Abstract
Background
A study was carried out in a village of the mainland region of Equatorial Guinea in order to ascertain a) which members of Anopheles gambiae complex could be involved in malaria transmission and b) the rate of infectivity for Anopheles melas comparing two different methods, a PCR able to detect sporozoite-DNA and an immunochromatographic assay MPR (Malaria Rapid Dipstick Panel Assay).
Methods
Mosquitoes were sampled at night by indoor captures in two houses of a coastal village in Equatorial Guinea (Ayantang). Collected mosquitoes were identified as An. gambiae s.l. These were individually dried into silica-gel. The head-thorax of the An. gambiae s.l. mosquitoes were analysed by PCR to verify that the species was of the gambiae complex. Individual head-thorax and pools (5 pools) of homogenized mosquitoes employed in Malaria Rapid Panel assay (MRP assay) were lysed and DNA was extracted. PCR was designed from the 753 base pair insert of pBRKl-14 and DNA was amplified. The relationship between dipstick and PCR to detect Plasmodium falciparum sporozoites was measured in terms of sensitivity, specificity and test association (Cohen's kappa value).
Results
Two hundred and sixty-four An. gambiae s.l. females were studied (214 individually and five pools with 10 mosquitoes in each). PCR analysis showed that 207 mosquitoes were An. melas, 3 An. gambiae s.s. and 4 could not be identified. By using PCR as the gold standard method when dipstick assay was compared, matching results were obtained for 6 mosquitoes and, in one case MRP was positive while PCR was not reactive. MRP assay showed a low sensitivity (3.3%) when compared with falciparum-DNA detection (17,7% and 14,3%, series A and B respectively). Agreement between the two test formats was low (κ = 0,224).
Conclusion
It was determined that An. melas is the main anopheline vector involved in malaria transmission in Ayantang, a coastal village in mainland Equatorial Guinea. A comparison of PCR and Vec-Test Assay®, concluded that the PCR method proved to be a more sensitive and useful tool than the dipstick assay to determine the malarial infection rate in mosquitoes in an area of stable and high malaria transmission like Equatorial Guinea.
Similar content being viewed by others
Background
Anopheles gambiae s.l. and Anopheles funestus are considered the main malaria vectors in the African-equatorial region [1, 2]. Both species are also involved in malaria transmission in Equatorial Guinea [3]. Malaria is hyperendemic throughout Equatorial Guinea and one of the three main causes of morbidity and mortality [4]. Previous taxonomic surveys carried out on Equatorial Guinea (Bioko, 3° 43' N, 8° 43' E; and Bata, mainland region, 1° 51' N, 9° 46' E) revealed the presence of five anopheline species: An. gambiae s.l., An. funestus, Anopheles lloreti, Anopheles cinctus and Anopheles smithi [5, 6]. The An. gambiae complex in Africa includes two species that breed in salt water (Anopheles melas in West Africa and Anopheles merus in East Africa) and three species which breed in freshwater (An gambiae, Anopheles arabiensis and An. quadriannulatus). A sixth member, Anopheles bwambae was described in the Semliki forest of Uganda and associated with water with a high mineral content [7].
On the other hand, to establish efficient control campaign, it is important to take into account local parameters of malaria epidemiology (sporozoite rates, entomological inoculation rate, parasitological parameters, etc) [8].
Traditionally, sporozoite rate in mosquitoes was assessed by dissection of the salivary glands of individual mosquitoes, but this is laborious [9]. In recent years different sporozoite detection methods have been used: serological (e.g. ELISA or immunochromatographic assay) [8, 10, 11] and molecular techniques (e.g. PCR and RAPD) [12].
Previously, a rapid method was used to determine Plasmodium in anophelines, with no need for special storage conditions. The malaria rapid panel (MRP) dipstick uses equivalent species-specific Mabs (monoclonal antibodies) as the enzyme-linked immunosorvent assay (ELISA) to detect parasite circumsporozoite protein (CSP) [13, 14].
In recent years, the polymerase chain reaction (PCR) has been used to amplify specific DNA sequences of P. falciparum for highly sensitive detection of the parasites in mosquitoes. It employs a set of primers derived from repetitive DNA sequences [12]. The PCR procedure can detect as few as 10 sporozoites in salivary glands (0,2 pg of parasite DNA) and is, therefore, an useful tool for screening small numbers of anophelines [15].
The objectives of this study were: a) to determine the anopheline species involved in malaria transmission, in a coastal village of mainland Equatorial Guinea and b) to assess the sensitivity and specificity of a MRP immunochromatographic assay in parallel with a PCR detection of P. falciparum in wild anophelines collected in the same village.
Material and Methods
Study area and station of capture
Equatorial Guinea is a small country located in the western coast of Central Africa, in the Gulf of Guinea (N 1° – 3° 47' E 5° – 11° 30'). Its overall area of 28.068 km2 is divided into an island and a continental region. The Bioko island (in the Gulf of Guinea), Annobón (south of the Republic of Sao Tomé and Príncipe) and some smaller islands closer to the coast (Corisco, Elobey Grande and Elobey Chico) represent the island regions, with a total surface of 2.051 km2. The mainland region, called Rio Muni, is located between Gabon and Cameroon, giving the country a total area of 26.017 km2.
Samples were taken during November 2001 and March–April 2002, in the coastal village of Ayantang (N 01° 43.927' E 009° 40.917'), 20 km from Bata, capital of the mainland region (see Figure 1). An. gambiae s.l. was the main vector collected and An. moucheti moucheti plays a minor role as malaria vector in this village. All anopheline vectors from An. gambiae s.l. were included in this study.
Mosquito collection
The samples were caught at night (19:00 to 6:00) in two houses of the village, by using CDC light-traps. People living in these houses were protected with non-impregnated bed-nets. Traps were attended daily and the collected mosquitoes were kept at ambient temperature, fed a 5% glucose solution on cotton wool and transported to the laboratory. Collected mosquitoes were identified using the key described by Hervy et al. [16] and then individually dried on silica-gel.
Differentiation of gambiae complex by PCR
The head-thorax of the An. gambiae s.l. mosquitoes were analysed by PCR to identify the species within the gambiae complex. PCR was carried out following a slightly modified version (in the "master mix" and the times of the amplification programme) of the protocol described by Scott et al. [17]. DNA of An. melas, An. gambiae s.s., An arabiensis, An quadriannulatus were used as positive controls. Sterilized water in the master mix was used for negative controls.
MRP Dipstick Assay
Two lots of Vec-Test™ kits were used; Serie A (Lot n° 10001) and Serie B (Lot n° 070.102) containing 25 test strips of the Plasmodium CSP panel, supplied by the manufacturer (Medical Analysis Systems, Camarillo, CA, USA). Monoclonal antibodies against the CSP of P. falciparum (2A10 produced by New York University), P. vivax variant 210 (NVS3 produced by the U.S. Naval Medical Research Center) and P. vivax variant 247 (2E10 produced by New York University) were incorporated into each MRP dipstick. The Mabs were immobilized in the test zone of the dipstick and combined with a gold sol particle label [18] to detect the species-specific proteins for P. falciparum, and the two Plasmodium vivax polymorphs.
Two readers examined dipsticks 15 minutes after immersion into the ground mosquito suspension. A test was interpreted as positive when at least two lines appeared on the strip and as negative if only the control line was present. Strips were also examined one month after testing to note retention of bands [14].
Detection of P. falciparum by PCR: preparation of samples, and DNA amplification
Homogenized samples of individual head-thorax and pools (5 pools) employed in MRP assay were lysed and DNA extracted by using the Bender Lysis Buffer method. PCR was designed from the 753 base pair insert of pBRKl-14 reported by Fucharoen et al. [12]. DNA amplification was performed following the protocol reported by Tassanakajon et al. 1993 [15]. Falciparum DNA used as positive control was obtained from strain 3D7 P. falciparum culture in vitro in the National Centre of Microbiology in Madrid. Three different DNA extractions were carried out from each mosquito sample (head-thorax). A mosquito sample was determined positive when the 206 bp fragment appeared in the three repeated process of amplification (three per head-thorax sample). The size of the amplified fragments was measured by MultiAnalyst Software – Gel Doc 1000 (BioRad®) (SE = ±15–20 bp) from 2% agarose gel electrophoresis stained with ethidium bromide. Sizes (base pairs) were calculated in accordance with their mobility relative to molecular size standards run on the gel.
Statistical analysis
The PCR was considered the reference method of choice or "gold standard" to determine reliability and test predictive value for the detection of native CSP from wild-caught anopheline mosquitoes. The relationships between dipstick and presence of infected mosquitoes was measured in terms of sensitivity (the proportion of mosquitoes that were PCR sporozoite-reactive with a positive dipstick), specificity (the proportion of mosquitoes that were PCR, sporozoite non-reactive with a negative dipstick) and a measure of test association (Cohen's kappa value). This is the probability of sporozoite infection given the dipstick results and provided the predictive value or discriminating power for both, positive and negative tests respectively [19].
Results
Differentiation of Anopheline mosquitoes
A total of two hundred and sixty-four female anophelines were studied, identified by entomological key as An. gambiae s.l., 214 individually and five pools with 10 mosquitoes each.
Analysis by PCR showed that 207 mosquitoes were An. melas, 3 An. gambiae s.s. and 4 could not be identified (see Figure 2).
Falciparum-infection rate in mosquitoes
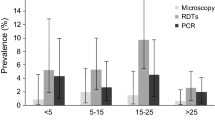
In both series of MRP dipstick assay, P. vivax CS antigen was not detected in any mosquitoes studied individually, confirming the expected results in the central-west African area (indigenous population without the Duffy receptor necessary for P. vivax invasion) and P. falciparum was detected in seven mosquitoes, giving a sporozoite prevalence of 3.27% by this diagnostic method. The PCR technique detected 38 falciparum infective mosquitoes (see Figure 3), a sporozoite prevalence of 17,75% (see Table 1). P. falciparum was only detected in An. melas.
Sensitivity and Specificity
PCR and dipstick assays showed concordant results for 6 mosquitoes (true positives) and, in one case, MRP was positive while PCR resulted not reactive (false positive). However, thirty-two mosquitoes were PCR positive and MRP negative (false negatives) (see Table 2). Taking into account that the PCR falciparum-diagnostic was considered the reference method of choice, the MRP assay presented a low sensitivity for falciparum-sporozoites detection (17,7% and 14,3%, series A and B respectively). On the other hand, the specificities were 100% and 99%, owing to the high number of true negative test strips.
The predictive positive value of the dipstick in both tests series was high (100% and 75,1%, series A and B respectively), because of a low number of false positive strips. Likewise, the predictive negative value of the dipstick was high (85,4% and 83,9%), due to the high number of true negative strips in spite of a relative high number of false negatives in both series (see Table 2). The agreement between the two test formats was low (κ = 0,224).
From the five anopheline pools tested with MRP dipstick assay, three positives were clearly observed. No mosquito pools resulted positive repeatedly in confirmatory testing using PCR, despite having made numerous dilutions of DNA extracted from homogenized pools.
Discussion and Conclusions
The first objective was to know which anopheline species could be involved in malaria transmission in the mainland coastal region of Equatorial Guinea inside of the activities of the National Malaria Control Programme. Although An. melas species has been described in Equatorial Guinea (Bioko island), data about its implication on malaria transmission in the mainland region had not been demonstrated. An. melas was identified as the major anopheline species collected in the village studied by using molecular markers and PCR technology.
The second objective was to evaluate the vectorial capacity of An. melas in malaria transmission by using two methods, VecTest™ and PCR.
Akogbeto et al. studied An. melas infectivity rates for P. falciparum compared to the rates of An. gambiae s.s., but low rates of infection were reported for An melas as compared with An. gambiae s.s. [20]. In these reports, the authors indicate that in areas where An. melas is the most abundant species (80–90%), the sporozoitic index and antigenCSP-falciparum rate are significantly lower for this species. This was not corroborated in the present study because similar rates of infections were found for An melas as compared with the infection rates reported for An gambiae s.s. in Equatorial Guinea [21] and other sub-Saharian countries, where both vectors cohabit.
The studies performed to evaluate the malaria sporozoite antigen panel assay (MRP dipstick assay or VecTest™) for detecting naturally infected mosquitoes show that it is an optimal diagnostic method for use in malaria endemic areas [13, 14]. These surveys compared MRP dipstick assay with another serological diagnostic probe (ELISA assay) for detecting CSP antigen.
It is recognized that the number of sporozoites found in individual mosquitoes can vary immensely in a naturally infected vector population [22], but the MRP panel assay demonstrated excellent sensitivity for detecting low levels of sporozoite density (200–400 sporozoites) [23]. On the other hand, soluble CSP can be found freely in the salivary gland and haemolymph as a result of sporozoite liberation from mature oocyst in the insect haemocoel [23, 24]. As a result, both, sporozoites and CS antigen, can be detected in mosquito thorax and other body parts, although sporozoites are not present in the salivary glands [22].
False positive results are expected in individual tests by using serological tests such as MRP assay or ELISA and, hence, an overestimate of infection rates. Low density infection would not be detected, thus underestimating the true sporozoite rate [23]. The MRP panel assay was compared with a molecular diagnostic test able to detect P. falciparum sporozoite DNA in homogenized samples of individual head-thorax. Amplified sporozoite-DNA by PCR had an excellent sensitivity for the detection of a low level of sporozoite density, useful for estimating entomological inoculation rates [21].
Although specificity of the VecTest™ is high, sensitivity of the dipstick was low when compared with PCR (17.7% and 14.3%, respectively in each series). In other studies, where VecTest™ has been compared with other immunological assays, sensitivities varied 88% and 100% [14, 23]. This fact could be attributed to the high sensitivity of PCR, able, as it is, to detect as few as ten sporozoites in salivary glands (0,2 pg/DNA parasite) [15], as compared with CS antigen detection probes for which the minimum detection rate was 200–400 sporozoites [23]. This difference suggests that the sporozoite index is conditioned by the kind of assay employed. In the present study, the sporozoite rate would be 3.27% when determined by MRP assay and 16% when determined by PCR.
Therefore, this kind of immunological assay could underestimate the infectivity rate when working with individual mosquitoes and may be improved by using pools [9, 10, 13, 14, 23]. In contrast, the PCR assay was unable to amplify P. falciparum DNA from pools, even when carrying out successive dilutions. The presence of PCR inhibitors in large pools of mosquitoes has been reported in several studies [25]. Optimazing pool sizes before PCR may solve this, but this is expensive and time-consuming.
Comparing PCR and Vec-Test Assay®, it was concluded that: a) the PCR method proved to be a sensitive and useful tool to determine malarial infection rates in mosquitoes in an area of stable and high malarial transmission (Equatorial Guinea) and b) although other authors have described An. melas in Bioko Island [21, 26], this survey suggests that An. melas could be playing the main role in malaria transmission in coastal areas of the mainland region of Equatorial Guinea.
References
Hamon J, Adam JP, Grjebine A: Observations sur le répartition et le comportement des anophèles de 1'Afrique Equatorial française, du Cameroun et de 1'Afrique occidentale. Bull World Health Organ. 1956, 15: 541-591.
White GB: Anopheles gambiae complex and disease transmission in Africa. Trans R Soc Trop Med Hyg. 1974, 68: 278-301. 10.1016/0035-9203(74)90035-2.
Molina R, Benito A, Roche J, Blanca F, Amela C, Sánchez A, Alvar J: Baseline entomological data for a pilot malaria control programme in Equatorial Guinea. J Med Entomol. 1993, 30: 622-624.
Roche J, Ayecaba S, Amela C, Alvar J, Benito A: Epidemiological characteristics of malaria in Equatorial Guinea. Res Rev Parasitol. 1996, 56: 99-104.
Gil-Collado J: Culícidos de la isla de Fernando Poo recogidos por la expedición J. Gil-F. Bonet. Revista Espanola de Entomologia. 1936, 11: 311-329.
Gil-Collado J: Los anofelinos de Fernando Poo. La Medicina Colonial. 1953, 2: 1-14.
White GB: Comparative studies on sibling species of Anopheles gambiae Giles complex (Dipt. Culicidae). III. The distribution, ecology, behaviour and vectorial importance of species D in Bwamba County, Uganda, with analysis of biological, ecological, morphological and cytogenetical relationships of Ugandan species D. Bull Entomol Resh. 1973, 63: 65-97.
Wirtz RA, Burkot TR: Detection of malarial parasites in mosquitoes. In Advances in disease vector research. Edited by: Harris KF. 1995, New York: Springer, 8: 77-106.
World Health Organization: Manual on practical entomology in malaria, part II. 1975, Offset Publication 13. WHO, Geneva
Wirtz RA, Burkot TR, Andre RG, Rosenberg R, Collins WE, Roberts DR: Identification of Plasmodium vivax sporozoites in mosquitoes using an enzyme-linked inmunosorbent assay. Am J Trop Med Hyg. 1985, 34: 1048-1054.
Wirtz RA, Sattabongkot J, Hall T, Burkot TR, Rosenberg R: Development and evaluation of an ELISA for Plasmodium vivax-VK247 sporozoites. J Med Entomol. 1992, 29: 854-857.
Fucharoen D, Tirawanchai N, Wilairat P, Panyim S, Thaithong S: Differentiation of Plasmodium falciparum clones by means of a repetitive DNA probe. Trans R Soc Trop Med Hyg. 1988, 82: 209-211. 10.1016/0035-9203(88)90413-0.
Ryan JR, Dave K, Emmerich E, Garcia L, Yi L, Coleman RE, Sattabongkot J, Dunton RF, Chan AST, Wirtz RA: Dipsticks for rapid detection of Plasmodium in vectoring Anopheles mosquitoes. Med Vet Entomol. 2001, 15: 225-230. 10.1046/j.1365-2915.2001.00296.x.
Bangs MJ, Rusmiarto S, Gionar YR, Chan AST, Dave K, Ryan JR: Evaluation of a dipstick malaria sporozoites panel assay for detection of naturally infected mosquitoes. J Med Entomol. 2002, 39: 324-330.
Tassanakajon A, Boonsaeng V, Wilairat P, Panyim S: Polymerase chain reaction detection of Plasmodium falciparum in mosquitoes. Trans R Soc Trop Med Hyg. 1993, 87: 273-275. 10.1016/0035-9203(93)90124-9.
Hervy JP, Le Goff G, Geoffroy JP, Hervé L, Manga L, Brunhes J: Les anophèles de la règion Afro-tropicale. Logiciel d'identification et d'enseignement. ORSTOM édition, série Didactiques. Paris, France (in French, English, Portuguese). 1998
Scott JA, Williams G, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993, 49: 520-529.
Hermanson GT: Preparation of colloidal-gold-labeled proteins. In Bioconjugate techniques. 1996, New York: Academic
Norman GR, Streiner DL: Biostatistics. 1994, Mosby, St. Louis, MO
Akogbeto M, Romano R: Infectivity of Anopheles melas vis-a-vis Plasmodium falciparum in the coastal lagoon area of Benin. Bull Soc Pathol Exot. 1999, 92: 57-61.
Cano J, Berzosa PJ, Roche J, Rubio JM, Moyano E, Guerra-Neira A, Brochero H, Mico M, Edu M, Benito A: Malaria vectors in Bioko Island (Equatorial Guinea): estimation of vector dynamics and transmission intensities. J Med Entomol. 2004, 41: 158-162.
Beier JC, Koros JK: Anatomical dissemination of circumsporozoite protein in wild Afrotropical Anopheles affects malaria sporozoites determinations by ELISA. Med Vet Entomol. 1991, 5: 81-85.
Ryan JR, Dave K, Collins KM, Hochberg L, Sattabongkot J, Coleman RE, Dunton RF, Bangs MJ, Mbogo CM, Cooper RD, Schoeler GB, Rubio-Palis Y, Magris M, Romero LI, Padilla N, Quakyi IA, Bigoga J, Leke RG, Akinpelu O, Evans B, Walsey M, Patterson P, Wirtz RA, Chan AST: Extensive multiple test centre evaluation of the VecTest™ malaria antigen assay. Med Vet Entomol. 2002, 16: 321-327. 10.1046/j.1365-2915.2002.00368.x.
Verhave JP, Leeuwenberg ADEM, Ponnudurai T, Meuwissen JHET, Van Druten JAM: The biotin-streptavadin system in a two-site ELISA for the detection of plasmodial sporozoite antigen in mosquitoes. Parasite Immunol. 1988, 10: 17-31.
Local transmission of Plasmodium vivax Malaria – Virginia, 2002. Morb Mortal Wkly Rep. 2002, 51: 921-923.
Berzosa PJ, Cano J, Roche J, Rubio JM, García L, Moyano E, Guerra A, Mateos JC, Petrarca V, do Rosario V, Benito A: Malaria vectors in Bioko Island (Equatorial Guinea): PCR determination of the members of Anopheles gambiae Giles complex (Diptera: Culicidae) and pyrethroid knockdown resistance (kdr) in An. gambiae sensu stricto. J Vect Ecology. 2002, 270: 102-106.
Acknowledgments
This work was supported by the Spanish Agency of International Cooperation (AECI), the Institute of Health Carlos III and by the Research Network of Centres for Tropical Medicine (RICET), supported by FIS. Nick Service made comments on the manuscript and helped with the language revision.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
MM carried out the molecular and immunological studies and she also collabored in the collection of anopheline; JC was the coordinator of the Project in Equatorial Guinea and participated on the design of the study and also participated on capture of mosquitoes; SN, LB, JNB, MO and FM were the entomologists from Malaria National Control Programme and the National Centre for Tropical Medicine which participated in the field study; AB designed the study and participated in its implementation and coordination. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Moreno, M., Cano, J., Nzambo, S. et al. Malaria Panel Assay versus PCR: detection of naturally infected Anopheles melas in a coastal village of Equatorial Guinea. Malar J 3, 20 (2004). https://doi.org/10.1186/1475-2875-3-20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-3-20