Abstract
Background
The knowledge on population structure of the parasite isolates has contributed greatly to understanding the dynamics of the disease transmission for designing and evaluating malaria vaccines as well as for drug applications. msp-1 and msp-3α genes have been used as a genetic marker in population studies of Plasmodium vivax isolates. In this study, msp-3α was compared and assessed with msp-1 marker in order to find whether msp-3α is a reliable genetic marker for P. vivax population studies.
Methods
This comparative study was designed and carried out as the first assessment of diversity in Pvmsp-3α gene by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) in the 50 northern and 94 southern P. vivax isolates from Iran, which had been analysed before for msp-1 gene.
Results
Three allele size as, Type A (1.8 kb), Type B (1.5 kb) and Type C (1.2 kb) have been detected among both northern and southern isolates based on PCR results. Type C (70%) and Type A (68.7%) were the predominant fragments among northern and southern parasites, respectively. 99 distinct Pvmsp-3α fragments defined by the size were detected in the 94 southern samples by PCR analysis. However, no mixed genotype infections have been detected among northern isolates. Based on restriction pattern from digestion with Hha I and Alu I 12 and 49 distinct allelic variants have been detected among 50 northern and 94 southern isolates. However, based on msp-1 gene, 30 distinct variants identified in all 146-sequenced Iranian P. vivax isolate.
Conclusion
The results suggested that PCR-RFLP on msp-3α gene is an adequate, applicable and easily used technique for molecular epidemiology studies of P. vivax isolates without the need for further sequencing analysis.
Similar content being viewed by others
Background
Knowledge of the population structure of the parasite isolates is important to understand the dynamics of disease transmission. Molecular epidemiological studies require reliable polymorphic markers [1]. Four polymorphic markers have been used to study the genetic structure of Plasmodium vivax, including Pvgam1, which encodes for a protein expressed during sexual stages [2, 3]Pvcs, the circumsporozoite protein [4–6], Pvmsp-1 coding for the merozoite surface protein 1 [7–11] and Pvmsp-3α, encoding for the merozoite surface proteins 3α [12–14].
A number of genes encoding for P. vivax MSPs have been identified, including Pvmsp-1 [8], Pvmsp-3α, Pvmsp-3β and Pvmsp-3γ [12, 15], Pvmsp-4 and Pvmsp-5 [16] and Pvmsp-9 [17]. Of these, Pvmsp-1 has been most extensively studied and the sequence of the Pvmsp-1 gene varies between isolates from different part of the world. Pvmsp-3α, Pvmsp-3β and Pvmsp-3γ are members of a multi-gene family of related MSPs [8, 15]. The three encoded proteins share only 35–38% identity and 48–53% similarity in pair-wise comparisons. They all contain similar structures including signal sequences and are expressed on the merozoite surface. The MSP-3α of P. vivax is a protein with a molecular weight ranging from 148 to 150 KD, an alanine-rich central domain and a series of heptad repeats that were predicted to form a coiled-coil tertiary peptide structure. Pvmsp-3α, like Pvmsp-1, is very polymorphic and has been used as a genetic marker in population studies of isolates from diverse geographic localities and origins [13, 14, 18].
In Iran, malaria is endemic in southeastern regions, including Hormozgan, Sistan and Baluchistan and the tropical part of Kerman provinces. In addition, malaria re-appeared in northern Iran in 1994 [19]. More than 80% of reported malaria cases in the south are due to P. vivax infection. Recent study on the 52 northern and 94 southern P. vivax parasite isolates carried out to determine the extent of genetic diversity and population structure of this parasite by using block 5 region of Pvmsp-1 gene [20]. A total of 7 and 27 distinct variants were detected among northern and southern isolates, respectively. Sequence alignments demonstrated the heterogeneity of P. vivax isolates in Iran and also showed that the parasites from the southern malaria endemic area were more polymorphic than those circulating in the northern area.
In this study, msp-3α was compared and assessed with the msp-1 marker in order to find out whether msp-3α is a reliable genetic marker for P. vivax population studies. Therefore, the same set of sequenced sample was examined here using the Pvmsp-3α gene by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Materials and methods
Study area and sample collection
P. vivax isolates were collected from two different malaria endemic areas as previously described [20]. This study was approved by the Ethical Review Committee of Research in Pasteur Institute of Iran. In this comparative study 50 northern and 94 southern isolates, which had been already sequenced for variable block 5 of Pvmsp-1, were selected for further analysis using Pvmsp-3α marker [20].
Extraction of DNA
P. vivax genomic DNA was extracted from 250 μl of infected venous blood, using standard phenol/phenol-chloroform methods, following washing of cells, saponin lysis of red blood cells and digestion with proteinase K [21]. The DNA sample was resuspended in 30–50 μl of sterile TE buffer, and stored at -20°C until use.
Polymerase chain reaction amplification of the Pvmsp-3α gene
Allelic diversity of the msp-3α gene was studied using the modified PCR-RFLP method described by Bruce and colleagues [13]. Briefly, the msp-3α gene was amplified by a nested PCR, using 2 μl of DNA extract in first reaction and 0.1 μl of the primary reaction in the second amplification. The oligonucleotide PCR primers were as described by Bruce and co-workers [13]. One unit of Taq polymerase, with 0.2 μM final concentration of both primers and 0.2 mM of each 4-deoxynucleotide triphosphates were used in reaction buffer containing 1.5 mM MgCl2. The reaction was carried out for 94°C for 3 min, 35 cycles at 94°C for 1 min, 56°C for 1 min, and 72°C for 2.5 min and a final primer extension at 72°C for 10 min. However, in second round reaction, 95°C for 2 min, 30 cycle at 94°C for 1 min, 62°C for 1 min, 72°C for 1.5 min and a final primer extension at 72°C. Two μl of the second PCR product was electrophoresed on a 1.5% agarose gel.
PCR-RFLP analysis of Pvmsp-3α gene
For RFLP analysis, both Hha I and Alu I were used for digestion [13]. Approximately 10 μl of the PCR product was digested individually with the both restriction enzymes in total 20 μl reaction volumes as described by manufacture (Roche Company). The DNA fragments were visualized under UV illumination after electrophoresis on 2% agarose gels. Major alleles were classified based on the differences in restriction banding patterns.
Results
PCR amplification of the Pvmsp-3α gene
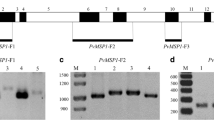
Fifty northern and 94 P. vivax southern isolates were successfully amplified for the msp-3α gene. A total of 99 distinct Pvmsp-3α fragments defined by the size were detected in the 94 southern samples by PCR analysis, showing that some patients (five of 94) had infections of more than one parasite genotype. No mixed genotype infections have been detected among northern isolates.
Based on the length variants of the PCR products, three allele sizes, Type A (1.8 kb), Type B (1.5 kb) and Type C (1.2 kb) have been detected among both northern and southern isolates (Figure 1). The predominant fragments among northern and southern parasites were Type C (70%) and Type A (68.7%) respectively. However, the remaining of 24% (12 of 50) of northern isolates was Type A, and 6% (3 of 50) were Type B. In addition, 7% (7/99) of southern samples were Type B, and 19% (18/99) were Type C (Figure 2).
PCR-RFLP analysis of Pvmsp-3α gene
Both Hha I and Alu I enzymes showed clear restriction patterns in all analyzed samples. In all northern and southern isolates the RFLP patterns showed 500–600 bp for Alu I digests (Figure 3A) and the largest fragments between 950–1100 bp for Hha I (Figure 3B), as showed by others [13, 22, 23]. These fragments were not included for distinguishing different alleles, as it was difficult to resolve, however, smaller fragments from 150–750 bp were applied for RFLP analysis in this study (Figure 3).
Based on restriction patterns from digestion of PCR products with Hha I and Alu I, 12 and 49 distinct variants have been detected among 50 northern and 94 southern isolates.
As we have shown before, based on msp-1 analysis, 30 distinct variants identified in all 146-sequenced Iranian P. vivax isolate. These variants were grouped in; Type 1 (B1–B9), Type 2 (S1–S16) and Type 3 (R1–R5) [20]. However, by using msp-3α gene, followed by RFLP analysis with Hha I and Alu I enzymes, more variation between and within different Type and sub Types of Iranian Pvmsp-1 were detected. Based on Pvmap-3α, the most diverse group of Pvmsp-1 was Type 2 S1 variants with 10 allelic forms (Table 1). Furthermore, the number of different allelic variants of Pvmsp-3α in Type 1 (B1–B9), Type 2 (S1–S16), and Type 3 (R1–R5) of Pvmsp-1 has been shown in Table 1. The two samples belonging to Type 3 R2 were not included in this analysis because there were no more blood and DNA samples available at the time of this study. The RFLP results showed high diversity among southern isolates in comparispn to northern isolates that was similar to the results obtained previously [20].
Discussion
The majority of researchers have used msp-1 gene as molecular markers for structural analysis of wild P. vivax isolates from different malaria endemic areas, which relies almost entirely on sequencing. However, the Pvmsp-3α gene has been recently applied and suggested as a polymorphic marker for molecular epidemiological studies [13, 14]. This survey has been carried out to evaluate the Pvmsp-3α marker for molecular epidemiology studies of P. vivax. Pvmsp-3α was applied to the same DNA samples of P. vivax isolates from Iran that had been previously analysed with the Pvmsp-1 marker. The analysis of Pvmsp-1 gene in Iranian isolates had been carried out by sequencing analysis while the Pvmsp-3α gene was only analysed using the PCR-RFLP method.
The PCR-RFLP analysis of the msp-3α gene demonstrated that P. vivax parasites were highly diverse in Iran. Furthermore, the parasite population was more diverse among southern isolates compared to northern isolates, which was in concordance with the result of the Pvmsp-1 sequencing analysis. In addition, sequencing analysis of the Pvmsp-1 gene showed 9 different allelic variant of Type 1 (B1–B9), 16 allelic variant of Type 2 (S1–S16) and 5 allelic variants of Type 3 (R1–R5) in 146 isolates. Within the same type and allelic variant group, all samples showed 100% similarity at both nucleotides and protein levels [20]. However, in the present study using a Pvmsp-3α analysis of the same samples, the results showed greater variation within each group. Therefore, these results support that this genetic marker may be more useful and sensitive than the msp-1 marker, without the need for further sequencing analysis.
Furthermore, in the previous msp-1 study, [20] none of the samples showed any multiple infections, while 5 of 144 isolates showed multiple infections by using the msp-3α marker.
Conclusion
The results demonstrated that the Pvmsp-3α is a powerful polymorphic marker, which could be applied for both genotyping, identification of mixed genotype parasite infections, and population structural analysis of P. vivax isolates. It is of particular importance to note that the msp-3α marker could be used with PCR-RFLP alone, without the need for further sequencing.
References
Read A, Day KP: The genetic structure of malaria parasite populations. Parasitol Today. 1992, 8: 239-242. 10.1016/0169-4758(92)90125-L.
Snewin VA, Khouri E, Wattavidanage J, Perera L, Premawansa S, Mendis KN, David PH: A new polymorphic marker for PCR typing of Plasmodium vivax parasites. Mol Biochem Parasitol. 1995, 71: 135-138. 10.1016/0166-6851(94)00040-T.
Snewin VA, Premawansa S, Udagama PV, Mattei DM, Khouri E, Del Giudice G, Mendis KN, David PH: Transmission blocking immunity in Plasmodium vivax malaria: Antibodies raised against a peptide block parasite development in the mosquito vector. J Exp Med. 1995, 181: 357-362. 10.1084/jem.181.1.357.
Arnot DE, Stewart MJ, Barnwell JW: Antigenic diversity in Thai Plasmodium vivax circumsporozoite proteins. Mol Biochem Parasitol. 1990, 43: 147-150. 10.1016/0166-6851(90)90140-H.
Mann VH, Huang T, Cheng Q, Saul AJ: Sequence variation in the circumsporozoite protein gene of Plasmodium vivax appears to be regionally biased. Mol Biochem Parasitol. 1994, 68: 45-52. 10.1016/0166-6851(94)00148-0.
Qari SH, Collins WE, Lobel HO, Taylor F, Lal AA: A study of Polymorphism in the circumsporozoite protein of human malaria parasites. Am J Trop Med Hyg. 1994, 50: 45-51.
Cheng Q, Stowers A, Huang TY, Bustos D, Huang YM, Rzepczyk C, Saul A: Polymorphism in Plasmodium vivax MSA-1 gene: The result of intragenic recombinations?. Parasitology. 1993, 106: 335-345. 10.1017/S003118200006707X.
Del Portillo HA, Longacre S, Khouri E, David PH: Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci USA. 1991, 88: 4030-4034. 10.1073/pnas.88.9.4030.
Pasay MCJ, Cheng Q, Rzepczyk CM, Saul AJ: Dimorphism of the C terminus of the Plasmodium vivax merozoite surface protein 1. Mol Biochem Parasitol. 1995, 70: 217-219. 10.1016/0166-6851(95)00015-S.
Premawansa S, Snewin VA, Khouri E, Mendis KN, David PH: Plasmodium vivax: recombination between potential allelic types of the merozoite surface protein MSP-1 in parasites isolated from patients. Exp Parasitol. 1993, 76: 192-199. 10.1006/expr.1993.1022.
Putaporntip C, Jongwutiwes S, Tanabe K, Thaithong S: Interallelic recombination in the merozoite surface protein (MSP-1) gene of Plasmodium vivax from Thai isolates. Mol Biochem Parasitol. 1997, 84: 49-56. 10.1016/S0166-6851(96)02786-7.
Galinski MR, Corredor-Medina C, Povoa M, Crosby J, Ingravallo P, Barnwell JW: Plasmodium vivax merozoite surface protein-3 contains coiled-coil motifs in an alanine-rich central domain. Mol Biochem Parasitol. 1999, 101: 131-147. 10.1016/S0166-6851(99)00063-8.
Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP: Polymorphism at the merozoite surface protein-3α locus of Plasmodium vivax. Am J Trop Med Hyg. 1999, 61: 518-525.
Bruce MC, Galinski MR, Barnwell JW, Donnelly CA, Walmsley M, Alpers MP, Walliker D, Day KP: Genetic diversity and dynamics of Plasmodium falciparum and Plasmodium vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology. 2000, 121: 275-272.
Galinski MR, Ingravallo P, Corredor-Medina C, Al-Khedery B, Povoa M, Barnwell JW: Plasmodium vivax merozoite surface proteins-3β and 3γ share structural similarities with P. vivax merozoite surface protein-3α and define a new gene family. Mol Biochem Parasitol. 2001, 115: 41-53. 10.1016/S0166-6851(01)00267-5.
Black CG, Barnwell JW, Huber CS, Galinski MR, Coppel RL: The Plasmodium vivax homologues of merozoite surface proteins 4 and 5 from Plasmodium falciparum are expressed at different locations in the merozoite. Mol Biochem Parasitol. 2002, 120: 215-224. 10.1016/S0166-6851(01)00458-3.
Vargas-Serrato E, Barnwell JW, Ingravallo P, Perler FB, Galinski MR: Merozoite surface protein-9 of Plasmodium vivax and related simian malaria parasites is orthologous to p101/ABRA of P. falciparum. Mol Biochem Parasitol. 2002, 120: 41-52. 10.1016/S0166-6851(01)00433-9.
Mueller I, Kaiok J, Reeder JC, Cortes A: The population structure of Plasmodium falciparum and Plasmodium vivax during an epidemic of malaria in the Eastern Highlands of Papua New Guinea. Am J Trop Med Hyg. 2002, 67 (5): 459-464.
Zakeri s, Mamaghani S, Mehrizi AA, Shahsavari Z, Raeisi A, Arshi S, Djadid ND: Molecular evidence of mixed P. vivax and P. falciparum infections in northern Islamic Republic of Iran. East Med Health J. 2004, 10: 336-342.
Zakeri S, Mehrizi AA, Mamaghani S, Noorizadeh S, Snounou G, Djadid ND: Population structure analysis of Plasmodium vivax in areas of Iran with different malaria endemicity. Am J Trop Med Hyg. 2006, 74: 394-400.
Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario WE, Thaithong S, Brown KN: High sensitivity of detection of human malaria parasites by use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993, 61: 315-320. 10.1016/0166-6851(93)90077-B.
Rayner JC, Corredor V, Feldman D, Ingravallo P, Iderabdullah F, Galinski MR, Barnwell JW: Extensive polymorphism in the Plasmodium vivax merozoite surface coat protein MSP-3α is limited to specific domains. Parasitology. 2002, 125: 393-405. 10.1017/S0031182002002317.
Cui L, Mascorro CN, Fan Q, Rzomp KA, Khuntirat B, Zhou G, Chen H, Yan G, Sattabongkot J: Genetic diversity and multiple infections of Plasmodium vivax malaria in western Thailand. Am J Trop Med Hyg. 2003, 68: 613-619.
Acknowledgements
This study was financially supported by Institut Pasteur of Iran.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
S. Zakeri designed the study and was responsible for both supervision of laboratory work, development of the protocols, analysis of the data and writing up the paper. H. Barjesteh carried out the laboratory work. N. D. Djadid helped with data analysis and editing the manuscript. All authors read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zakeri, S., Barjesteh, H. & Djadid, N.D. Merozoite surface protein-3α is a reliable marker for population genetic analysis of Plasmodium vivax. Malar J 5, 53 (2006). https://doi.org/10.1186/1475-2875-5-53
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-5-53