2003.01034.x.
Diabate A, Brengues C, Baldet T, Dabire KR, Hougard JM, Akogbeto M, Kengne P, Simard F, Guillet P, Hemingway J, Chandre F: The spread of the Leu-Phe kdr mutation through Anopheles gambiae complex in Burkina Faso: genetic introgression and de novo phenomena. Trop Med Int Health. 2004, 9 (12): 1267-1273. 10.1111/j.1365-3156.2004.01336.x.
Stump AD, Atieli FK, Vulule JM, Besansky NJ: Dynamics of the pyrethroid knockdown resistance allele in western Kenyan populations of Anopheles gambiae in response to insecticide-treated bed net trials. American Journal of Tropical Medicine and Hygiene. 2004, 70 (6): 591-596.
Hastings IM, D'Alessandro U: Modelling a predictable disaster: the rise and spread of drug-resistant malaria. Parasitology Today. 2000, 16 (8): 340-347. 10.1016/S0169-4758(00)01707-5.
Roush RT, McKenzie JA: Ecological genetics of insecticide and acaricide resistance. Annual Review of Entomology. 1987, 32 (1): 361-380. 10.1146/annurev.en.32.010187.002045.
Kolaczinski JH, Fanello C, Herve JP, Conway DJ, Carnavale P, Curtis CF: Experimental and molecular genetic analysis of the impact of pyrethroid and non-pyrethroid insecticide impregnated bednets for mosquito control in an area of pyrethroid resistance. Bulletin of Entomological Research. 2000, 90: 125-132.
Lynd A, Ranson H, McCall P, Randle N, Black W, Walker E, Donnelly M: A simplified high-throughput method for pyrethroid knock-down resistance (kdr) detection in Anopheles gambiae. Malaria Journal. 2005, 4 (1): 16-10.1186/1475-2875-4-16.
Alifrangis M, Enosse S, Pearce RJ, Drakeley C, Roper C, Khalil IF, Nkya WMMM, Ronn AM, Theander TG, Bygbjerg IC: A simple, high-throughput method to detect Plasmodium falciparum single nucleotide polymorphisms in the dihydrofolate reductase, dihydropteroate synthase, and P. falciparum chloroquine resistance transporter genes using polymerase chain reaction- and enzyme-linked immunosorbent assay-based technology. American Journal of Tropical Medicine and Hygiene. 2005, 72 (2): 155-162.
WHO: Test procedures for insecticide resistance monitoring in malaria vectors, bio-efficacy and persistence of insecticides on treated surfaces. 1998, Geneva, Switzerland , World Health Organization
Wirtz RA, Zavala F, Charoenvit Y, Campbell GH, Burkot TR, Schneider I, Esser KM, Beaudoin RL, Andre RG: Comparative testing of monoclonal antibodies against Plasmodium falciparum sporozoites for ELISA development. Bulletin of the World Health Organization. 1987, 65 (1): 39-45.
Collins FH, Mendez MA, Rasmussen MO, Mehaffrey PC, Besansky NJ, Finnerty V: A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. American Journal of Tropical Medicine and Hygiene. 1987, 37: 37-41.
Scott JA, Brogdon WG, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. American Journal of Tropical Medicine and Hygiene. 1993, 49: 520-529.
Weill M, Chandre F, Brengues C, Manguin S, Akogbeto M, Pasteur N, Guillet P, Raymond M: The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Molecular Biology. 2000, 9 (5): 451-455. 10.1046/j.1365-2583.2000.00206.x.
Darriet F, N'Guessan R, Koffi AA, Konan LY, Doannio JMC, Chandre F, Carnevale P: Impact de la resistance aux pyrethrinoides sur l'efficacite des moustiquaires impregnee dans la prevention du paludisme: resultats des essais en cases experimentales avec la deltamethrine SC. Bulletin de la Societe de Pathologie Exotique. 2000, 93: 131-134.
Henry MC, Assi SB, Rogier C, Dossou-Yovo J, Chandre F, Guillet P, Carnevale P: Protective efficacy of lambda-cyhalothrin treated nets in Anopheles gambiae pyrethroid resistance areas of Cote d'Ivoire. American Journal of Tropical Medicine and Hygiene. 2005, 73 (5): 859-864.
Kulkarni MA, Matowo J, Lyimo I, Malima R, Rau ME, Mosha FW, Alifrangis M, Rowland M, Drakeley C: Abstract for the 4th MIM Pan-African Malaria Conference. Characterization of permethrin tolerance in Anopheles arabiensis in Tanzania using a novel high-throughput method to detect knockdown resistance (kdr). Acta Tropica. 2005, Suppl 95S: S64-S65.
Mwerinde O, Oesterholt M, Harris C, Lushino P, Masokoto A, Bousema T, Mwerinde H, Targett G, Drakeley C: Abstract for the 4th MIM Pan-African Malaria Conference. Monitoring seasonal variations in malaria transmission in a low endemicity area in Tanzania. Acta Tropica. 2005, Suppl 95S: S183-
Acknowledgements
The study was conducted within the Joint Malaria Programme, a collaboration between the National Institute for Medical Research in Tanzania, Kilimanjaro Christian Medical Centre, London School of Hygiene and Tropical Medicine, and the University of Copenhagen. The field work in Mabogini was supported by a grant from Gates Malaria Partnership (GMP) to MR & FM and the field work in Msitu wa Tembo by a grant from the Wellcome Trust to CD. The assay development was supported by a grant from GMP to MR and CD. CD is supported by a research fellowship in tropical medicine (#063516) from the Wellcome Trust. MAK and MER are supported by the Natural Sciences and Engineering Research Council of Canada (NSERC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
Manisha Kulkarni organized and coordinated the field work, developed and validated the assay and wrote the first and subsequent drafts of the manuscript.
Michael Alifrangis and Ali Salanti developed the assay and performed the validatory sequencing.
Robert Malima, Eliningaya Kweka, Issa Lyimo and Stephen Magesa performed the field work and mosquito collections.
Johnson Matowo and Justin Peter processed mosquitoes and perfomed SSOP-ELISA assays.
Mark Rowland, Manfred Rau, Frank Mosha and Chris Drakeley conceived the project, oversaw its implementation and contributed to the writing of the manuscript.
All authors have seen and contributed to revisions of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kulkarni, M.A., Rowland, M., Alifrangis, M. et al. Occurrence of the leucine-to-phenylalanine knockdown resistance (kdr) mutation in Anopheles arabiensis populations in Tanzania, detected by a simplified high-throughput SSOP-ELISA method. Malar J 5, 56 (2006). https://doi.org/10.1186/1475-2875-5-56
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-5-56