Abstract
Background
A detailed analysis of the expression of 440 cancer-related genes was performed after the combined treatment of medulloblastoma cells with all-trans retinoic acid (ATRA) and inhibitors of lipoxygenases (LOX) and cyclooxygenases (COX). The combinations of retinoids and celecoxib as a COX-2 inhibitor were reported to be effective in some regimens of metronomic therapy of relapsed solid tumors with poor prognosis. Our previous findings on neuroblastoma cells using expression profiling showed that LOX/COX inhibitors have the capability of enhancing the differentiating action of ATRA. Presented study focused on the continuation of our previous work to confirm the possibility of enhancing ATRA-induced cell differentiation in these cell lines via the application of LOX/COX inhibitors. This study provides more detailed information concerning the mechanisms of the enhancement of the ATRA-induced differentiation of medulloblastoma cells.
Methods
The Daoy and D283 Med medulloblastoma cell lines were chosen for this study. Caffeic acid (an inhibitor of 5-LOX) and celecoxib (an inhibitor on COX-2) were used in combined treatment with ATRA. The expression profiling was performed using Human Cancer Oligo GEArray membranes, and the most promising results were verified using RT-PCR.
Results
The expression profiling of the selected cancer-related genes clearly confirmed that the differentiating effects of ATRA should be enhanced via its combined administration with caffeic acid or celecoxib. This effect was detected in both cell lines. An increased expression of the genes that encoded the proteins participating in induced differentiation and cytoskeleton remodeling was detected in both cell lines in a concentration-dependent manner. This effect was also observed for the CDKN1A gene encoding the p21 protein, which is an important regulator of the cell cycle, and for the genes encoding proteins that are associated with proteasome activity. Furthermore, our results showed that D283 Med cells are significantly more sensitive to treatment with ATRA alone than Daoy cells.
Conclusions
The obtained results on medulloblastoma cell lines are in accordance with our previous findings on neuroblastoma cells and confirm our hypothesis concerning the common mechanism of the enhancement of ATRA-induced cell differentiation in various types of pediatric solid tumors.
Similar content being viewed by others
Background
Medulloblastoma (MBL), an embryonal neuroectodermal tumor of the cerebellum, is the most common type of malignant brain tumor in children. Although recent advances in MBL therapy have led to a dramatic increase in survival rate, the mortality rate is currently approximately 20–40% [1]. Moreover, MBL survivors are often affected by treatment-related side effects such as growth hormone deficiency, gonadal alterations, hypo- or hyperthyroidism, and long-term cognitive, neuropsychological and academic impairments, etc. New approaches are thus needed to improve the survival rate and to reduce the negative side effects of MBL treatment [2].
The induced differentiation of tumor cells has become a promising strategy in modern antineoplastic therapy. Retinoids, which are derivatives of vitamin A, are the most frequently used group of cell differentiation inducers. The regulation of relevant cell signaling pathways via retinoids is based on the activation of two groups of nuclear receptors, RAR and RXR [3, 4]. These activated receptors can influence the transcription either directly by binding to the DNA or indirectly by interacting with other transcription factors [5, 6].
In general, retinoids play an important role in cell proliferation and differentiation, and their efficacy in the treatment of various types of tumor cells has been described both in vivo and in vitro[7–11]. However, the toxicity of and intrinsic or acquired resistance to retinoids substantially limit their use in clinical protocols [12].
Therefore, special attention has been paid to treating cancer cells with a combination of retinoids and other compounds that may enhance or prolong their antineoplastic effects. The enhancing effects of these modulators were described in several clinical trials focused on the treatment of leukemia [13–16]; they were also demonstrated under in vitro conditions using tumor cells of a neurogenic origin [17–20].
To date, many studies on various cancer cell lines have reported the additive or synergistic effects of combined treatment with retinoids and inhibitors of lipoxygenases (LOX) [21–24] or cyclooxygenases (COX) [25–27]. The molecular mechanisms of this modulation remain unknown, but the published data suggest the inhibition of the retinoid degradation pathways [28] or the cooperation of compounds that are utilized in cell signaling inhibition (through the PI3K/Akt pathway) or the induction of the mitochondrial apoptotic pathway [25].
Our previous studies were also focused on how to enhance the differentiation effect of all-trans retinoic acid (ATRA) through its combination with LOX/COX inhibitors in neuroblastoma cell lines [20, 29]. In these experiments, we used caffeic acid (CA) as an inhibitor of 5-LOX and celecoxib (CX) as an inhibitor of COX-2. Our results clearly confirmed that the ATRA-induced differentiation of neuroblastoma cells can be enhanced via the combined application of these inhibitors [20, 29]. Furthermore, data from the expression profiling of the treated cells showed an increase in the expression of the genes involved in the process of retinoid-induced neuronal differentiation, especially in cytoskeleton remodeling after combined treatment [20].
To verify these findings, we used a different type of neurogenic tumor cells, i.e. established medulloblastoma cell lines but the same experimental design for the treatment of these cells. Our new data from the gene expression profiling of medulloblastoma cells also demonstrated the capability of CA or CX to enhance the cell differentiation induced via ATRA.
Methods
Cell lines and cell culture
Daoy (ATCC HTB-186™) and D283 Med (ATCC HTB-185™) medulloblastoma cell lines were used in this study. These cell lines were chosen according their different origin (primary tumor vs. metastatic site) and their different biological features as described by supplier in the documentation to cover known heterogeneity in this disease. Both of these cell lines are widely used in medulloblastoma research. These cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F-12 medium mixture (1:1) supplemented with 20% fetal calf serum, 1% non-essential amino acids, 2 mM L-glutamine, 100 IU.ml-1 penicillin, and 100 mg.ml-1 streptomycin (all purchased from PAA Laboratories, Linz, Austria). Cell culture was performed under standard conditions at 37°C in a humidified atmosphere containing 5% CO2. Both of these cell lines were subcultured 1–2 times weekly. The Research Ethics Committee of the University Hospital Brno approved the study protocol.
Chemicals
ATRA (Sigma-Aldrich, St. Louis, MO, USA), CA (Sigma), and CX (LKT Laboratories, St. Paul, MN, USA) were prepared as stock solutions at concentrations of 100 mM in dimethyl sulfoxide (DMSO) (Sigma). For experiments, these stock solutions were diluted in fresh cell culture medium to obtain final concentrations as follow: 0.05 and 0.1 μM of ATRA for the treatment of D283 Med cells, 1 and 10 μM of ATRA for the treatment of Daoy cells, 13 and 52 μM of CA, as well as 10 and 50 μM of CX for the treatment of both cell lines.
Experimental design
The experimental design was the same as that in our previous studies [20, 29]: the cell populations were treated with ATRA alone or with ATRA and an inhibitor (CA or CX) at the concentrations mentioned above. The concentrations of ATRA and inhibitors were chosen on the basis of previously published data, and they corresponded to the plasma levels obtained using these compounds therapeutically [23, 30–33]. However, lower concentrations of 0.05 and 0.1 μM ATRA were used for treating the D283 Med cells due to the predominant cytotoxic effect on the cell populations at higher concentrations [34]. In all experiments, the cells were seeded into Petri dishes or culture flasks 24 h prior to the treatment. Untreated cells were used as controls in all experiments.
Expression profiling
The total RNA of the treated cell populations was isolated using the GenElute™ Mammalian Total RNA Miniprep Kit (Sigma), and its concentration and integrity were determined using a spectrophotometer. This isolation of RNA was performed at day 3 after treatment. The conversion of the experimental RNA into cDNA and its further transcription and biotin-UTP labeling was performed using TrueLabeling-AMP™ 2.0 cRNA (SABiosciences, Frederick, MD, USA). After purification with the SuperArray ArrayGrade cRNA Cleanup Kit, the labeled target cRNA was hybridized to Human Cancer OHS-802 Oligo GEArray membranes that profile 440 genes (both SABiosciences). The expression levels of each gene were detected via chemiluminescence using the alkaline phosphatase-conjugated streptavidin substrate. The membranes were then recorded using the MultiImage™ II Light Cabinet DE- 500 (Alpha Innotech, CA, USA). The image data were processed and analyzed using the GEArray Expression Analysis Suite software (SABiosciences) with background subtraction. All data were standardized as a ratio of the gene expression intensity to the mean expression intensity of the selected HSP90AB1 housekeeping gene, which was chosen using the GeNorm [35] and NormFinder [36] software tools. Standardized spot intensity ratios (treated/control samples) were calculated and data filtering criteria were as follows: genes with ratio higher than 2 were rated as upregulated and genes with ratio lower than 0.5 were rated as downregulated. The expression of the specific gene was evaluated as changed if the same trend of change, i.e. upregulation or downregulation was detected at least in four experimental variants (of six in total) regardless the concentrations used for treatment. Cluster analyses were performed using the GEArray Expression Analysis Suite software according to the design of the experiments, i.e., separately for each cell line and inhibitor type. DAVID software tool [37] was used for primary detection of relevant pathways.
RT-PCR
The changes in the expression of the two selected candidate genes were evaluated using RT-PCR. The RNA was isolated as described above. A total of 0.25 μg RNA was then reverse transcribed using M-MLV reverse transcriptase (Top-Bio, Prague, Czech Republic) according to the manufacturer’s protocol. RT-PCR was performed on 4 μl cDNA using Taq DNA polymerase 1.1 (Top-Bio) with human primers for the CDKN1A and GDF15 candidate genes as well as the HSP90AB1 housekeeping gene (Table 1) in 20 μl of the reaction volume. The PCR reaction was performed with denaturation at 94°C for 4 min, annealing at 60°C for 30 s, and elongation at 72°C for 1 min (35 cycles for all primers) (Table 1). A total of 10 μl of the PCR product was loaded on the 1% agarose gel and examined using electrophoresis. The optical density was stained and quantified using ImageJ software [38], and the data were normalized to HSP90AB1 expression.
Results
The present study was focused on a detailed analysis of Daoy and D283 Med medulloblastoma cells after the combined application of ATRA and LOX/COX inhibitors. CA as the specific inhibitor of 5-LOX and CX as the specific inhibitor of COX-2 were used in these experiments. The changes in the expression of cancer-related genes were evaluated using expression profiling. Furthermore, a detailed analysis of the expression of five candidate genes was performed using RT-PCR to verify the microarray results. We used the same experimental design as our previous studies on neuroblastoma cells [20, 29].In Daoy cells, changes in the expression of 80 cancer-related genes were detected after combined treatment with ATRA and inhibitors (Figure 1A). A total of 29 of these genes demonstrated changed expressions after combinations of ATRA and CA as well as of ATRA and CX. The expressions of another 29 genes were changed only after the combined treatment of ATRA and CA. A total of 22 different genes showed changes in expression after undergoing combined treatment with ATRA and CX (Figure 1A).In D283 Med cells, the expressions of 37 genes were changed (Figure 1B). Of these, 22 showed changes after combined treatment with ATRA and CA as well as with ATRA and CX. Changes in the expression of another 11 genes were identified after treatment with ATRA and CA only. Similarly, the expressions of 4 different genes were changed after treatment with ATRA and CX only (Figure 1B).The data achieved via expression profiling are presented after cluster analysis, which grouped the genes or gene groups by the type of changes in their expressions (Figure 2). Based on this analysis, three typical patterns of changes in gene expression are described:
Summary of detected changes in the expression of 440 cancer-related genes after the combined treatment of the Daoy (A) and D283 Med (B) cell lines. The black circles indicate the total number of genes with changed expression after combinations of ATRA with CA or CX. The green areas indicate the number of genes with changed expression after combined treatment with ATRA and CA only. Similarly, the violet areas indicate the number of genes with changed expression after combined treatment with ATRA and CA only. The gray overlays indicate the number of genes that demonstrated changed expressions after treatment with ATRA in both combinations; i.e., with CA as well as with CX.
Results of gene cluster analysis after the expression profiling of treated cells. The genes were clustered according to the type of changes in the expression in the respective cell line (Daoy or D283 Med) after the combined treatment with ATRA and inhibitors (CA or CX). The cells were treated with ATRA alone or in combination with CA (an inhibitor of 5-LOX) or CX (an inhibitor of COX-2); numbers indicate the concentration in μM. The green color at the farthest left end of the color scale corresponds to the minimal value; the red color at the farthest right end of the color scale corresponds to the maximal value; the black color in the middle corresponds to the average value. Each of the other values corresponds to a certain color according to its magnitude. The colors are assigned according to the value of the particular gene expression in all samples in the respective experimental variant (I, II, III or IV). Genes discussed in the text in detail are highlighted by red dots.
(i.) Genes with strong concentration-dependent changes in their expressions. This pattern was typical in the I.A, II.A, III.A, and IV.A groups, in which the upregulation of gene expression was detected. This concentration-dependent pattern was also detected in the groups with downregulated gene expressions, i.e., in the I.E., II.D, III.D, and IV.C groups.
(ii.) Genes for which the expression was upregulated (groups I.B and II.C) or downregulated (group III.C) after treatment with lower concentrations of reagents. The use of higher concentrations had no or minimal influence on the expression of these genes.
(iii.) Genes with changes in their expressions after treatment with higher concentrations of reagents only. This pattern was characteristic in the II.B and III.B groups; however, such trends were also obvious in the I.A and IV.A groups.
Similarities in the gene expression changes are summarized in Table 2 and Table 3. The genes with changed expression in a particular cell line after combined treatment with ATRA and both inhibitors (CA or CX) are given: 8 genes were upregulated and 8 genes were downregulated in the Daoy cells; 16 genes were upregulated and 3 were downregulated in the D283 Med cells (Table 2). Two of these genes, GDF15 and UBE2L6, were upregulated in both cell lines after all types of combined treatment (Table 2). Changes in gene expression after the same type of combined treatment (ATRA with CA or ATRA with CX) were identified in both cell lines; 2 of the genes were upregulated and 1 was downregulated after treatment with ATRA and CA, while 8 were upregulated and 1 was downregulated after treatment with ATRA and CX (Table 3).
The analysis of the detected changes in gene expressions showed a concentration-dependent increase in the expression of genes known to be involved in the process of retinoid-induced differentiation and especially of the genes generally associated with the regulation of the cell cycle, of genes involved in mitochondrial metabolism, etc. The observed changes in the expressions of two selected candidate genes - GDF15 (Figure 3) and CDKN1A (Figure 4) - were subsequently verified via RT-PCR.
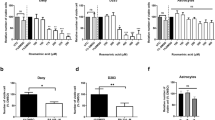
Changes in the expression of the GDF15 gene in Daoy (A) and D283 Med (B) cells analyzed using RT-PCR. The cells were treated with ATRA alone or in combination with CA (an inhibitor of 5-LOX) or CX (an inhibitor of COX-2); numbers indicate the concentrations of these compounds in μM. Respective numerical data from densitometry for three independent experiments are shown in the graphs: X-axis, treatment condition; Y-axis, relative gene expression. The data represent the means ± SD; they were normalized to the expression of the HSP90AB1 housekeeping gene and were related to the untreated control cells. Representative electrophoresis is presented for each cell line.
Changes in the expression of the CDKN1A gene in Daoy (A) and D283 Med (B) cells analyzed using RT-PCR. The cells were treated with ATRA alone or in combination with CA (an inhibitor of 5-LOX) or CX (an inhibitor of COX-2); numbers indicate the concentrations of these compounds in μM. Respective numerical data from densitometry for three independent experiments are shown in the graphs: X-axis, treatment condition; Y-axis, relative gene expression. The data represent the means ± SD; they were normalized to the expression of the HSP90AB1 housekeeping gene and were related to the untreated control cells. Representative electrophoresis is presented for each cell line.
Discussion
At present, retinoids (powerful inducers of cell differentiation) have become a part of many therapeutic regimens in pediatric oncology, especially in the treatment of solid tumors of neurogenic origin [39–43]. In our previous experiments, we demonstrated the enhancement of the ATRA-induced differentiation of neuroblastoma cells via the combined application of LOX/COX inhibitors [20, 29].
In our present study, we profiled the expression of 440 cancer-related genes in medulloblastoma cells after the same type of combined treatment (see above). Our results clearly confirmed our previous finding that ATRA-induced cell differentiation is enhanced via combined treatment with CA (inhibitor of 5-LOX) or with CX (inhibitor of COX-2) because the expression of the genes involved in the process of induced differentiation is increased in a concentration-dependent manner. A comparison of two established medulloblastoma cell lines showed a higher sensitivity in Daoy cells (compared with D283 Med cells) to the combined treatment with ATRA and inhibitors; nevertheless, our pilot experiments indicated that D283 Med cells were apparently more sensitive to the ATRA alone [34, 44]. The same difference in sensitivity of Daoy and D283 Med cell lines to the ATRA was already described and the higher sensitivity of D283 Med cells is explained by expression of OTX2, a transcription factor, which was reported as a suppressor of neuronal differentiation in medulloblastoma cells [45].
The three patterns of changes in gene expression described here were very similar to our previous results on neuroblastoma cells [20]. In terms of the individual genes, we noted significant changes in the expression of the GDF15, RHOA, and RHOC genes. Although the expression of the GDF15 gene was enhanced in both cell lines via combined treatment with ATRA and both inhibitors (Figures 2 and 3), changes in the expression of the RHOA and RHOC genes were detected in the D283 Med cells only (Figure 2). These two genes are members of the Rho GTPases family and are known to participate in cytoskeleton rearrangement and regulation processes such as changes in cell morphology during cell differentiation, proliferation and motility [46, 47]. This finding is in accordance with our pilot study in which the decrease in proliferation activity and in the formation of multicellular aggregates was reported after the combined treatment (using CA or CX) of D283 Med cells [44].
The protein encoded by the GDF15 gene is an important regulator of cell differentiation during embryonal development, especially in neural tissues [48]. This protein also has other important functions in the regulation of immune response or response to stress factors [49], and some studies have described its role as a paracrine mediator in the p53 cell signaling pathway that can inhibit cell proliferation and induce apoptosis [50, 51]. Furthermore, the expression of GDF15 correlated with the amount of the p21 protein that was encoded by the CDKN1A gene [50, 51]. Although the overexpression of GDF15 was detected in both cell lines after combined treatment with both inhibitors (Tables 2 and 3), we observed some differences between these cell lines. The expression of GDF15 increased in Daoy cells in a concentration-dependent manner (Figures 2 and 3A), but the changes in GDF15 expression in D283 Med cells were not so obvious (Figures 2 and 3B).
As mentioned above, some previous studies have demonstrated a correlation between the expressions of the GDF15 and CDKN1A genes [50, 51]. The CDKN1A gene encodes the p21 protein, which serves as an important regulator of the cell cycle progression via the inhibition of cyclin-dependent kinases. In our experiments, the expression of CDKN1A was enhanced in both cell lines after combined treatment with ATRA and CX, whereas the combination of ATRA and CA did not show a similar effect (Table 3, Figure 4). The increase in CDKN1A expression after treatment with retinoids was described in many human malignancies under in vivo and in vitro conditions: acute promyelocytic leukemia [52], acute T-lymphoblastic leukemia [53], pre-B lymphoma [54], hepatoblastoma [55, 56], and neuroblastoma [57–60]. Our in vitro data confirmed these findings (especially in D283 Med cells) and also clearly showed that this effect of ATRA can be enhanced through its combined administration with CX (Figure 4B).
Furthermore, we detected the upregulated expression of the UBE2L6 gene, which encodes a member of the E2 ubiquitin-conjugating enzyme family in both cell lines after combined treatment with both inhibitors (Tables 2 and 3). The overexpression of the ACP2 gene encoding a beta-subunit of lysosomal acid phosphatase was observed in Daoy cells after combined treatment with both inhibitors (Table 2). The upregulated expression of both of these genes indicated an increased activity of proteasome in the treated cells. Higher proteasome activity was reported in breast carcinoma cells [61] as well as in acute promyelocytic leukemia cells [62, 63] after treatment with retinoids. However, the relationship between retinoids and proteasome activity should be associated with resistance to retinoids [64]. This mechanism of resistance to retinoids, i.e., the increased activity of proteasome in Daoy cells, could be one of the possible explanations for the differing sensitivity demonstrated by these cell lines to ATRA treatment, as reported in our pilot study [44]. This hypothesis is also supported by the fact that the upregulation of the UCHL1 gene encoding a deubiquitinating enzyme was detected in more sensitive D283 Med cells after combined treatment with both inhibitors (Table 2). Moreover, the protein encoded by this gene was found solely in mature neurons [65]; its increased expression in D283 Med, which is apparently enhanced via a combined treatment with ATRA and inhibitors, should indicate the neuronal differentiation of D283 Med after experimental treatment.
Higher sensitivity of D283 Med cells to the ATRA-induced neuronal differentiation is also indicated by the overexpression of two genes encoding subunits of cytochrome C oxidase, COX6C and COX7A2, after both types of combined treatment (Table 2). An increased mitochondrial activity after treatment with retinoids was reported in neuroblastoma cell lines [66], and it was demonstrated that more differentiated neuronal cells exhibit higher oxygen consumption rates as well as metabolic rates [67]. These findings thus support the presumed neuronal differentiation of D283 Med cells treated with ATRA and the capability of CA and CX to enhance this effect. Furthermore, a similar upregulation of these two genes was previously detected in neuroblastoma cell lines after the same type of experimental treatment [20].
The presented results are closely connected to our previously published studies reporting encouraging treatment responses to metronomic therapy in children suffering from some types of relapsed solid tumors with poor prognosis [42, 43, 68]. In these protocols, retinoids are administered in combination with celecoxib (as an anti-angiogenic agent) and several cytotoxic agents. The usefulness of this type of metronomic therapy was also demonstrated in patients with MBL [43, 69, 70]. In light of this, our experimental data clearly demonstrated that the combined administration of retinoids and celecoxib should also be beneficial in enhancing tumor cell differentiation. Furthermore, a very similar effect could be achieved through the dietary uptake of plant phenolic compounds including caffeic acid [71, 72].
Conclusion
To summarize, our results on two established medulloblastoma cell lines – Daoy and D283 Med – confirmed our previous findings in leukemia and neuroblastoma cells that the differentiating effects of ATRA should be enhanced in its combined administration with caffeic acid (an inhibitor of 5-LOX) or celecoxib (an inhibitor of COX-2). This effect was apparently achieved in both cell lines via the increased expression of genes encoding proteins participating in inducing the differentiation and cytoskeleton remodeling (GDF-15, Rho GTPases) or the p21 protein, which is an important regulator of the cell cycle and of proteins associated with proteasome activity. Furthermore, our results showed an important difference between the established MBL cell lines: the Daoy cells showed the same sensitivity as the cell lines that were derived from other types of pediatric solid tumors, but the D283 Med cells were significantly more sensitive to the treatment with ATRA alone (this effect was further enhanced via combined treatment with LOX/COX inhibitors). To clarify detailed mechanisms of such difference, additional experiments concerning more cell lines derived from various MBL subtypes are needed. Nevertheless, the obtained results confirmed our initial hypothesis regarding the common mechanism of enhancement in ATRA-induced cell differentiation in various types of pediatric solid tumors.
Abbreviations
- ATRA:
-
All-trans retinoic acid
- COX-2:
-
Cyclooxygenase 2
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- DMSO:
-
Dimethyl sulfoxide
- 5-LOX:
-
5-lipoxygenase
- MBL:
-
Medulloblastoma.
References
Gerber NU, Mynarek M, von Hoff K, Friedrich C, Resch A, Rutkowski S: Recent developments and current concepts in medulloblastoma. Cancer Treat Rev. 2014, 40: 356-365. 10.1016/j.ctrv.2013.11.010.
Massimino M, Giangaspero F, Garre ML, Gandola L, Poggi G, Biassoni V, Gatta G, Rutkowski S: Childhood medulloblastoma. Crit Rev Oncol Hematol. 2011, 79: 65-83. 10.1016/j.critrevonc.2010.07.010.
Dragnev KH, Petty WJ, Dmitrovsky E: Retinoid targets in cancer therapy and chemoprevention. Cancer Biol Ther. 2003, 2: S150-S156.
Brtko J, Dvorak Z: Role of retinoids, rexinoids and thyroid hormone in the expression of cytochrome p450 enzymes. Curr Drug Metab. 2011, 12: 71-88. 10.2174/138920011795016881.
Dedieu S, Lefebvre P: Retinoids interfere with the AP1 signalling pathway in human breast cancer cells. Cell Signal. 2006, 18: 889-898. 10.1016/j.cellsig.2005.08.001.
Masia S, Alvarez S, de Lera AR, Barettino D: Rapid, nongenomic actions of retinoic acid on phosphatidylinositol-3-kinase signaling pathway mediated by the retinoic acid receptor. Mol Endocrinol. 2007, 21: 2391-2402. 10.1210/me.2007-0062.
Cruz FD, Matushansky I: Solid tumor differentiation therapy - is it possible?. Oncotarget. 2012, 3: 559-567.
Hallahan AR, Pritchard JI, Chandraratna RA, Ellenbogen RG, Geyer JR, Overland RP, Strand AD, Tapscott SJ, Olson JM: BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat Med. 2003, 9: 1033-1038. 10.1038/nm904.
Garattini E, Gianni M, Terao M: Retinoids as differentiating agents in oncology: a network of interactions with intracellular pathways as the basis for rational therapeutic combinations. Curr Pharm Des. 2007, 13: 1375-1400. 10.2174/138161207780618786.
Andres D, Keyser BM, Petrali J, Benton B, Hubbard KS, McNutt PM, Ray R: Morphological and functional differentiation in BE(2)-M17 human neuroblastoma cells by treatment with Trans-retinoic acid. BMC Neurosci. 2013, 14: 49-10.1186/1471-2202-14-49.
Maurer BJ, Kang MH, Villablanca JG, Janeba J, Groshen S, Matthay KK, Sondel PM, Maris JM, Jackson HA, Goodarzian F, Shimada H, Czarnecki S, Hasenauer B, Reynolds CP, Marachelian A: Phase I trial of fenretinide delivered orally in a novel organized lipid complex in patients with relapsed/refractory neuroblastoma: a report from the New Approaches to Neuroblastoma Therapy (NANT) consortium. Pediatr Blood Cancer. 2013, 60: 1801-1808. 10.1002/pbc.24643.
Patatanian E, Thompson DF: Retinoic acid syndrome: a review. J Clin Pharm Ther. 2008, 33: 331-338. 10.1111/j.1365-2710.2008.00935.x.
Kuendgen A, Gattermann N: Valproic acid for the treatment of myeloid malignancies. Cancer. 2007, 110: 943-954. 10.1002/cncr.22891.
Tomita A, Kiyoi H, Naoe T: Mechanisms of action and resistance to all-trans retinoic acid (ATRA) and arsenic trioxide (As2O 3) in acute promyelocytic leukemia. Int J Hematol. 2013, 97: 717-725. 10.1007/s12185-013-1354-4.
Nowak D, Stewart D, Koeffler HP: Differentiation therapy of leukemia: 3 decades of development. Blood. 2009, 113: 3655-3665. 10.1182/blood-2009-01-198911.
Lo-Coco F, Avvisati G, Vignetti M, Thiede C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, di Bona E, Specchia G, Sica S, Divona M, Levis A, Fiedler W, Cerqui E, Breccia M, Fioritoni G, Salih HR, Cazzola M, Melillo L, Carella AM, Brandts CH, Morra E, von Lilienfeld-Toal M, Hertenstein B, Wattad M, Lübbert M, Hänel M, Schmitz N: Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013, 369: 111-121. 10.1056/NEJMoa1300874.
Haque A, Banik NL, Ray SK: Emerging role of combination of all-trans retinoic acid and interferon-gamma as chemoimmunotherapy in the management of human glioblastoma. Neurochem Res. 2007, 32: 2203-2209. 10.1007/s11064-007-9420-z.
De los Santos M, Zambrano A, Aranda A: Combined effects of retinoic acid and histone deacetylase inhibitors on human neuroblastoma SH-SY5Y cells. Mol Cancer Ther. 2007, 6: 1425-1432. 10.1158/1535-7163.MCT-06-0623.
Frumm SM, Fan ZP, Ross KN, Duvall JR, Gupta S, VerPlank L, Suh BC, Holson E, Wagner FF, Smith WB, Paranal RM, Bassil CF, Qi J, Roti G, Kung AL, Bradner JE, Tolliday N, Stegmaier K: Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol. 2013, 20: 713-725. 10.1016/j.chembiol.2013.03.020.
Chlapek P, Redova M, Zitterbart K, Hermanova M, Sterba J, Veselska R: Enhancement of ATRA-induced differentiation of neuroblastoma cells with LOX/COX inhibitors: an expression profiling study. J Exp Clin Cancer Res. 2010, 29: 45-10.1186/1756-9966-29-45.
Avis I, Martinez A, Tauler J, Zudaire E, Mayburd A, Abu-Ghazaleh R, Ondrey F, Mulshine JL: Inhibitors of the arachidonic acid pathway and peroxisome proliferator-activated receptor ligands have superadditive effects on lung cancer growth inhibition. Cancer Res. 2005, 65: 4181-4190. 10.1158/0008-5472.CAN-04-3441.
Kuo HC, Kuo WH, Lee YJ, Wang CJ, Tseng TH: Enhancement of caffeic acid phenethyl ester on all-trans retinoic acid-induced differentiation in human leukemia HL-60 cells. Toxicol Appl Pharmacol. 2006, 216: 80-88. 10.1016/j.taap.2006.04.007.
Veselska R, Zitterbart K, Auer J, Neradil J: Differentiation of HL-60 myeloid leukemia cells induced by all-trans retinoic acid is enhanced in combination with caffeic acid. Int J Mol Med. 2004, 14: 305-310.
Bell E, Ponthan F, Whitworth C, Westermann F, Thomas H, Redfern CP: Cell Survival Signalling through PPARdelta and Arachidonic Acid Metabolites in Neuroblastoma. PLoS One. 2013, 8: e68859-10.1371/journal.pone.0068859.
Schroeder CP, Kadara H, Lotan D, Woo JK, Lee HY, Hong WK, Lotan R: Involvement of mitochondrial and Akt signaling pathways in augmented apoptosis induced by a combination of low doses of celecoxib and N-(4-hydroxyphenyl) retinamide in premalignant human bronchial epithelial cells. Cancer Res. 2006, 66: 9762-9770. 10.1158/0008-5472.CAN-05-4124.
Simeone AM, Li YJ, Broemeling LD, Johnson MM, Tuna M, Tari AM: Cyclooxygenase-2 is essential for HER2/neu to suppress N- (4-hydroxyphenyl) retinamide apoptotic effects in breast cancer cells. Cancer Res. 2004, 64: 1224-1228. 10.1158/0008-5472.CAN-03-2188.
Liu JP, Wei HB, Zheng ZH, Guo WP, Fang JF: Celecoxib increases retinoid sensitivity in human colon cancer cell lines. Cell Mol Biol Lett. 2010, 15: 440-450. 10.2478/s11658-010-0016-2.
Soda M, Hu D, Endo S, Takemura M, Li J, Wada R, Ifuku S, Zhao HT, El-Kabbani O, Ohta S, Yamamura K, Toyooka N, Hara A, Matsunaga T: Design, synthesis and evaluation of caffeic acid phenethyl ester-based inhibitors targeting a selectivity pocket in the active site of human aldo-keto reductase 1B10. Eur J Med Chem. 2012, 48: 321-329.
Redova M, Chlapek P, Loja T, Zitterbart K, Hermanova M, Sterba J, Veselska R: Influence of LOX/COX inhibitors on cell differentiation induced by all-trans retinoic acid in neuroblastoma cell lines. Int J Mol Med. 2010, 25: 271-280.
Nardini M, Scaccini C, Packer L, Virgili F: In vitro inhibition of the activity of phosphorylase kinase, protein kinase C and protein kinase A by caffeic acid and a procyanidin-rich pine bark (Pinus marittima) extract. Biochim Biophys Acta. 2000, 1474: 219-225. 10.1016/S0304-4165(00)00009-X.
Dandekar DS, Lopez M, Carey RI, Lokeshwar BL: Cyclooxygenase-2 inhibitor celecoxib augments chemotherapeutic drug-induced apoptosis by enhancing activation of caspase-3 and -9 in prostate cancer cells. Int J Cancer. 2005, 115: 484-492. 10.1002/ijc.20878.
Kang KB, Zhu C, Yong SK, Gao Q, Wong MC: Enhanced sensitivity of celecoxib in human glioblastoma cells: Induction of DNA damage leading to p53-dependent G1 cell cycle arrest and autophagy. Mol Cancer. 2009, 8: 66-10.1186/1476-4598-8-66.
Graff J, Skarke C, Klinkhardt U, Watzer B, Harder S, Seyberth H, Geisslinger G, Nusing RM: Effects of selective COX-2 inhibition on prostanoids and platelet physiology in young healthy volunteers. J Thromb Haemost. 2007, 5: 2376-2385. 10.1111/j.1538-7836.2007.02782.x.
Gumireddy K, Sutton LN, Phillips PC, Reddy CD: All-trans-retinoic acid-induced apoptosis in human medulloblastoma: activation of caspase-3/poly(ADPribose) polymerase 1 pathway. Clin Cancer Res. 2003, 9: 4052-4059.
Vandesompele J, de Preter K, Pattyn F, Poppe B, van Roy N, de Paepe A, Speleman F: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3: RESEARCH0034-
Andersen CL, Jensen JL, Orntoft TF: Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64: 5245-5250. 10.1158/0008-5472.CAN-04-0496.
Huang DW, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009, 4: 44-57.
Schneider CA, Rasband WS, Eliceiri KW: NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012, 9: 671-675. 10.1038/nmeth.2089.
Reynolds CP, Lemons RS: Retinoid therapy of childhood cancer. Hematol Oncol Clin North Am. 2001, 15: 867-910. 10.1016/S0889-8588(05)70256-2.
Reynolds CP, Matthay KK, Villablanca JG, Maurer BJ: Retinoid therapy of high-risk neuroblastoma. Cancer Lett. 2003, 197: 185-192. 10.1016/S0304-3835(03)00108-3.
Sterba J: Contemporary therapeutic options for children with high risk neuroblastoma. Neoplasma. 2002, 49: 133-140.
Sterba J, Valik D, Mudry P, Kepak T, Pavelka Z, Bajciova V, Zitterbart K, Kadlecova V, Mazanek P: Combined biodifferentiating and antiangiogenic oral metronomic therapy is feasible and effective in relapsed solid tumors in children: single-center pilot study. Onkologie. 2006, 29: 308-313. 10.1159/000093474.
Zapletalova D, Andre N, Deak L, Kyr M, Bajciova V, Mudry P, Dubska L, Demlova R, Pavelka Z, Zitterbart K, Skotakova J, Husek K, Martincekova A, Mazanek P, Kepak T, Doubek M, Kutnikova L, Valik D, Sterba J: Metronomic chemotherapy with the COMBAT regimen in advanced pediatric malignancies: a multicenter experience. Oncology. 2012, 82: 249-260. 10.1159/000336483.
Redova M: Enhancement of differentiation inductors’ effect on the solid tumors model in vitro. PhD thesis. Brno 2010, Masaryk University, Department of Experimental Biology
Bai RY, Staedtke V, Lidov HG, Eberhart CG, Riggins GJ: OTX2 represses myogenic and neuronal differentiation in medulloblastoma cells. Cancer Res. 2012, 72: 5988-6001. 10.1158/0008-5472.CAN-12-0614.
David M, Petit D, Bertoglio J: Cell cycle regulation of Rho signaling pathways. Cell Cycle. 2012, 11: 3003-3010. 10.4161/cc.21088.
Bustelo XR, Sauzeau V, Berenjeno IM: GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007, 29: 356-370. 10.1002/bies.20558.
Strelau J, Strzelczyk A, Rusu P, Bendner G, Wiese S, Diella F, Altick AL, von Bartheld CS, Klein R, Sendtner M, Unsicker K: Progressive postnatal motoneuron loss in mice lacking GDF-15. J Neurosci. 2009, 29: 13640-13648. 10.1523/JNEUROSCI.1133-09.2009.
Mimeault M, Batra SK: Divergent molecular mechanisms underlying the pleiotropic functions of macrophage inhibitory cytokine-1 in cancer. J Cell Physiol. 2010, 224: 626-635. 10.1002/jcp.22196.
Li PX, Wong J, Ayed A, Ngo D, Brade AM, Arrowsmith C, Austin RC, Klamut HJ: Placental transforming growth factor-beta is a downstream mediator of the growth arrest and apoptotic response of tumor cells to DNA damage and p53 overexpression. J Biol Chem. 2000, 275: 20127-20135. 10.1074/jbc.M909580199.
Yang H, Filipovic Z, Brown D, Breit SN, Vassilev LT: Macrophage inhibitory cytokine-1: a novel biomarker for p53 pathway activation. Mol Cancer Ther. 2003, 2: 1023-1029.
Ryningen A, Stapnes C, Paulsen K, Lassalle P, Gjertsen BT, Bruserud O: In vivo biological effects of ATRA in the treatment of AML. Expert Opin Investig Drugs. 2008, 17: 1623-1633. 10.1517/13543784.17.11.1623.
Luo P, Lin M, Chen Y, Yang B, He Q: Function of retinoid acid receptor alpha and p21 in all-trans-retinoic acid-induced acute T-lymphoblastic leukemia apoptosis. Leuk Lymphoma. 2009, 50: 1183-1189. 10.1080/10428190902934936.
Bao GC, Wang JG, Jong A: Increased p21 expression and complex formation with cyclin E/CDK2 in retinoid-induced pre-B lymphoma cell apoptosis. FEBS Lett. 2006, 580: 3687-3693. 10.1016/j.febslet.2006.05.052.
Lim JS, Park SH, Jang KL: All-trans retinoic acid induces cellular senescence by up-regulating levels of p16 and p21 via promoter hypomethylation. Biochem Biophys Res Commun. 2011, 412: 500-505. 10.1016/j.bbrc.2011.07.130.
Park SH, Lim JS, Jang KL: All-trans retinoic acid induces cellular senescence via upregulation of p16, p21, and p27. Cancer Lett. 2011, 310: 232-239. 10.1016/j.canlet.2011.07.009.
Wainwright LJ, Lasorella A, Iavarone A: Distinct mechanisms of cell cycle arrest control the decision between differentiation and senescence in human neuroblastoma cells. Proc Natl Acad Sci U S A. 2001, 98: 9396-9400. 10.1073/pnas.161288698.
Liu Y, Encinas M, Comella JX, Aldea M, Gallego C: Basic helix-loop-helix proteins bind to TrkB and p21(Cip1) promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol Cell Biol. 2004, 24: 2662-2672. 10.1128/MCB.24.7.2662-2672.2004.
Marzinke MA, Clagett-Dame M: The all-trans retinoic acid (atRA)-regulated gene Calmin (Clmn) regulates cell cycle exit and neurite outgrowth in murine neuroblastoma (Neuro2a) cells. Exp Cell Res. 2012, 318: 85-93. 10.1016/j.yexcr.2011.10.002.
Qiao J, Paul P, Lee S, Qiao L, Josifi E, Tiao JR, Chung DH: PI3K/AKT and ERK regulate retinoic acid-induced neuroblastoma cellular differentiation. Biochem Biophys Res Commun. 2012, 424: 421-426. 10.1016/j.bbrc.2012.06.125.
Son SH, Yu E, Ahn Y, Choi EK, Lee H, Choi J: Retinoic acid attenuates promyelocytic leukemia protein-induced cell death in breast cancer cells by activation of the ubiquitin-proteasome pathway. Cancer Lett. 2007, 247: 213-223. 10.1016/j.canlet.2006.04.005.
Isakson P, Bjoras M, Boe SO, Simonsen A: Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010, 116: 2324-2331. 10.1182/blood-2010-01-261040.
Trocoli A, Mathieu J, Priault M, Reiffers J, Souquere S, Pierron G, Besancon F, Djavaheri-Mergny M: ATRA-induced upregulation of Beclin 1 prolongs the life span of differentiated acute promyelocytic leukemia cells. Autophagy. 2011, 7: 1108-1114. 10.4161/auto.7.10.16623.
Ferry C, Gaouar S, Fischer B, Boeglin M, Paul N, Samarut E, Piskunov A, Pankotai-Bodo G, Brino L, Rochette-Egly C: Cullin 3 mediates SRC-3 ubiquitination and degradation to control the retinoic acid response. Proc Natl Acad Sci U S A. 2011, 108: 20603-20608. 10.1073/pnas.1102572108.
Day IN, Thompson RJ: UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog Neurobiol. 2010, 90: 327-362. 10.1016/j.pneurobio.2009.10.020.
Schneider L, Giordano S, Zelickson BR, SJ M, AB G, Ouyang X, Fineberg N, Darley-Usmar VM, Zhang J: Differentiation of SH-SY5Y cells to a neuronal phenotype changes cellular bioenergetics and the response to oxidative stress. Free Radic Biol Med. 2011, 51: 2007-2017. 10.1016/j.freeradbiomed.2011.08.030.
Xun Z, Lee DY, Lim J, Canaria CA, Barnebey A, Yanonne SM, McMurray CT: Retinoic acid-induced differentiation increases the rate of oxygen consumption and enhances the spare respiratory capacity of mitochondria in SH-SY5Y cells. Mech Ageing Dev. 2012, 133: 176-185. 10.1016/j.mad.2012.01.008.
Malik PS, Raina V, Andre N: Metronomics as maintenance treatment in oncology: time for chemo-switch. Front Oncol. 2014, epub ahead of print, doi: 10.3389/fonc.2014.00076
Choi LMR, Rood B, Kamani N, La Fond D, Packer RJ, Santi MR, MacDonald TJ: Feasibility of metronomic maintenance chemotherapy following high-dose chemotherapy for malignant central nervous system tumors. Pediatr Blood Cancer. 2008, 50: 970-975. 10.1002/pbc.21381.
Sterba J, Pavelka Z, Andre N, Ventruba J, Skotakova J, Bajciova V, Bronisova D, Dubska L, Valik D: Second complete remission of relapsed medulloblastoma induced by metronomic chemotherapy. Pediatr Blood Cancer. 2010, 54: 616-617.
Jaganathan SK, Mandal M: Antiproliferative effects of honey and of its polyphenols: a review. J Biomed Biotechnol. 2009, 2009: 830616-
Korkina LG: Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell Mol Biol (Noisy-le-Grand). 2007, 53: 15-25.
Acknowledgements
This study was supported by grant IGA NR9341-3/2007, by the European Regional Development Fund, as well as by the State Budget of the Czech Republic – project RECAMO CZ.1.05/2.1.00/03.0101 and the project CEB CZ.1.07/2.3.00/20.0183. We are grateful to Mrs. Johana Maresova for her technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PC carried out the experiments with the cell lines, performed the expression profiling and RT-PCR and drafted the manuscript. JN participated in manuscript preparation and the experiments concerning RT-PCR and expression profiling and also in manuscript preparation. MR participated in the experiments with the cell lines. KZ participated in the analysis of results and in manuscript preparation. JS coordinated this study and participated in manuscript preparation. RV conceived the study, participated in the analysis of results and drafted the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Chlapek, P., Neradil, J., Redova, M. et al. The ATRA-induced differentiation of medulloblastoma cells is enhanced with LOX/COX inhibitors: an analysis of gene expression. Cancer Cell Int 14, 51 (2014). https://doi.org/10.1186/1475-2867-14-51
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2867-14-51