Abstract
Background
Within affected communities, Plasmodium falciparum infections may be skewed in distribution such that single or small clusters of households consistently harbour a disproportionate number of infected individuals throughout the year. Identifying these hotspots of malaria transmission would permit targeting of interventions and a more rapid reduction in malaria burden across the whole community. This study set out to compare different statistical methods of hotspot detection (SaTScan, kernel smoothing, weighted local prevalence) using different indicators (PCR positivity, AMA-1 and MSP-1 antibodies) for prediction of infection the following year.
Methods
Two full surveys of four villages in Mwanza, Tanzania were completed over consecutive years, 2010-2011. In both surveys, infection was assessed using nested polymerase chain reaction (nPCR). In addition in 2010, serologic markers (AMA-1 and MSP-119 antibodies) of exposure were assessed. Baseline clustering of infection and serological markers were assessed using three geospatial methods: spatial scan statistics, kernel analysis and weighted local prevalence analysis. Methods were compared in their ability to predict infection in the second year of the study using random effects logistic regression models, and comparisons of the area under the receiver operating curve (AUC) for each model. Sensitivity analysis was conducted to explore the effect of varying radius size for the kernel and weighted local prevalence methods and maximum population size for the spatial scan statistic.
Results
Guided by AUC values, the kernel method and spatial scan statistics appeared to be more predictive of infection in the following year. Hotspots of PCR-detected infection and seropositivity to AMA-1 were predictive of subsequent infection. For the kernel method, a 1 km window was optimal. Similarly, allowing hotspots to contain up to 50% of the population was a better predictor of infection in the second year using spatial scan statistics than smaller maximum population sizes.
Conclusions
Clusters of AMA-1 seroprevalence or parasite prevalence that are predictive of infection a year later can be identified using geospatial models. Kernel smoothing using a 1 km window and spatial scan statistics both provided accurate prediction of future infection.
Similar content being viewed by others
Background
Malaria transmission in endemic countries is heterogeneous over multiple spatial scales [1, 2]. At the micro scale, P. falciparum infections are frequently clustered in relatively few households that consistently have significantly more infections than others [3, 4]. Many factors can contribute to this increased risk of malaria exposure, including design of housing, the proximity to mosquito breeding sites, host genetic factors, poor access to treatment, maternal education, wealth, and other as yet undefined characteristics [3, 5–8]. At sites with very low levels of transmission, such as those found in Swaziland, cases of symptomatic malaria detected at health facilities can help in identification of a hotspot, as additional asymptomatic cases can be found living in close proximity to the index case [9]. In areas of moderate transmission intensity, malaria hotspots may provide a reservoir of infected human hosts that can maintain some transmission year round. The individuals in such hotspots are thus likely to have acquired anti-parasite immunity and to carry parasites without clinical symptoms. In the wet season, when the mosquito population increases, these clusters of asymptomatic carriers may be responsible for seeding transmission to the rest of the community, including less immune people who are more likely to suffer symptomatic infections [7]. Thus in these settings, hotspots are difficult to identify using the distribution of clinical (symptomatic) malaria cases alone.
The most used geospatial method to detect clusters of infection is the spatial scan statistic [10–12]. Measures of exposure which have been explored using spatial scan statistics include prevalence of infection, incidence of clinical malaria and serological markers of malaria exposure [13–18]. While this approach allows identification of clusters using statistical hypothesis testing, it may ignore more subtle small-scale spatial heterogeneity and clusters that do not fit within circular or elliptical windows [19]. An alternative method that has been used to detect clustering of infection is distance-weighted prevalence of infection, whereby infection prevalence in neighbours is used as a proxy measure for household level exposure [20, 21]. This method allows for a smoother estimation of risk in space than spatial scan statistics.
This study seeks to determine which geospatial method best describes a malaria transmission hotspot by comparing methodologies using cross-sectional data collected during the first year of the study to predict the distribution of infections found in the second year.
Methods
Study site
Misungwi district (lat 2.85000 S, long 33.08333 E) is located 60 km from Mwanza town in the north-west of Tanzania at an altitude of 1,178 m above sea level (see Figure 1). The district is rural with moderately intense malaria transmission; the overall prevalence of infection in the region is estimated to be 31.4% by microscopy in children 6 -59 months (Tanzania HIV and Malaria Indicator Survey 2008). The district has two annual rainy seasons, the long rains between February and May, and the short rains between November and December. The dry and relatively hot season falls between June and September. Malaria incidence peaks one to two months after the rains start. The National Malaria Control Programme (NMCP) carried out indoor residual spraying (IRS) in the study area during the period from late November 2010 to late January 2011.
Data collection
A census of four villages in a single ward was carried out in the dry season, between August and early November 2010. All data were collected using personalized digital assistants and every household was visited and mapped using a global positioning system (GPS). All individuals in the ward were invited to participate in the study. The head of household gave information on the age, sex and insecticide-treated net (ITN) use of those who were not present. Individuals who consented to join the study were asked to provide a finger-prick sample of blood which was spotted onto Whatman® standard 3 mm filter paper for parasite detection and serological analysis. Subjects who reported having had fever within the previous 24 hours were tested for malaria using a histidine-rich protein 2 (HRP2) rapid malaria diagnostic test (RDT, Paracheck-Pf®, Orchid Biomedical Systems, Goa, India) and referred to a study clinician for management of their febrile illness.
A follow-up survey was carried out in the same study villages during August to November 2011, one year after the initial study. The same procedures were carried out during the second survey as during the baseline survey.
Molecular estimation of P. falciparum infection
DNA was extracted from filter papers using the Chelex® (Sigma, USA) extraction method described previously [22] in 96 deep-well plates. Parasite DNA was detected using nested PCR (nPCR) targeting the 18S rRNA gene as previously described [23].
Serology
Antibodies were eluted from filter paper spots and assayed for specific IgG responses to P.falciparum AMA-1 and MSP-119 by ELISA as described by Corran et al. [24]. Samples were tested in duplicate. Duplicate optical density (OD) values OD values that differed by more than 1.5-fold were rejected and, if possible, rerun. For each plate a standard curve was generated from a known positive control and blank wells were included and OD values normalised to these. To define seroprevalence a mixture model was applied to the OD data which assumed two inherent Gaussian distributions; a narrow distribution or sero-negatives and a broader distribution of seropositives. A cut-off was calculated as the mean plus 3 standard deviations of the narrow distribution and was calculated separately for each antigen [25].
Cluster analysis
While there are a range of different methodological approaches to identifying clusters of infection [12, 26], here we focus on three geospatial cluster detection methods to explore baseline clustering of infection and serological markers and their ability to predict infection in the second year of the study. The unit of analysis was the individual, meaning that clustering of infected individuals was assessed rather than clustering of households with infection. Infection in the second year was defined as a positive nPCR result recorded as a binary variable.
Satscan analysis
Spatial analysis was performed to assess possible clustering of nPCR-positive individuals. A spatial scan statistic was obtained using the Bernoulli model [11] and SaTScan software (SaTScan, version 8.2.1). This software applies multiple circular windows, which are plastic in both position and size, across the study area. Each distinct circle represents a possible cluster. For each circle, the number of observed and expected infected individuals are counted, with expected numbers calculated assuming an even distribution of infections across the population. As multiple infected and non-infected individuals can be specified at each household, the spatial distribution of households is accounted for. A likelihood ratio test is used to compare the prevalence of infection within the circle to that outside it to identify significant clusters of higher than expected (hotspot) or lower than expected (coldspot) prevalence. The statistical significance of this hotspot is evaluated taking into account the multiple tests for the many potential cluster locations and sizes evaluated as well as the distribution of the population [10]. The maximum proportion of the population that a cluster could contain was set at 50%. This method has been extensively explored in studies of the micro-epidemiology of malaria [12, 13, 27–29].
Households were grouped into three categories: 1) hotspots (clusters of significantly higher than expected malaria prevalence); 2) coldspots (clusters of significantly lower than expected malaria prevalence); and, 3) all other households. Clusters were defined using three measures: 1) nPCR positivity; 2) antibody seropositivity to AMA-1; 3) antibody sero-positivity to MSP-119; and, 4) antibody seropositivity to AMA-1and/or MSP-119. So as to make results from analyses using different clustering methods comparable, hotspots were assigned a score of 1, coldspots 0 and all remaining households a score of 0.5. Households for which data were only available in the second year were assigned a hotspot score according to whether the household lay within the radius of the hot or coldspot.
Kernel analysis
Kernel density estimation is a statistical procedure used to produce a smoothed estimate of density of events, such as individuals, across space [26]. For any given point, the density of events within a predefined window is estimated, with the influence of events weighted according to the distance from the centre of the window. The weight assigned to each event is derived from the kernel function applied. In this analysis a quadratic kernel function was used with an initial window radius of 1 km. A quadratic function allows importance of data from neighbouring households to be relative to the distance to the index household. To obtain a smoothed estimate of infection prevalence over the study region, a kernel density surface of numbers nPCR positive was divided by a kernel density surface of numbers examined. This resulted in each household having a value between 0 (least exposed households) and 1 (most exposed households). Households for which data were only available in the second year were assigned a prevalence value based on infection in neighbouring households only.
Weighted local prevalence analysis
This method calculates parasite prevalence amongst all neighbours within 1 km of the index house, weighting the prevalence estimate according to the inverse of the distance of the neighbouring house to the index house [20]. While a form of spatial smoothing, an important distinction between weighted local prevalence and kernel smoothing is that individuals in the index household are not included in the weighted prevalence estimate. As for kernel prevalence estimates, the weighted local prevalence for each household ranged from 0 (least exposed households) to 1 (most exposed households). As this method does not include infection status of individuals in the index household in the calculation of prevalence, no further action was required for those households with data from only the second year.
Statistical analysis
To compare the ability of different cluster detection methods to predict infection in the second year, mixed effect logistic regression models was used. The outcome of interest was infection status by nPCR (0/1) in the second year. The risk factors explored were nPCR, AMA-1, MSP-119 and AMA-1 and/or MSP-119 (hereon termed combined seroprevalence) cluster score in the first year (generated via each of the three cluster detection methods). Simple summary contingency tables, graphs and scatter plots with Lowess curves were used to explore the relationship with potential risk factors and their associations with age. To explore the possibility of a non-linear relationship, risk factors were categorized into quartiles and a likelihood ratio test was used to assess which model (linear or categorical) was better. A household level random effect was included in the models to take account of correlation between individuals within the same household. All models were controlled for potential confounding by age, which due to an obvious non-linear relationship with infection was categorized before analysis into –zero to four years, five to nine years, ten to 15 years, 16-25 year, 26-35 years and over 36 years (Table 1).
To establish the effect of radius size on results obtained with the kernel and weighted local prevalence methods, models using different radii were built. In addition to the initial 1 km radius, radii of 500 m, 100 m and 0 m (i e, household) were explored. Models assuming individual level infection and serological status were also compared. Similarly, for the SaTScan analysis, maximum population sizes of 20 and 10% were explored. To compare the predictive performance of using different methods and radii, the area under the receiver operating curve (AUC) was calculated for each model. AUC values were compared using DeLong’s test for paired ROC curves [30]. Statistical analysis was performed using STATA (version 12, College Station, TX, USA) and R (version 3.0.1) [31].
Results
Study subjects
In 2010, 668 households from randomly selected sub-villages participated in the first year survey, comprising a total of 3,801 individuals, 3,057 (80.4%) of whom were seen, consented to participate and provided a blood specimen. Approximately half of the participants (n = 1,612, 52.7%) were male. The median age of the study population was 13 years (IQR = 5-30 years; range 1-99 years). The overall prevalence of P. falciparum by nPCR was 34.3%. In the second year survey, 697 households participated in the survey with 3,246 (85.4%) of eligible individuals providing a blood specimen, 51.6% of whom were male. Distribution of age was similar to that of the first year survey. P. falciparum prevalence by nPCR was significantly higher at 51.9% than during the baseline survey (OR 1.95; 95% CI, 1.76-2.17; p <0.001).
Association of age and other individual factors with PCR positivity and seropositivity
Individuals aged 10 to 15 years had the highest nPCR prevalence of P. falciparum at baseline and at follow-up (Table 1). Seropositivity to AMA-1 similarly peaked in the age group ten to 15 years. This age group had more than eight times the odds of being seropositive to AMA-1 compared to individuals aged zero to four years (OR 8.87, 95% CI 6.29-12.5; P < 0.001). Seropositivity to MSP-119 showed a different relationship with age, displaying a steady increase with age, with those aged >36 years having roughly five times the odds of being seropositive compared to those aged zero to four years (OR 5.10 95%, CI 3.66-7.10) (Table 1).
Prediction of infection in the second year survey
nPCR prevalence in the baseline survey
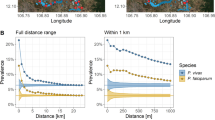
Fifty-seven per cent of individuals who were nPCR positive in the first year were also nPCR positive in the second year whilst 47% who were negative in the first year were also negative in the second year (χ2 = 27.2; P <0.001). Guided by AUC values, clustering estimated using kernel analysis appeared to predict infection by nPCR in the second year more accurately than the weighted local prevalence method (p = 0.016) (Table 2). While clustering estimated by SaTScan gave a higher AUC value than clustering by the weighted local prevalence method, there was no evidence for a difference in AUC (p = 0.12).
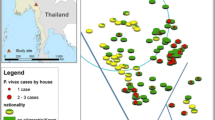
Using SaTScan analysis to detect nPCR hotspots, one large cluster was identified with a radius of 2.88 km, covering 141 households and one small cluster was identified with a radius of 0.1 km covering five households (Figure 1A). SaTScan analysis showed that individuals who were residing in a nPCR hotspot cluster in the first year had four times the odds of testing positive for malaria by nPCR in the second year than those residing in nPCR coldspots (OR 4.54 95% CI 2.68-7.72). The kernel and weighted local prevalence analyses showed a more complex distribution of hotspots (Figure 1B and C). Both clearly show the central hotspot detected by SaTScan, but also show numerous other high transmission areas, more consistent with the micro-epidemiology of malaria. The kernel analysis also showed that individuals who were residing in the top quartile (areas with a high prevalence of infection by nPCR) had three times the odds of testing positive for malaria by nPCR in the second year compared to those living in the lowest quartile (OR 3.45, 95% CI 2.06-5.75).
Seropositivity to AMA-1 and MSP-119 antibodies
Defining clusters of seroprevalence using AMA-1 and MSP-119 antibodies separately improved prediction of nPCR positivity in the second year compared to using combined seroprevalence. SaTScan analysis revealed that individuals living in areas of high AMA-1 seroprevalence (hotspots) in the first year had five times the odds of being nPCR positive in the second year compared to those who lived in AMA-1 coldspots (OR 5.84 95% CI 3.75-9.10), adjusting for age (Table 2). SaTScan could not identify any significant clusters using combined seroprevalence.
When clusters were identified by kernel analysis, those individuals living in households with the highest quartile of AMA-1 seroprevalence (hotspots) had a more than five times the odds of being nPCR positive in the second year than those in the lowest quintile (OR 5.16 95% CI 3.06-8.69), adjusting for age (Table 2). Using weighted local prevalence scores to distinguish clusters showed a similar pattern, those residing in the households in the top quartile of AMA-1 seroprevalence (hotspots) had more than three times the odds of being nPCR positive than those residing in lowest quartile (OR 3.33 95% CI 1.97-5.62) (Table 2). Likewise the kernel analyses showed a more complex distribution of AMA-1 hotspots than SaTScan analysis (Figure 2). A comparison of the predictive ability of different clustering methods showed that both SaTScan and kernel analysis yielded higher AUC values than the weighted prevalence method, however, only the SaTScan method produced a significantly different result (p = 0.002 and p = 0.27 respectively).
Antibody responses to MSP-119 showed a less clear association with infection in the second year, with individual age-adjusted seroprevalence at baseline showing no relationship with infection status in the second year. SaTScan analysis suggested that individuals living in MSP-119 hotspots were at lower risk of infection in the second year. Both kernel and distance weighted prevalence analysis also suggested individuals living in areas of highest MSP-1 seroprevalence were at lower risk of infection, however those living in areas of intermediate seroprevalence (third quartile) were at higher risk of subsequent infection.
Individual seropositivity at baseline to the combined seroprevalence of AMA-1 and/ or MSP-119 antibodies showed no relationship with infection in the second year. Similar to results using just AMA-1, kernel analysis of combined seroprevalence showed that those individuals living in the highest quartile had more than two times the odds of being nPCR positive in the second year than those residing in the lowest quintile (OR 2.44 95% CI 1.44-4.14). While a similar relationship was seen if hotspots were determined by weighted local prevalence, overall predictive ability using this method was worse than when using kernels with an AUC value of 0.530 (Table 2). SaTScan was not able to find any hotspots or coldspots using combined seroprevalence.
Sensitivity analysis of kernel and SaTScan methods for determining the best radius to predict malaria in the second year of follow-up
Based on AUC values, the weighted local prevalence method to identify clusters was generally less predictive of infection in the second year than the SaTScan and kernel methods. Sensitivity analyses of these two methods were therefore conducted to determine the radius size that best predicted infection in the second year. For the kernel method, using larger radii to identify clusters of nPCR tended to produced similar AUC values than smaller radii (Table 3). Using larger radii of 500 m and 1 km to identify clusters of AMA-1 seroprevalence, MSP-119 or the antigens combined, generally produced higher AUC values. Similar sensitivity analyses were done for SaTScan, whereby the maximum population size allowable was set to 20 and 10%. As for the kernel analysis, there was a general trend to suggest that a larger maximum population size of 50%, which allows for larger geographic clusters, was more predictive of subsequent infection than smaller maximum population sizes (Table 3).
Discussion
It has been suggested that if malaria transmission hotspots can be identified, targeting interventions can have a improved impact on transmission [7]. A number of previous studies have explored the use of geospatial techniques to identify clusters of transmission markers such as infection or seropositivity to selected antigens [13, 14, 18, 28, 32, 33]. These studies show that households with active and historic exposure tend to cluster together geographically. It is less clear however, whether these clusters predict future infection and if so, which geospatial techniques and transmission indicators should be used for their detection. Using two consecutive years’ data, this study shows that clusters of infection and seropositivity to AMA-1 are predictive of future infection and that kernel analysis and SaTScan are superior to the weighted local prevalence method of cluster detection.
Several authors have identified the existence of hotspots at single time points, using a variety of different measures of transmission [13, 18, 28]. Fewer studies have shown that hotspots are stable over time. Using data from multiple years in Kenya, Bejon et al. applied spatial scan statistics to identify infection hotspots that were predictive of future hotspots up to seven years later [14]. Another study done in a highland of Kenya by Ernst et al. identified stable spatial clusters of malaria cases by SaTScan statistics over a period of four years [33]. Again using spatial scan statistics, Bousema et al. showed that over the period of two years, clinical episodes of malaria cluster into hotspots [13]. This study is consistent with these findings, showing that hotspots of infection are predictive of future infection. The study also shows that being seropositive to AMA-1 or being in a hotspot of AMA-1 seroprevalence is predictive of future infection. As seropositivity to AMA-1 is indicative of recent exposure to P. falciparum, this finding adds further evidence that hotspots of transmission are stable over several years. The relatively low AUC values do, however, suggest the importance of other factors related to risk of infection that were not accounted for. In addition, the higher prevalence of infection seen in the second year, likely due to higher rainfall observed that year, led to some infections in non-hotspot households, which negatively impacts the AUC.
The relationship between hotspots of seropositivity to MSP-119 and future infection was less clear. Clusters with high MSP-1 seroprevalence were found to be at lower risk of infection suggesting some protection at the neighbourhood level. However, whilst some studies have demonstrated a protective effect of antibodies to MSP-119,[34–37] at the individual level, this was not observed in this study. The reasons for these observations and the differences in the patterns seen with AMA-1 require further investigation but they may relate to the differing immunogenicity and half-life of the antibody response to these two antigens [38].
In terms of methods to detect clusters, this study suggests that using spatial scan statistics or kernel analysis allows better characterization of hotspots than the weighted local prevalence method. This may be due to the fact that estimates of weighted local prevalence for each household are made using infection status of neighbours only. This likely leads to an inferior indication of hotspot location as individual or household level factors play an important role in risk of subsequent infection in that household. Sensitivity analyses, varying both the window size and maximum population size for kernel and SaTScan analysis respectively, suggests that generally hotspots form over larger (1-3 km) scales. While this likely varies by setting, similarly sized hotspots have been detected by previous studies in similar transmission settings [13, 14, 20]. In lower transmission settings, transmission appears to cluster over increasingly small scales. A recent study by Searle et al. in Zambia, where infection prevalence was estimated to be 23% by rapid diagnostic test (RDT), showed that active case detection within a 500-m radius could identify 76% of all RDT-positive individuals [39]. A study in Swaziland, where transmission is extremely low (PCR-derived parasite prevalence <1%), suggested that infections tend to cluster within households of passively detected cases [9].
This study has several potential operational implications for malaria control. Firstly, given the apparent stability of hotspots, targeting clusters of infection and seropositivity to AMA-1 (and/or antigens with similar properties) with complete cure treatment and vector control could have a dramatic impact on transmission [7]. Secondly, kernel analysis and SaTScan appear to be optimal methods to detect hotspots. Currently, establishment of seropositivity to AMA-1 can only be done using assays that require samples to be processed in the laboratory. Equally, while RDTs exist for determining infection status, these miss a large fraction of infections, most of which are likely to be subpatent [40–42]. Previous work has shown that these subpatent infections tend to cluster in hotspots, making RDTs inappropriate methods to detect hotspots [43]. In order to target interventions at hotspots, therefore, the development of sensitive rapid diagnostics for infection and seropositivity to AMA-1 (or similar) is required. Alternatively, it may be possible to identify hotspots in the field by clustering of particular risk factors or passively detected cases. This is the focus of further research. In the meantime, in the setting of moderate malaria transmission around Lake Victoria, mass drug administration of entire villages may be required to interrupt transmission [43].
Limitations
This study used indirect measures to define household malaria exposure. Using more direct measures, such as entomological inoculation rate (EIR) and other vector measures, may have led to different results. However, EIR can be challenging to measure in low-endemic settings. Thus, individual parasite prevalence was chosen as the measure of subsequent transmission for this study. In addition, indoor residual spraying (IRS) was applied between survey periods throughout the study area. While there is no supporting data, it is likely that households that did not receive IRS were randomly distributed and therefore unlikely to introduce bias into the results. Lastly, the study continued for only two years, thus stability of malaria hotspots could only be predicted for that time period. However, as stated, the fact that hotspots of AMA-1 seroprevalence were predictive of future infection suggests transmission hotspots are stable over a longer time frame.
Conclusions
This study supports previous work showing that hotspots can be defined using geospatial methods and are stable over a period of at least one year. Hotspots can be detected either by using parasite prevalence or seroprevalence of AMA-1 antibodies. It was also found that spatial scan statistics and kernel analysis were better at characterizing hotspots of transmission than the weighted local prevalence method. Given the lack of highly sensitive rapid diagnostic tests for infection and AMA-1 seropositivity, routine detection of hotspots is challenging. Further work exploring simple methods to identify hotspots with existing tools is therefore required. Furthermore, while theorized, it has yet to be shown in the field that targeting interventions does indeed lead to greater reductions in transmission over an untargeted approach. Studies linking methods of hotspot detection with assessments of the subsequent impact of targeted interventions would be extremely valuable.
References
Woolhouse ME, Dye C, Etard JF, Smith T, Charlwood JD, Garnett GP, Hagan P, Hii JL, Ndhlovu PD, Quinnell RJ, Watts CH, Chandiwana SK, Anderson RM: Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proc Natl Acad Sci USA. 1997, 94: 338-342. 10.1073/pnas.94.1.338.
Mwangi TW, Fegan G, Williams TN, Kinyanjui SM, Snow RW, Marsh K: Evidence for over-dispersion in the distribution of clinical malaria episodes in children. PLoS One. 2008, 3: e2196-10.1371/journal.pone.0002196.
Gamage-Mendis AC, Carter R, Mendis C, De Zoysa AP, Herath PR, Mendis KN: Clustering of malaria infections within an endemic population: risk of malaria associated with the type of housing construction. Am J Trop Med Hyg. 1991, 45: 77-85.
Carter R, Mendis KN, Roberts D: Spatial targeting of interventions against malaria. Bull World Health Organ. 2000, 78: 1401-1411.
Ghebreyesus TA, Haile M, Witten KH, Getachew A, Yohannes M, Lindsay SW, Byass P: Household risk factors for malaria among children in the Ethiopian highlands. Trans R Soc Trop Med Hyg. 2000, 94: 17-21. 10.1016/S0035-9203(00)90424-3.
Lindsay SW, Jawara M, Paine K, Pinder M, Walraven GEL, Emerson PM: Changes in house design reduce exposure to malaria mosquitoes. Trop Med Int Health. 2003, 8: 512-517. 10.1046/j.1365-3156.2003.01059.x.
Bousema T, Griffin JT, Sauerwein RW, Smith DL, Churcher TS, Takken W, Ghani A, Drakeley C, Gosling R: Hitting hotspots: spatial targeting of malaria for control and elimination. PLoS Med. 2012, 9: e1001165-10.1371/journal.pmed.1001165.
Midega JT, Smith DL, Olotu A, Mwangangi JM, Nzovu JG, Wambua J, Nyangweso G, Mbogo CM, Christophides GK, Marsh K, Bejon P: Wind direction and proximity to larval sites determines malaria risk in Kilifi District in Kenya. Nat Comm. 2012, 3: 674-
Sturrock HJW, Novotny JM, Kunene S, Dlamini S, Zulu Z, Cohen JM, Hsiang MS, Greenhouse B, Gosling RD: Reactive case detection for malaria elimination: real-life experience from an ongoing program in Swaziland. PLoS ONE. 2013, 8: e63830-10.1371/journal.pone.0063830.
Kulldorff M: A spatial -scan statistic. Community Statistic Theory Methods. 1997, 26: 1481-1496. 10.1080/03610929708831995.
Kulldorff M: SaTScan -Software for the spatial, temporal, and space-time scan statistics. 2010, Boston: Harvard Medical School and Harvard PilgrimHealth Care
Pullan RL, Sturrock HJW, Soares Magalhaes RJ, Clements ACA, Brooker SJ: Spatial parasite ecology and epidemiology: a review of methods and applications. Parasitology. 2012, 139: 1870-1887. 10.1017/S0031182012000698.
Bousema T, Drakeley C, Gesase S, Hashim R, Magesa S, Mosha F, Otieno S, Carneiro I, Cox J, Msuya E, Kleinschmidt I, Maxwell C, Greenwood B, Riley E, Sauerwein R, Chandramohan D, Gosling R: Identification of hot spots of malaria transmission for targeted malaria control. J Infect Dis. 2010, 201: 1764-1774. 10.1086/652456.
Bejon P, Williams TN, Liljander A, Noor AM, Wambua J, Ogada E, Olotu A, Osier FHA, Hay SI, Farnert A, Marsh K: Stable and unstable malaria hotspots in longitudinal cohort studies in Kenya. PLoS Med. 2010, 7: e1000304-10.1371/journal.pmed.1000304.
Kreuels B, Kobbe R, Adjei S, Kreuzberg C, von Reden C, Bater K, Klug S, Busch W, Adjei O, May J: Spatial variation of malaria incidence in young children from a geographically homogeneous area with high endemicity. J Infect Dis. 2008, 197: 85-93. 10.1086/524066.
Cook J, Kleinschmidt I, Schwabe C, Nseng G, Bousema T, Corran PH, Riley EM, Drakeley CJ: Serological markers suggest heterogeneity of effectiveness of malaria control interventions on Bioko Island, equatorial Guinea. PLoS ONE. 2011, 6: e25137-10.1371/journal.pone.0025137.
Stone W, Bousema T, Jones S, Gesase S, Hashim R, Gosling R, Carneiro I, Chandramohan D, Theander T, Ronca R, Modiano D, Arca B, Drakeley C: IgG responses to Anopheles gambiae salivary antigen gSG6 detect variation in exposure to malaria vectors and disease risk. PLoS ONE. 2012, 7: e40170-10.1371/journal.pone.0040170.
Bejon P, Turner L, Lavstsen T, Cham G, Olotu A, Drakeley CJ, Lievens M, Vekemans J, Savarese B, Lusingu J, von Seidlein L, Bull PC, Marsh K, Theander TG: Serological evidence of discrete spatial clusters of Plasmodium falciparum parasites. PLoS ONE. 2011, 6: e21711-10.1371/journal.pone.0021711.
Aamodt G, Samuelsen SO, Skrondal A: A simulation study of three methods for detecting disease clusters. Int J Health Geogr. 2006, 5: 15-10.1186/1476-072X-5-15.
Olotu A, Fegan G, Wambua J, Nyangweso G, Ogada E, Drakeley C, Marsh K, Bejon P: Estimating individual exposure to malaria using local prevalence of malaria infection in the field. PLoS ONE. 2012, 7: e32929-10.1371/journal.pone.0032929.
Olotu A, Fegan G, Wambua J, Nyangweso G, Awuondo KO, Leach A, Lievens M, Leboulleux D, Njuguna P, Peshu N, Marsh K, Bejon P: Four-year efficacy of RTS, S/AS01E and its interaction with malaria exposure. N Engl J Med. 2013, 368: 1111-1120. 10.1056/NEJMoa1207564.
Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE: Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995, 52: 565-568.
Snounou G, Singh B: Nested PCR analysis of Plasmodium parasites. Meth Mol Med. 2002, 72: 189-203.
Corran PH, Cook J, Lynch C, Leendertse H, Manjurano A, Griffin J, Cox J, Abeku T, Bousema T, Ghani AC, Drakeley C, Riley E: Dried blood spots as a source of anti-malarial antibodies for epidemiological studies. Malar J. 2008, 7: 195-10.1186/1475-2875-7-195.
Stewart L, Gosling R, Griffin J, Gesase S, Campo J, Hashim R, Masika P, Mosha J, Bousema T, Shekalaghe S, Cook J, Corran P, Ghani A, Riley EM, Drakeley C: Rapid assessment of malaria transmission using age-specific sero-conversion rates. PLoS One. 2009, 4: e6083-10.1371/journal.pone.0006083.
Pfeiffer DU, Robinson TP, Stevenson M, Stevens KB, Rogers DJ, Clements ACA: Analysis in Epidemiology. 2008, New York: Oxford University Press
Oesterholt MJAM, Bousema JT, Mwerinde OK, Harris C, Lushino P, Masokoto A, Mwerinde H, Mosha FW, Drakeley CJ: Spatial and temporal variation in malaria transmission in a low endemicity area in northern Tanzania. Malar J. 2006, 5: 98-10.1186/1475-2875-5-98.
Brooker S, Clarke S, Njagi JK, Polack S, Mugo B, Estambale B, Muchiri E, Magnussen P, Cox J: Spatial clustering of malaria and associated risk factors during an epidemic in a highland area of western Kenya. Trop Med Int Health. 2004, 9: 757-766. 10.1111/j.1365-3156.2004.01272.x.
Shabani J, Lutambi AM, Mwakalinga V, Masanja H: Clustering of under-five mortality in Rufiji Health and Demographic Surveillance System in rural Tanzania. Glob Health Action. 2010, 3: doi: 10.3402/gha.v3i0.5264
DeLong ER, DeLong DM, Clarke-Pearson DL: Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988, 44: 837-845. 10.2307/2531595.
Team RC: R: A language and enviroment for statistical computing. 2013, Vienna, Australia: R foundation for statistical computing, 3-900051-07-0, URL
Drakeley CJ, Corran PH, Coleman PG, Tongren JE, McDonald SLR, Carneiro I, Malima R, Lusingu J, Manjurano A, Nkya WMM, Lemnge MM, Cox J, Reyburn H, Riley E: Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci USA. 2005, 102: 5108-5113. 10.1073/pnas.0408725102.
Ernst KC, Adoka SO, Kowuor DO, Wilson ML, John CC: Malaria hotspot areas in a highland Kenya site are consistent in epidemic and non-epidemic years and are associated with ecological factors. Malar J. 2006, 5: 78-10.1186/1475-2875-5-78.
Al-Yaman F, Genton B, Kramer KJ, Chang SP, Hui GS, Baisor M, Alpers MP: Assessment of the role of naturally acquired antibody levels to Plasmodium falciparum merozoite surface protein-1 in protecting Papua New Guinean children from malaria morbidity. Am J Trop Med Hyg. 1996, 54: 443-448.
Egan AF, Morris J, Barnish G, Allen S, Greenwood BM, Kaslow DC, Holder AA, Riley EM: Clinical immunity to Plasmodium falciparum malaria is associated with serum antibodies to the 19-kDa C-terminal fragment of the merozoite surface antigen, PfMSP-1. J Infect Dis. 1996, 173: 765-769. 10.1093/infdis/173.3.765.
Conway DJ, Cavanagh DR, Tanabe K, Roper C, Mikes ZS, Sakihama N, Bojang KA, Oduola AM, Kremsner PG, Arnot DE, Greenwood BM, McBride JS: A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med. 2000, 6: 689-692. 10.1038/76272.
Perraut R, Marrama L, Diouf B, Sokhna C, Tall A, Nabeth P, Trape J-F, Longacre S, Mercereau-Puijalon O: Antibodies to the conserved C-terminal domain of the Plasmodium falciparum merozoite surface protein 1 and to the merozoite extract and their relationship with in vitro inhibitory antibodies and protection against clinical malaria in a Senegalese village. J Infect Dis. 2005, 191: 264-271. 10.1086/426398.
Corran P, Coleman P, Riley E, Drakeley C: Serology: a robust indicator of malaria transmission intensity?. Trends Parasitol. 2007, 23: 575-582. 10.1016/j.pt.2007.08.023.
Searle KM, Shields T, Hamapumbu H, Kobayashi T, Mharakurwa S, Thuma PE, Smith DL, Glass G, Moss WJ: Efficiency of household reactive case detection for malaria in rural southern Zambia: simulations based on cross-sectional surveys from two epidemiological settings. PLoS One. 2013, 8: e70972-10.1371/journal.pone.0070972.
Okell LC, Ghani AC, Lyons E, Drakeley CJ: Submicroscopic infection in Plasmodium falciparum-endemic populations: a systematic review and meta-analysis. J Infect Dis. 2009, 200: 1509-1517. 10.1086/644781.
Okell LC, Bousema T, Griffin JT, Ouedraogo AL, Ghani AC, Drakeley CJ: Factors determining the occurrence of submicroscopic malaria infections and their relevance for control. Nat Comm. 2012, 3: 1237-
Manjurano A, Okell L, Lukindo T, Reyburn H, Olomi R, Roper C, Clark TG, Joseph S, Riley EM, Drakeley C: Association of sub-microscopic malaria parasite carriage with transmission intensity in north-eastern Tanzania. Malar J. 2011, 10: 370-10.1186/1475-2875-10-370.
Mosha JF, Sturrock HJ, Greenhouse B, Greenwood B, Sutherland CJ, Gadalla N, Atwal S, Drakeley C, Kibiki G, Bousema T, Chandramohan D, Gosling R: Epidemiology of subpatent Plasmodium falciparum infection: implications for detection of hotspots with imperfect diagnostics. Malar J. 2013, 12: 221-10.1186/1475-2875-12-221.
Acknowledgements
This study was supported by Malaria Capacity development consortium (MCDC), which is funded by Welcome Trust (Grant number WT084289MA) and Bill & Melinda Gates Foundation, Grand Challenge for Exploration no: 01916000035 supported the field work and OPP1013170 supported the analysis. We thank Ramadhani Hashim for excellent technical assistance in data management. We acknowledge support of John Changalucha and the management team of NIMR Mwanza Centre.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
JFM was involved in the study design, supervised the implementation of the study and data collection, analysed data, drafted and revised the manuscript. HJWS was involved in data analysis, interpretation of the data, drafted and revised the manuscript. DC and RDG were involved in overall study design and supervision, interpretation of the data and revisions of the manuscript. TB, CJS and CD were involved in supervision of laboratory work, interpretation of the data and revision of the manuscript. BG, JMB and KG were involved in interpretation of the data and revisions of the manuscript. NG, SA and SH performed the real time PCR testing, serology testing and revised the manuscript. All authors have read and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Mosha, J.F., Sturrock, H.J., Greenwood, B. et al. Hot spot or not: a comparison of spatial statistical methods to predict prospective malaria infections. Malar J 13, 53 (2014). https://doi.org/10.1186/1475-2875-13-53
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-13-53