Abstract
Background
Although several studies have investigated the impact of reduced malaria transmission due to insecticide-treated bed nets (ITNs) on the patterns of morbidity and mortality, there is limited information on their effect on parasite diversity.
Methods
Sequencing was used to investigate the effect of ITNs on polymorphisms in two genes encoding leading Plasmodium falciparum vaccine candidate antigens, the 19 kilodalton blood stage merozoite surface protein-1 (MSP-119kDa) and the Th2R and Th3R T-cell epitopes of the pre-erythrocytic stage circumsporozoite protein (CSP) in a large community-based ITN trial site in western Kenya. The number and frequency of haplotypes as well as nucleotide and haplotype diversity were compared among parasites obtained from children <5 years old prior to the introduction of ITNs (1996) and after 5 years of high coverage ITN use (2001).
Results
A total of 12 MSP-119kDa haplotypes were detected in 1996 and 2001. The Q-KSNG-L and E-KSNG-L haplotypes corresponding to the FVO and FUP strains of P. falciparum were the most prevalent (range 32–37%), with an overall haplotype diversity of > 0.7. No MSP-119kDa 3D7 sequence-types were detected in 1996 and the frequency was less than 4% in 2001. The CSP Th2R and Th3R domains were highly polymorphic with a total of 26 and 14 haplotypes, respectively detected in 1996 and 34 and 13 haplotypes in 2001, with an overall haplotype diversity of > 0.9 and 0.75 respectively. The frequency of the most predominant Th2R and Th3R haplotypes was 14 and 36%, respectively. The frequency of Th2R and Th3R haplotypes corresponding to the 3D7 parasite strain was less than 4% at both time points. There was no significant difference in nucleotide and haplotype diversity in parasite isolates collected at both time points.
Conclusion
High diversity in these two genes has been maintained overtime despite marked reductions in malaria transmission due to ITNs use. The frequency of 3D7 sequence-types was very low in this area. These findings provide information that could be useful in the design of future malaria vaccines for deployment in endemic areas with high ITN coverage and in interpretation of efficacy data for malaria vaccines based on 3D7 parasite strains.
Similar content being viewed by others
Background
Malaria caused by Plasmodium falciparum continues to place a heavy burden on the health and economic development of populations in endemic areas [1]. Over the last decade, increased funding for malaria control has resulted in rapid scale-up of key interventions including insecticide-treated bed nets (ITNs) in many areas [2] ITNs have been shown to reduce malaria transmission by 70-90%, leading to significant reductions in human-vector contact, the number of infectious mosquito bites [3, 4] and the force of infection [5]. Previous studies have shown that the intensity of malaria transmission is an important determinant of the malaria burden in the population, the rates of acquisition of protective immunity and the dynamics of parasite dispersal in mosquito vectors [6]. Based on these observations, fears have been expressed that reduced transmission and exposure to malaria parasites could modify the vector-host-parasite interaction, delay the acquisition of protective immunity and result in a shift in the burden of severe disease and mortality [7]. Studies designed to investigate whether sustained use of ITNs during infancy leads to an increase in malaria-related morbidity and mortality in older children demonstrated that there is no shift in the malaria burden [8, 9]. The few studies that have assessed the impact of ITNs on acquired immunity by comparing antibody responses to malaria antigens in children with or without ITNs have been inconclusive [10–14].
However, there is limited information on the impact of transmission reduction as a result of sustained use of ITNs on the genetic diversity of P. falciparum parasites. Malaria transmission intensity has been shown to play an important role in determining the genetic diversity of malaria parasites [15]. This is because malaria parasites undergo an obligate sexual reproduction in the mosquito vector, thereby creating an opportunity for generation of parasite diversity through intragenic recombination and re-assortment of genetic material during meiosis [16]. In areas of high transmission, both the numbers of infected hosts and the carriage of multiple-clone infections per individual are generally higher, therefore increasing the probability for mosquito vectors to obtain genetically diverse gametocytes during blood feeding [16, 17]. This in turn increases the chance of out-breeding and higher rates of genetic recombination. These observations have lead to the hypothesis that significant reduction in the intensity of malaria transmission could lead to a decrease in the mean number of multiple-genotype infections in humans, which could decrease the frequency of recombination resulting in a reduction in the level of genetic diversity [16].
A previous study conducted by this same group using neutral microsatellite (MS) markers has shown an overall stability in genetic diversity of P. falciparum after five years of high ITN coverage except for a significant MS locus specific change associated with gametocytes [18]. The current study was designed to investigate the impact of sustained use of ITNs on the genetic diversity of two leading P. falciparum vaccine candidate antigens during the same study period and in the same study area in western Kenya. This study assessed single nucleotide polymorphisms (SNPs) in genes encoding the relatively conserved B-cell epitopes of the blood-stage 19-kDa region of the merozoite surface protein (MSP-1 19kDa) and the highly polymorphic pre-erythrocytic-stage circumsporozoite protein (CSP) Th2R and Th3R T-helper cell epitopes. These two genes were chosen for assessment because their polymorphism is thought to be partly driven by the host immune pressure and are likely to be subjected to the diversifying effects of changes in malaria transmission [15]. Monitoring the genetic diversity of leading P. falciparum vaccine candidate antigens over time will help better understand the relationship between transmission intensity as malaria interventions are scaled up and the diversity of parasite antigens that are under selection pressure. In addition, these data will be useful in interpreting efficacy data of malaria vaccines that are based on 3D7 sequences.
The merozoite surface protein-1 (MSP-1) is the major protein on the surface of asexual blood stages of the malaria parasites [19]. The MSP-1 195-kDa protein is synthesized during schizont maturation and undergoes two rounds of proteolytic cleavage leaving a 19-kDa fragment (MSP-1 19kDa) adhering on the surface of merozoites during erythrocyte invasion [20]. The MSP-1 19kDa contains two cysteine-rich epidermal growth factor (EGF)-like motifs that play an important role in erythrocyte invasion, making it an ideal target of inhibitory antibodies for blocking parasite invasion of erythrocytes [20] and therefore an ideal candidate for a malaria vaccine. Although MSP-1 is characterized by extensive polymorphisms, the 19-kDa region is relatively conserved apart from five key non-synonymous SNPs corresponding to amino-acid positions 1644 (Q/E), 1691 (K/T), 1699 (S/N), 1700 (N/S), and 1701 (G/R). Intragenic recombination and single-nucleotide mutations are thought to play a role in generation of allelic variation in MSP-1 19kDa[21, 22]. Several malaria blood-stage vaccine candidates containing MSP-1 19kDa have undergone field trials with varied results [23].
The CSP is the predominant protein found on the surface of sporozoites [24]. The P. falciparum csp gene encodes a protein of approximately 420 amino acid residues with a molecular weight of 58 kDa consisting of two non-repetitive regions at the 5' and 3' ends and a variable central region that encodes immunodominant B-cell epitopes with multiple repeats of four amino acid motifs [25]. The C-terminal end of the gene contains two highly polymorphic T-helper cell epitopes, Th2R and Th3R, flanking the conserved RII region [25]. The CSP has been the subject of extensive research for the development of a vaccine against pre-erythrocytic stages of malaria parasites [26]. The central repeat and C-terminal regions of the CSP forms part of the most clinically advanced malaria vaccine candidate RTS,S that is currently undergoing a phase III clinical trial in several African countries [27, 28]. Studies conducted in areas of different transmission settings have shown an association between the genetic diversity in the csp gene and malaria transmission, with the highest diversity observed in parasite populations from Africa [15].
The current study investigated the impact of transmission reduction due to the use of ITNs on the genetic diversity of genes encoding two P. falciparum polymorphic vaccine candidate antigens in 1996 before and 5 years after the deployment of ITNs in an area of western Kenya holoendemic for malaria. Prior to the ITN trial, malaria transmission in this area was intense and perennial with an estimated entomologic inoculation rate (EIR) of 60 to 300 infective bites per person per year [29]. It was estimated that malaria transmission in this area reduced by 95% after the introduction of ITNs [9].
Methods
Study area and population
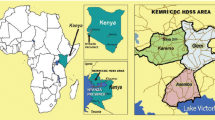
This study was conducted in the context of a community-based cluster randomized controlled trial of ITNs in western Kenya that consisted of two phases. Phase 1 was designed to assess the impact of ITNs on all-cause mortality in young children while phase 2 consisted of an extended surveillance to monitor whether sustained use of ITNs during infancy increased all-cause morbidity and mortality in older children. The characteristics of the study area, design of the ITN trial and the two-year surveillance have been described in detail elsewhere [9, 30]. Briefly, a randomized ITN trial was conducted between January 1997 and March 1999 in Rarieda Division (Asembo) where malaria is holoendemic and transmission occurs throughout the year.
At the end of the two-year trial period, ITNs were also distributed to all control villages and monitoring continued for an additional two years until March 2001. Throughout the two phases, annual cross-sectional surveys were conducted in 60 villages to collect information on malaria morbidity indicators. During the surveys finger stick blood samples were taken among children aged below five years. In addition, entomologic indices were collected throughout the two phases of the trial to estimate malaria transmission. During phase 1 of the trial there was a 72% reduction in the density of indoor-resting blood-fed Anopheles in the houses with ITNs and in the second phase, villages with ITNs experienced a 77% reduction in Anopheles densities compared with villages in an adjacent area [9]. In addition, parasite prevalence in children under the age of five decreased from 70% prior to the ITN trial to 34% in 2001 [9, 31]. The use of ITNs before the trial was less than 5% but increased to 65.9% and 82.5% in phase 1 and 2 respectively [9].
The blood samples used in the current study were collected in October and November 1996, before phase 1 of the ITN trial and in May and June in 2001, 5 years after introduction of ITNs in phase 1. Microscopically confirmed malaria positive blood samples were randomly selected from those collected in the 1996 cross-sectional survey and matched by village with samples collected in the 2001 survey.
Laboratory procedures
Sample collection
During each survey, 250-500 μl of blood was collected from each child by finger-stick into EDTA microtainers (Becton Dickinson, Franklin Lakes, NJ). Blood samples were transported in cool boxes to the central laboratory in Kisian, about 50 km from the field site and stored at 4°C until further processing. All samples were labelled with unique identification numbers that could not be linked to the randomization status of the sampled children. Erythrocytes were separated from plasma by centrifugation at 700 G for 5 min. Packed red blood cells (RBCs) were aliquoted into sterile vials and stored at -70°C until the day of DNA isolation.
DNA extraction and PCR amplification
Parasite genomic DNA was extracted using the QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The CSP gene was amplified by polymerase chain reaction (PCR) in a 25 μl reaction mixture containing 1 μl of DNA, 0.2 μM of forward and reverse primers CSP-F: ACAATCAAGG TAATGGACAAGG and CSP-R:GGATTAATAATGGTATTATCCTTCT, 1× TBE buffer, 2.0 mM MgCl2, 250 μM dNTPs, and 2.5 U Taq DNA polymerase (Promega Corporation, Madison, Wisconsin, USA). The cycling conditions were as follows: 95°C for 5 min, 58°C for 2 min and 72°C for 2 min for 1 cycle, then 94°C for 1 min, 58°C for 2 min and 72°C for 1 min for 32 cycles followed by 10 min extension at 72°C. The primers were designed to hybridize to conserved regions covering the Th2R, Th3R and the universal T-cell epitopes, resulting in a 354 base-pair fragment. The MSP-119-kDa gene was amplified in a 25 μl reaction mixture containing 1 μl of DNA, 0.2 μM of two forward primers- 5’ CGTTTTATCTAATTTACTTGATGGAA 3’(K16F), specific for K1 and 5’ CCTAATACAATAATATCAAAATTAATTGA 3’ (M16F), specific for MAD-20 and a common reverse primer 5’ TTAAGGTAACATATTTTAACTCCTAC 3’ (C3flaR), 2.0 mM MgCl2, 200 μM dNTPs, and 1.25U Taq DNA polymerase with the following cycling conditions: 91°C for 1.5 min, then 91°C for 30 sec, 50°C for 40 sec and 70°C for 40 sec for 40 cycles followed by 5 min extension at 72°C. PCR was carried out in a GeneAmp® PCR System 9700 (Biosystems, PE, USA).
Sequencing
The PCRs products were cleaned using CentriSep spin columns (Princeton Separations, Adelphia, NJ) according to the manufacturer instructions. Sequencing reactions were carried out using forward and reverse primers in separate reactions by the BigDye® Terminator v3.1 Cycle Sequencing Kit and analysed in an ABI 3100 DNA Sequencer (Applied Biosystems, Foster City, CA). The ChromasPro, version 2.31 software (Technelysium Pty Ltd, Queensland, AU) was used to align the sequences. Each sequence was manually checked for true base calling and samples with doubtful sequencing results were repeated.
Data analysis
The MSP-119-kDa sequences were aligned with known sequences from the P. falciparum genome database at the National Center for Biotechnology Information (NCBI) using the P. falciparum clone 3D7 as the reference sequence while the CSP Th2R and Th3R sequences were aligned using the 3D7 and 7G8 clones. Molecular analyses were conducted using MEGA version 4 [32] and DNA Sequence Polymorphism, version 4.0 [33] softwares to create haplotypes. Differences in the frequency of haplotypes between baseline and post-ITN intervention surveys were compared by Chi-square test. The genetic diversity parameters including nucleotide diversity (π), number of haplotypes (NHap) and haplotype diversity (HapDiv) in parasite populations at baseline and at post ITN intervention were determined using the sequence polymorphism softwares, DnaSP v5 [34], MEGA version 4 [32] and ARLEQUINS suite ver 3.5 [35]. Nucleotide sequences reported here are available in GeneBank under accession numbers BankIt1630040: KF158421 - KF158555 (Pfmsp-119-kDa) and BankIt1630320: KF158556 - KF158712 (Pfcsp C-terminal domain).
Ethical approval
The study was approved by the Ethical Review Committee of the Kenya Medical Research Institute, Nairobi, Kenya and the Institutional Review Board of the Centers for Disease Control and Prevention (CDC) Atlanta, Georgia, USA.
Consent
Written informed consent was obtained from the patient’s guardian/parent/next of kin for the publication of this report and any accompanying images.
Results
Frequency of MSP-119-kDa haplotypes
A total of 136 parasite samples were successfully sequenced for the gene encoding MSP-119-kDa, 58 at baseline and 78 in 2001. Overall, seven non-synonymous amino acid replacements were detected at positions 1644 (Q-E), 1691 (K-T), 1695 (E-K), 1699 (S-N), 1700 (S-N), 1701 (G-R) and 1716 (L-F). Based on the amino acid replacements, a total of 12 MSP-119-kDa haplotypes were detected in the study area in 1996 and 2001. Out of these, 3 haplotypes, Q-TESNGL, E-TESSGL and E-TESNGF are new and have not been reported before. Out of the 12 haplotypes identified, 8 haplotypes were observed in the 1996 and 2001 surveys while 4 haplotypes were observed at only one time point, 2 in 1996 and 2 in 2001. The E-KSNGL and Q-KSNGL haplotypes corresponding to the FUP-Uganda PA and FVO-Wellcome strains of P. falciparum had the highest prevalence in 1996, 37 and 32% respectively, and in 2001 it was, 37% for both strains (Figure 1). The frequency of all the other haplotypes was low, ranging from 2-7% at both time points. The frequency of the MSP-1 haplotype (E-TESSRL) corresponding to the 3D7 strain, the sequence contained in malaria vaccine candidates that are at various stages of development was less than 4% in 2001 and none was detected in 1996.
Genetic diversity of the MSP-119-kDa haplotypes
The genetic diversity of the MSP-119-kDa in parasite populations in 1996 and 2001 were comparable, with a nucleotide diversity (π) of 0.0038 and 0.0033 respectively (Table 1). Likewise, there was no significant difference in the number of haplotypes (NHap) or haplotype diversity (HapDiv) in parasite isolates collected in 1996 and 2001.
Frequency of CSP Th2R and Th3R haplotypes
At both surveys, a total of 157 (68 in 1996 and 89 in 2001) out of 175 parasite samples yielded interpretable sequences and were included in the analysis. Overall, there was a high diversity on the CSP C-terminal domain containing immunodominant T-cell Th2R and Th3R epitopes (Figures 2 and Figure 3). At the Th2R domain, a total of 26 and 34 haplotypes were detected in 1996 and 2001 surveys respectively, and for the Th3R domain, these were 14 and 13 haplotypes. Overall, there was a high fluctuation in the frequency of both the Th2R and Th3R haplotypes at both time points with some haplotypes detected at baseline and not in 2001 and vice versa (Figures 3A and B). Two Th2R haplotypes, QHIEKYLKTIQNSL and QHIEQYLKTIQNSL, had the highest frequency in 1996; 14 and 13% respectively; and 2001, 7 and 12% respectively. The frequency of all the other Th2R haplotypes was less than 5% at both time points. At the Th3R domain, the NKPKDEQDYEND and NKPKDEQDYAND haplotypes had the highest frequencies, in 1996, 30% for both haplotypes; and in 2001, 31 and 36% respectively. The frequency of all the other Th3R haplotypes was less than 10%. At both Th2R and Th3R domains, the frequencies of haplotypes corresponding to the 3D7 strain of P. falciparum were less than 4% at both time points.
Genetic diversity of the CSP Th2R and Th3R haplotypes
The genetic diversity of the CSP gene at the Th2R and Th3R domains are shown in Tables 2 and Table 3. Similar to the high number of haplotypes detected in 1996 and 2001, there was a high diversity in both domains at the two time points. The genetic diversity was higher in Th2R than the Th3R region as seen in the higher haplotype and nucleotide diversity (HapDiv and π) at both time points. There was no significant difference in the genetic diversity at both T cell epitopes between parasite isolates collected in the two surveys.
Discussion
For the gene encoding the blood stage MSP-119-kDa antigen, a total of 12 haplotypes were identified at both baseline and 5 years after distribution of ITNs in this area. This number is consistent with findings from previous studies conducted in our study area and in other malaria holoendemic areas of Africa showing a higher number of MSP-119-kDa haplotypes (range 6-15) in areas of high transmission and high EIRs ranging from 50 to 600 infective bites person per year, [21, 22, 36] compared with fewer haplotypes (range 1-3) in areas of lower transmission and EIRs of less than 10 infective bites per person per year [37, 38]. The E-KSNGL haplotype corre-sponding to the FUP-Uganda PA parasite strain and the Q-KSNGL corresponding to the FVO-Wellcome strain were the most predominant MSP-119-kDa haplotypes in 1996 and 2001. These two haplotypes have been reported at high frequencies in previous studies conducted in this area and other regions in Africa. In a study conducted in the same area in 1998, Qari et al reported frequencies of 42 and 21% for the E-KSNGL and Q-KSNGL haplotypes respectively [21]. Similarly, frequencies of 36 and 38% for the E-KSNGL and Q-KSNGL haplotypes respectively have been reported in a site near this study area [39]. In the current study, the frequency of other MSP-119-kDa haplotypes was very low.
There was no significant change overtime in the frequency, the number of MSP-119-kDa haplotypes, haplotype diversity and π. The stability in the frequency of haplotypes and the genetic diversity overtime, suggests that the MSP-119-kDa gene could be under balancing selection as documented in previous studies [40–42]. Similar levels of genetic stability have been reported in a malaria holoendemic area of Tanzania where polymorphic sites of the gene encoding the MSP-119-kDa gene was relatively stable for a period of 15 years [22]. This contrasts with significant temporal variations in the frequencies of MSP-119-kDa haplotypes reported in a study conducted in an area of low malaria transmission [43]. This difference has been explained to be due to the relatively low rates of meiotic recombination or strong selection pressure that maintain a clonal structure in parasite populations from areas of low transmission, while the reverse is true in areas of high transmission [43]. Therefore, the relative stability of MSP-119-kDa haplotypes observed in this study over a period of 5 years suggests that the rates of meiotic recombination in parasite populations in this area remain high despite the significant reduction in transmission associated with the deployment of ITNs.
The frequency of the MSP-1 haplotype (E-TESSRL) corresponding to the 3D7 strain of P. falciparum was not detected at baseline and was less than 4% in 2001. Low frequencies of MSP-1 3D7 sequence-types has been reported in previous studies conducted in different malaria transmission areas [21, 36, 43, 44]. It has been suggested that the low frequency of haplotypes corresponding to the 3D7 strain of P. falciparum could compromise the efficacy of MSP-1 vaccines that are based on the 3D7 clone. For example, a recent phase IIb trial of an MSP-142-based vaccine candidate derived from the 3D7 clone of P. falciparum in children aged 12-27 months in a nearby study area reported an overall vaccine efficacy of 5.1% [45]. The poor efficacy was partly attributed to the high multiplicity of infections and diversity of MSP-142 gene in this area [45]. Similarly, the variable success of MSP-1 vaccines that are at various stages of development including those based on FVO alleles has been attributed to polymorphisms in P. falciparum antigens [46]. Therefore, data on the genetic diversity of parasite populations circulating in area similar to that obtained in the current study provide important baseline information that could be useful in planning and interpreting data on the efficacy of vaccines that are based on polymorphic antigens such as MSP-1.
For the highly polymorphic Th2R and Th3R T-helper cell epitopes, there were 26 and 34 Th2R haplotypes at baseline and 5 years post-ITNs respectively and 14 and 13 Th3R haplotypes at the two time points. These findings are consistent with results of an earlier study conducted in the same area that reported 29 and 16 haplotypes for the Th2R and Th3R epitopes respectively [47]. This suggests that the CSP haplotypes circulating in this area have remained relatively stable for an extended period. This contrasts sharply with findings from areas of low malaria transmission which show that 1 or 2 haplotypes are predominant with frequencies as high as 80% [48]. In this study, the most frequent Th2R and Th3R haplotypes were 14% and 30% at baseline and 13 and 36% in 2001. Other haplotypes were rare with a prevalence of less than 10%. The frequency of haplotypes corresponding to the 3D7 parasite strain was less than 4%. The RTS,S malaria vaccine candidate that is currently undergoing a phase 3 efficacy trial in areas of different transmission intensities in Africa [27, 28] is derived from the 3D7 strain of P. falciparum and comprises part of the repeat, Th2R and Th3R regions of the CSP. Although there is no evidence that the observed efficacy of the RTS,S vaccine is sequence-dependent [49, 50] the information obtained in this study could be useful in the interpretation efficacy data of the RTS,S vaccine and other CSP-based malaria vaccines that are based on the 3D7 parasite strain in this area. This information could also inform the design of future vaccines for deployment in areas of high and perennial malaria transmission.
The high genetic diversity in the T-cell epitopes of the CSP gene was also apparent when measured by haplotype diversity and π. These findings are consistent with previous studies that have documented high genetic diversity in ThR2 and Th3R regions of CSP in parasite populations from regions of high malaria transmission [47, 51, 52]. This contrasts with low genetic diversity in parasite populations from areas of low malaria transmission of Brazil [52], Papua New Guinea [48, 53], India [54] and Iran. In addition, similar to previous studies, the current study also shows that the Th2R region is more diverse than the Th3R region, suggesting that the Th2R region could be under strong selective pressure unrelated to the intensity of malaria transmission. Similar to MSP-119-kDa gene, there was no significant difference in genetic diversity of the two T-cell epitopes in parasite populations at baseline and 5 years post-ITN intervention. The relative stability in the diversity of Th2R and Th3R regions has also been documented over a period of 5 years in a region of low transmission in Vietnam [55]. Taken together, our results indicate that transmission reduction due to sustained use of ITNs over a period of five years had no impact on the genetic diversity of the highly polymorphic Th2R and Th3R epitopes of the CSP gene.
Several factors have been suggested to play a role in the generation and maintenance of parasite diversity including transmission intensity, immune-mediated pressure, parasite positive rates, rates of multiple-genotype infections and patterns of antimalarial drug use [22]. In the current study, the sustained use of ITNs over a period of five years in a region of western Kenya with intense and perennial malaria transmission had no significant impact on parasite diversity of the two polymorphic P. falciparum genes. A few factors could explain this observation. Firstly, despite a decline in transmission intensity following the deployment of ITNs, parasite prevalence remained relatively high and 50% of children sleeping under ITNs were parasitaemic during phase 1 of the ITN trial (1996-1998) [31], and > 30% of post neonatal infants (one to 11 months) were parasitaemic during phase 2 of the trial (1999-2001) [9]. Similarly, cross-sectional surveys conducted 2-4 years after introduction of ITNs showed that up to one third of infants sleeping under ITNs were parasitaemic [9]. Secondly, although the target of the ITN trial was to cover all the sleeping spaces and efforts were made to monitor adherence, a significant proportion of people did not sleep under ITNs and remained exposed to malaria [56]. In phase 1 of the trial which distributed ITNs only to half of the villages, adherence to the use of ITNs was 65.9% and increased to 82.5% during the second phase when ITNs were distributed to the villages that acted as control in phase 1 [9]. Collectively, these epidemiological findings and the results obtained in the current study on stability of parasite genetic diversity suggest that exposure to malaria parasites in western Kenya remains at a level that allows genetic mixing of diverse parasite genes despite transmission reduction due to ITNs. Furthermore, it is possible that any observable impact of vector control interventions on genetic diversity of malaria parasites might require reduction of transmission intensity by more than the 90% reported in the ITN trial [4]. It is also conceivable that in areas of intense transmission, the impact of transmission reduction on parasite diversity will take greater reduction in transmission and for a much longer period than the 5-year period used to select sample for analysis in the current study or 15 years as shown in a previous study in Tanzania [22]. The short time period used in comparing parasite diversity after introduction of ITNs is a limitation in this study. The antigens used in this study could also be a limitation. Although the antigens used have been shown in previous studies to be under strong immunological pressure, it is likely that detection of changes in parasite diversity as a result of transmission reduction requires antigens that are highly sensitive to transmission dynamics. Lastly, although the study compared parasite diversity before and after introduction of ITNs, the absence of a contemporaneous area without ITNs for comparison is also a limitation in the current study.
Conclusions
Data obtained in this study show that the MSP-119kDa haplotypes have remained stable overtime and transmission reduction due to ITNs had no impact on the diversity of MSP-119kDa and the Th2R and Th3R domains of the CSP. The results also show that the CSP Th2R region is more diverse than the Th3R region and could be under strong selective pressure unrelated to the intensity of malaria transmission. The information on low frequency of 3D7 clone could be useful in interpreting data on the efficacy of malaria vaccines based on the 3D7 parasite strain in areas of stable malaria transmission and in informing the development of future malaria vaccines for deployment in areas of high transmission.
References
WHO: World malaria report. 2010, Geneva: World Health Organization,http://wwwwhoint/malaria/world_malaria_report_2010/en/indexhtml,
Lengeler C: Insecticide-treated bed nets and curtains for preventing malaria. Cochrane Database Syst Rev. 2004, 2: 363-
Lindsay S, Snow RW, Broomfield GL, Janneh MS, Wirtz RA, Greenwood BM: Impact of permethrin-treated bednets on malaria transmission by the Anopheles gambiae complex in The Gambia. Med Vet Entomol. 1989, 3: 263-271. 10.1111/j.1365-2915.1989.tb00226.x.
Gimnig JE, Vulule JM, Lo TQ, Kamau L, Kolczak MS, Phillips-Howard PA, Mathenge EM, Ter Kuile FO, Nahlen BL, Hightower AW, Hawley W: Impact of permethrin-treated bed nets on entomologic indices in an area of intense year-round malaria transmission. Am J Trop Med Hyg. 2003, 68 (Suppl 4): 16-22.
Ter Kuile FO, Terlouw DJ, Kariuki SK, Phillips-Howard PA, Mirel LBHW, Friedman JF, Shi YP, Kolczak MS, Lal AA, Vulule JM, Nahlen BL: Impact of permethrin-treated bed nets on malaria, anemia, and growth in infants in an area of intense perennial malaria transmission in western Kenya. Am J Trop Med Hyg. 2003, 68 (Suppl 4): 68-77.
Arnot DE: The influence of the genetic complexity of Plasmodium falciparum infections on the epidemiology of malaria. Trans R Soc Trop Med Hyg. 2002, 96 (Suppl 1): S131-136.
Marsh K, Snow RW: Host-parasite interaction and morbidity in malaria endemic areas. Philos Trans R Soc Lond B Biol Sci. 1997, 352: 1385-1394. 10.1098/rstb.1997.0124.
Binka FN, Hodgson A, Adjuik M, Smith T: Mortality in a seven-and-a-half-year follow-up of a trial of insecticide-treated mosquito nets in Ghana. Trans R Soc Trop Med Hyg. 2002, 96: 597-599. 10.1016/S0035-9203(02)90321-4.
Lindblade KA, Eisele TP, Gimnig JE, Alaii JA, Odhiambo F, Ter Kuile FO, Hawley WA, Wannemuehler KA, Phillips-Howard PA, Rosen DH, Nahlen BL, Terlouw DJ, Adazu K, Vulule JM, Slutsker L: Sustainability of reductions in malaria transmission and infant mortality in western Kenya with use of insecticide-treated bednets: 4 to 6 years of follow-up. JAMA. 2004, 291: 2571-2580. 10.1001/jama.291.21.2571.
Snow RW, Shenton FC, Lindsay SW, Greenwood BM, Bennett S, Wheeler J, Del Giudice G, Verdini AS, Pessi A: Sporozoite antibodies and malaria in children in a rural area of The Gambia. Ann Trop Med Parasitol. 1989, 83: 559-568.
Genton B, Hii J, al-Yaman F, Paru R, Beck HP, Ginny M, Dagoro H, Lewis D, Alpers MP: The use of untreated bednets and malaria infection, morbidity and immunity. Ann Trop Med Parasitol. 1994, 88: 263-270.
Snow RW, Molyneux CS, Warn PA, Omumbo J, Nevill CG, Gupta S, Marsh K: Infant parasite rates and immunoglobulin M seroprevalence as a measure of exposure to Plasmodium falciparum during a randomized controlled trial of insecticide-treated bed nets on the Kenyan coast. Am J Trop Med Hyg. 1996, 55: 144-149.
Askjaer N, Maxwell C, Chambo W, Staalsoe T, Nielsen M, Hviid L, Curtis C, Theander TG: Insecticide-treated bed nets reduce plasma antibody levels and limit the repertoire of antibodies to Plasmodium falciparum variant surface antigens. Clin Diagn Lab Immunol. 2001, 8: 1289-1291.
Kariuki SK, Lal AA, Terlouw DJ, Ter Kuile FO, Ong'echa JM, Phillips-Howard PA, Orago AS, Kolczak MS, Hawley WA, Nahlen BL, Shi YP: Effects of permethrin-treated bed nets on immunity to malaria in western Kenya II. Antibody responses in young children in an area of intense malaria transmission. Am J Trop Med Hyg. 2003, 68 (Suppl 4): 108-114.
Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP: Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000, 17: 1467-1482. 10.1093/oxfordjournals.molbev.a026247.
Paul RE, Packer MJ, Walmsley M, Lagog M, Ranford-Cartwright LC, Paru R, Day KP: Mating patterns in malaria parasite populations of Papua New Guinea. Science. 1995, 269: 1709-1711. 10.1126/science.7569897.
Babiker HA, Ranford-Cartwright LC, Currie D, Charlwood JD, Billingsley P, Teuscher T, Walliker D: Random mating in a natural population of the malaria parasite Plasmodium falciparum. Parasitology. 1994, 109: 413-421. 10.1017/S0031182000080665.
Gatei W, Kariuki S, Hawley W, Ter Kuile F, Terlouw D, Phillips-Howard P, Nahlen B, Gimnig J, Lindblade K, Walker E, Hamel M, Crawford S, Williamson J, Slutsker L, Shi YP: Effects of transmission reduction by insecticide-treated bed nets (ITNs) on parasite genetics population structure: I. The genetic diversity of Plasmodium falciparum parasites by microsatellite markers in western Kenya. Malar J. 2010, 9: 353-
Cooper JA: Merozoite surface antigen-I of Plasmodium. Parasitol Today. 1993, 9: 5-54.
Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA: A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med. 1990, 172: 379-382. 10.1084/jem.172.1.379.
Qari SH, Shi YP, Goldman IF, Nahlen BL, Tibayrenc M, Lal AA: Predicted and observed alleles of Plasmodium falciparum merozoite surface protein-1 (MSP-1), a potential malaria vaccine antigen. Mol Biochem Parasitol. 1998, 92: 241-252. 10.1016/S0166-6851(98)00010-3.
Tanabe K, Sakihama N, Rooth I, Björkman A, Färnert A: High frequency of recombination-driven allelic diversity and temporal variation of Plasmodium falciparum msp1 in Tanzania. Am J Trop Med Hyg. 2007, 76: 1037-1045.
Good MF: Towards a blood-stage vaccine for malaria: are we following all the leads?. Nat Rev Immunol. 2001, 1: 117-125. 10.1038/35100540.
Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M: Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. 1980, 207: 71-73. 10.1126/science.6985745.
Enea V, Ellis J, Zavala F, Arnot DE, Asavanich A, Masuda A, Quakyi I, Nussenzweig RS: DNA cloning of Plasmodium falciparum circumsporozoite gene: amino acid sequence of repetitive epitope. Science. 1984, 225: 628-630. 10.1126/science.6204384.
Good MF: T cells, T sites, and malaria immunity–further optimism for vaccine development. J Immunol. 1988, 140: 1715-1716.
The RTS: SCTP: First results of phase 3 trial of RTS, S/AS01 malaria vaccine in African children. N Engl J Med. 2011, 365: 1863-1875.
The RTS: SCTP: A phase 3 trial of RTS, S/AS01 malaria vaccine in African infants. N Engl J Med. 2012, 367: 2284-2295.
Beier JC, Oster CN, Onyango FK, Bales JD, Sherwood JA, Perkins PV, Chumo DK, Koech DV, Whitmire RE, Roberts CR, Diggs CL, Hoffman SK: Plasmodium falciparum incidence relative to entomologic inoculation rates at a site proposed for testing malaria vaccines in western Kenya. Am J Trop Med Hyg. 1994, 50: 529-536.
Phillips-Howard PA, Nahlen BL, Kolczak MS, Hightower AW, Ter Kuile FO, Alaii JA, Gimnig JE, Arudo J, Vulule JM, Odhacha A, Kachur SP, Schoute E, Rosen DH, Sexton JD, Oloo AJ, Hawley WA: Efficacy of permethrin-treated bed nets in the prevention of mortality in young children in an area of high perennial malaria transmission in western Kenya. Am J Trop Med Hyg. 2003, 68 (Suppl 4): 23-29.
Ter Kuile FO, Terlouw DJ, Phillips-Howard PA, Hawley WA, Friedman JF, Kolczak MS, Kariuki SK, Shi YP, Kwena AM, Vulule JM, Nahlen BL: Impact of permethrin-treated bed nets on malaria and all-cause morbidity in young children in an area of intense perennial malaria transmission in western Kenya: cross-sectional survey. Am J Trop Med Hyg. 2003, 68 (Suppl 4): 100-107.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24: 1596-1599. 10.1093/molbev/msm092.
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R: DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003, 19: 2496-2497. 10.1093/bioinformatics/btg359.
Librado PJR: DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009, 25: 1451-1452. 10.1093/bioinformatics/btp187.
Excoffier L, H.E L: Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010, 10: 564-567. 10.1111/j.1755-0998.2010.02847.x.
Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, Guindo AB, Kone AK, Traore K, Ouattara A, Djimde AA, Sehdev PS, Lyke KE, Diallo DA, Doumbo OK, Plowe CV: Dynamics of polymorphism in a malaria vaccine antigen at a vaccine-testing site in Mali. PLoS Med. 2007, 4: e93-10.1371/journal.pmed.0040093.
Chenet SM, Branch OH, Escalante AA, Lucas CM, Bacon DJ: Genetic diversity of vaccine candidate antigens in Plasmodium falciparum isolates from the Amazon basin of Peru. Malar J. 2008, 7: 93-10.1186/1475-2875-7-93.
Sakihama N, Ohmae H, Bakote'e B, Kawabata M, Hirayama K, Tanabe K: Limited allelic diversity of Plasmodium falciparum merozoite surface protein 1 gene from populations in the Solomon Islands. Am J Trop Med Hyg. 2006, 74: 31-40.
Dent AE, Yohn CT, Zimmerman PA, Vulule J, Kazura JW, Moormann AM: A polymerase chain reaction/ligase detection reaction fluorescent microsphere assay to determine Plasmodium falciparum MSP-119 haplotypes. Am J Trop Med Hyg. 2007, 77: 250-255.
Hughes AL: Positive selection and interallelic recombination at the merozoite surface antigen-1 (MSA-1) locus of Plasmodium falciparum. Mol Biol Evol. 1992, 9: 381-393.
Tanabe K, Sakihama N, Nakamura Y, Kaneko O, Kimura M, Ferreira MU, Hirayama K: Selection and genetic drift of polymorphisms within the merozoite surface protein-1 gene of Plasmodium falciparum. Gene. 2000, 241: 325-331. 10.1016/S0378-1119(99)00472-2.
Barry AE, Schultz L, Buckee CO, Reeder JC: Contrasting population structures of the genes encoding ten leading vaccine-candidate antigens of the human malaria parasite. Plasmodium falciparum. PLoS One. 2009, 4: e8497-10.1371/journal.pone.0008497.
Da Silveira LA, Ribeiro WL, Kirchgatter K, Wunderlich G, Matsuoka H, Tanabe K, Ferreira MU: Sequence diversity and linkage disequilibrium within the merozoite surface protein-1 (Msp-1) locus of Plasmodium falciparum: a longitudinal study in Brazil. J Eukaryot Microbiol. 2001, 48: 433-439. 10.1111/j.1550-7408.2001.tb00176.x.
Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM: Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene. 2003, 304: 65-75.
Ogutu BR, Apollo OJ, McKinney D, Okoth W, Siangla J, Dubovsky F, Tucker K, Waitumbi JN, Diggs C, Wittes J, Malkin E, Leach A, Soisson LA, Milman JB, Otieno L, Holland CA, Polhemus M, Remich SA, Ockenhouse CF, Cohen J, Ballou WR, Martin SK, Angov E, Stewart VA, Lyon JA, Heppner DG, Withers M: MSP-1 MVWG: Blood stage malaria vaccine eliciting high antigen-specific antibody concentrations confers no protection to young children in Western Kenya. PLoS One. 2009, 4: e4708-10.1371/journal.pone.0004708.
Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GA, Su XZ: Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007, 39: 126-130. 10.1038/ng1924.
Escalante AA, Grebert HM, Isea R, Goldman IF, Basco L, Magris M, Biswas S, Kariuki S, Lal AA: A study of genetic diversity in the gene encoding the circumsporozoite protein (CSP) of Plasmodium falciparum from different transmission areas–XVI. Asembo Bay Cohort Project Mol Biochem Parasitol. 2002, 125: 83-90. 10.1016/S0166-6851(02)00216-5.
Shi YP, Alpers MP, Povoa MM, Lal AA: Diversity in the immunodominant determinants of the circumsporozoite protein of Plasmodium falciparum parasites from malaria-endemic regions of Papua New Guinea and Brazil. Am J Trop Med Hyg. 1992, 47: 844-851.
Enosse S, Dobaño C, Quelhas D, Aponte JJ, Lievens M, Leach A, Sacarlal J, Greenwood B, Milman J, Dubovsky F, Cohen J, Thompson R, Ballou WR, Alonso PL, Conway DJ, Sutherland CJ: RTS, S/AS02A malaria vaccine does not induce parasite CSP T cell epitope selection and reduces multiplicity of infection. PLoS Clin Trials. 2006, 1: e5-10.1371/journal.pctr.0010005.
Waitumbi JN, Anyona SB, Hunja CW, Kifude CM, Polhemus ME, Walsh DS, Ockenhouse CF, Heppner DG, Leach A, Lievens M, Ballou WR, Cohen JD, Sutherland CJ: Impact of RTS, S/AS02(A) and RTS, S/AS01(B) on genotypes of P. falciparum in adults participating in a malaria vaccine clinical trial. PLoS One. 2009, 4: e7849-10.1371/journal.pone.0007849.
Alloueche A, Silveira H, Conway DJ, Bojang K, Doherty T, Cohen J, Pinder M, Greenwood BM: High-throughput sequence typing of T-cell epitope polymorphisms in Plasmodium falciparum circumsporozoite protein. Mol Biochem Parasitol. 2000, 106: 273-282. 10.1016/S0166-6851(99)00221-2.
Yoshida N, Di Santi SM, Dutra AP, Nussenzweig RS, Nussenzweig V, Enea V: Plasmodium falciparum: restricted polymorphism of T cell epitopes of the circumsporozoite protein in Brazil. Exp Parasitol. 1990, 71: 386-392. 10.1016/0014-4894(90)90064-J.
Doolan DL, Saul AJ, Good MF: Geographically restricted heterogeneity of the Plasmodium falciparum circumsporozoite protein: relevance for vaccine development. Infect Immun. 1992, 60: 675-682.
Bhattacharya PR, Bhatia V, Pillai CR: Genetic diversity of T-helper cell epitopic regions of circumsporozoite protein of Plasmodium falciparum isolates from India. Trans R Soc Trop Med Hyg. 2006, 100: 395-400. 10.1016/j.trstmh.2005.07.012.
Jalloh A, Van Thien H, Ferreira MU, Ohashi J, Matsuoka H, Kanbe T, Kikuchi A, Kawamoto F: Sequence variation in the T-cell epitopes of the Plasmodium falciparum circumsporozoite protein among field isolates is temporally stable: a 5-year longitudinal study in southern Vietnam. J Clin Microbiol. 2006, 44: 1229-1235. 10.1128/JCM.44.4.1229-1235.2006.
Alaii JA, Hawley WA, Kolczak MS, Ter Kuile FO, Gimnig JE, Vulule JM, Odhacha A, Oloo AJ, Nahlen BL, Phillips-Howard PA: Factors affecting use of permethrin-treated bed nets during a randomized controlled trial in western Kenya. Am J Trop Med Hyg. 2003, 68 (Suppl 4): 137-141.
Acknowledgements
We thank all the people of Rarieda Division, the field workers and study supervisors. This investigation received financial support under the Multilateral Initiative on Malaria (MIM) Project A40046 through the UNICEF/UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases (TDR) and Malaria Branch, Division of Parasitic Diseases and Malaria, Centers for Disease Control and Prevention (CDC). This paper is published with the permission of the Director KEMRI.
Disclaimer
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC) or the Kenya Medical Research Institute (KEMRI).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
SK, YPS, FK, DT, WH, PP-H, BL KL, MH, and LS designed the study. SK, JN, AM, and GM collected the molecular biology data. JW, SK, YPS, WG, JN, AM participated in the data analysis. SK and YPS wrote the paper. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kariuki, S.K., Njunge, J., Muia, A. et al. Effect of malaria transmission reduction by insecticide-treated bed nets (ITNs) on the genetic diversity of Plasmodium falciparum merozoite surface protein (MSP-1) and circumsporozoite (CSP) in western Kenya. Malar J 12, 295 (2013). https://doi.org/10.1186/1475-2875-12-295
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-12-295