Abstract
Background
The Notch signaling pathway is crucial in T-cell development, Notch 1 mutations are frequently present in T-cell acute lymphoblastic leukemia (T-ALL). To investigate the feature of Notch 1 mutation and its corresponding expression level in Chinese patients with T-ALL, detection of mutation and the expression level of Notch 1 gene was preformed using RT-PCR, sequencing and real-time PCR respectively.
Results
Two Notch 1 point mutations (V1578E and L1593P) located on HD-N domain were identified in three cases out of 13 T-ALL patients. The mutation on 4733 position (V1578E) found in two cases was a novel mutation. The overexpression of Notch 1 was detected in all samples with T-ALL, moreover, significantly higher expression of Notch 1 was detected in the T-ALL with Notch 1 mutation group compared with T-ALL with WT Notch 1 group (p = 0.0192).
Conclusions
Higher expression of Notch 1 was associated with Notch 1 mutation, more novel mutation of this gene might be identified in different populations and its contribution to the molecular pathogenesis of T-ALL is needed further research.
Similar content being viewed by others
Background
T-cell acute lymphoblastic leukemia (T-ALL) which occurs mainly via the proliferation of malignant T cell clones, accounts for 15% of newly diagnosed ALL cases in children and 20-25% of ALL cases in adults [1]. Overall, these are aggressive malignancies that do not respond well to chemotherapy and have a poorer prognosis than their B-cell counterparts [2]. Complex acquired genetic aberrations include chromosomal translocations (frequently involving TCR), as well as gene rearrangements and mutations resulting in abnormal expression of oncogenes like Notch 1 may be associated with the advance and resistance to treatment of this disease [3].
Notch 1 was discovered in 1991 through analysis of rare T-cell lymphoblastic leukemia/lymphoma with balanced (7;9) translocation [4]. Acquired Notch 1 mutations are present in about 50% of T-ALL [5, 6]. More than hundred different mutations frequently involved in heterodimerization domain (HD), transactivation domain (TAD) and praline, glutamic acid, serine, threonine-rich (PEST) domains of Notch 1 were reported in patients with T-ALL from a lot of researcher groups in different countries [5–10]. Little is known the incidence and feature of Notch 1 mutations in Chinese T-ALL patients [10], in this study, we detected the Notch 1 mutations in 13 Chinese patients with T-ALL and analyzed the corresponding expression level of Notch 1 gene.
Materials and methods
Samples
Thirteen newly diagnosed and untreated cases of T-ALL, 11 males and 2 females (6-55 years old; median age: 23.5 years) were included in this study, along with 20 healthy individuals as controls. The samples were collected with informed consent. All procedures were conducted in accordance with the guidelines of the medical ethics committees of the Health Bureau of Guangdong Province, China. The peripheral blood mononuclear cells (PBMCs), RNA extraction using Trizol reagent (Trizol®, Invitrogen, Carlsbad, CA, USA) and cDNA synthesis using random hexamer primers and reverse transcriptase (SuperScript® III, Invitrogen, Carlsbad, CA, USA) were performed according to the manufacturer's instructions.
RT-PCR and sequencing
To amplify different domain and exon of Notch 1 according the structure of the Notch 1 gene, 4-pair primers were purchased, which covered different exons, where the mutations happen frequently (Table 1) [5, 6]. RT-PCR was performed as our previous study, positive control (Jurkat cell line) and negative control (non-template) were included in each reaction [11, 12]. The PCR products were directly sequenced using a BigDye Terminator v3.1 Cycle Sequencing kit (Perkin Elmer, ABI) and the ABI PRISM 3100-Avant genetic analyzer. The sequences from different samples of T-ALL were analyzed with the BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify the mutations of Notch 1 gene.
Real-time quantitative RT-PCR (qRT-PCR)
Expression levels of Notch1 and the reference gene β2M were determined by SYBR Green I real-time PCR. PCR was performed as our previous description [12, 13]. The 2(-ΔΔT) method was used to present the data of the genes of interest relative to an internal control gene [12–14]. The sequences of primers used in qRT-PCR were listed in Table 1.
Statistical analysis
Univariate analyses were done using the Mann-Whitney test to compare manes of Notch1 expression level between T-ALL with Notch 1 mutations or with wild-type (WT) Notch 1 status. P < 0.05 was considered as statistically significant.
Results
Notch 1 mutation and polymorphirsm in T-ALL
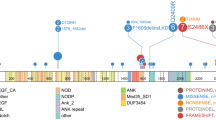
Notch 1 expression was detected in all of 13 cases with T-ALL by RT-PCR (Figure 1). The search for Notch 1 mutation was performed in exon 26, 27 and 28 (HD), TAD and PEST domain. The PCR products were direct sequenced and revealed two mutations at nucleotide position 4733 and 4778 respectively. The mutation on 4733 position was identified in two different cases, the nucleotide sequence was changed from GTG to GAG (codon 1578), thereby leading to an amino acid exchange (Valine to Glutamic), this was a novel mutation. While the mutation on 4778 position was identified in one case, the nucleotide sequence was changed from CTG to CCG (codon 1593), leading to an amino acid exchange from Leucine to Proline. All of mutations presented were heterozygous genetic alteration. And the mutations were located on HD N-terminus (Figure 2). Sequence analysis of Notch 1 segments revealed further a nucleotide exchange in all samples analyzed at position 5094, where we found a cytosine instead of a thymine, the alteration affected the third position of the codon 1698 and did not alter the amino acid sequence (data not shown).
Expression level of Notch 1 in T-ALL
In order to compare the expression level of Notch 1 gene in different T-ALL samples with Notch 1 mutations or with wild-type (WT) Notch 1 status, the expression level of Notch 1 gene was analyzed by real-time PCR, in comparison with healthy controls (0.37 ± 0.33), significantly higher expression of Notch1 was found in both T-ALL with WT Notch 1 (6.34 ± 7.03) (p = 0.0006) or with Notch 1 mutations (Mut Notch 1) (20.47 ± 10.81) (p < 0.0001) (Figure 3). Moreover, significantly higher expression of Notch 1 was detected in the T-ALL with Mut Notch 1 group compared with WT Notch 1 group (p = 0.0192) (Figure 3). Interesting, the expression level of Notch1 in two cases with WT Notch 1 T-ALL (24.28 and 9.06 respectively), which were not identified any mutation in the present study, was as high as the samples with Notch 1 mutation.
Discussion
Notch 1 signaling is crucial for T-cell differentiation and proliferation, mutational activation of Notch 1 is an important factor in T-ALL pathogenesis [5, 7]. Translocation and mutations of Notch 1 may alter its function resulting in overexpression and independent activation [15]. In this study, Notch 1 mutations were identified in 3 Chinese patients with T-ALL, the incidence of Notch 1 mutation was only 23.08% (3/13), it seemed relatively low in comparison with previous studies from different European and American countries [5, 6, 16]. There are rare studies described the incidence of Notch 1 mutation in Chinese cases, one report by Zhu et al showed that Notch 1 mutation was found in 29 patient out of 77 cases (37.7%) with Chinese T-ALL [10]. Similar incidence (22%) of Notch 1 mutations was reported by a research group from Turkey [8]. However, further research is needed to collect and investigate more samples and find out the representational results.
The higher frequency of Notch 1 mutation is found in HD, TAD and PEST domains [5–10]. In the present study, we used the 4 pair primers covered all the HD, TAD and PEST domains to amplify and sequence. Two mutations were identified in three cases, one was the mutation on 4778 position (L1593P) which was reported in previous studies [10, 17], while the another mutation was identified on 4733 position (V1578E) in two different cases with T-ALL, to our best knowledge, this is a novel identified mutation. The effect of the novel mutation is needed to evaluate by further functional analysis. Both mutations were located at HD N-terminus (HD-N) domain. The Notch 1 HD-N mutation may destabilize the subunit interaction and do not require the ligand-binding domain to activate signaling, resulting in constitutive Notch 1 activation and subsequent cell transformation [9, 17]. Based on the results, we compared the expression level of Notch 1 in T-ALL with WT Notch 1 or with Mut Notch 1 group, definitive result indicated that the expression level of Notch 1 was significant associated with Notch 1 mutation in HD-N domain, significantly higher expression of Notch 1 was detected in the T-ALL with Mut Notch 1 group compared with WT Notch 1 T-ALL group. However, overexpression of Notch 1 was a common feature in all T-ALL patients, whether the mechanism of Notch 1 overexpression in mutation or WT samples is different, it remains an open question. Although no mutation was detected in HD, TAD or PEST domains, high expression level of Notch1 in two cases with T-ALL in the present study was thought that might has potential mutation in the other domains, whole notch 1 gene sequence analysis for these cases is needed to follow up.
In the present study, we were unable to identify mutation of Notch 1 in PEST domain which regulates protein turnover by targeting proteins to the ubiquitin-proteosome complex for subsequent degradation [9, 15]. However, a high incidence (4/15 cases) of Notch 1 mutation in PEST domain was reported in Indian T-ALL patients [9], while lower incidence was described from a study in Chinese T-ALL patients (5/77, 6.5%) [15], as well as in Turkish patients (7%) and German patients (8.2%) [7, 18], the difference may due to the racial diversify.
In summary, Notch 1 mutations including a novel mutation were identified in a small cohort of Chinese T-ALL cases, and concomitant significantly higher expression level of Notch 1 was found. More ongoing study was performed to follow up its predictive value and to elucidate its contribution to the molecular pathogenesis of T-ALL.
References
Pui CH, Relling MV, Downing JR: Acute lymphoblastic leukemia. N Engl J Med. 2004, 350: 1535-48. 10.1056/NEJMra023001.
Morris JC, Waldmann TA, Janik JE: Receptor-directed therapy of T-cell leukemias and Lymphomas. J Immunotoxicol. 2008, 5: 235-48. 10.1080/15476910802129661.
Vanura K, Vrsalovic MM, Le T, Marculescu R, Kusec R, Jäger U, Nadel B: V(D)J targeting mistakes occur at low frequency in acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009, 48: 725-36. 10.1002/gcc.20677.
Aster JC, Blacklow SC, Pear WS: Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011, 223: 262-73.
Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, Beldjord K, Reman O, Witz F, Fagot T, Tavernier E, Turlure P, Leguay T, Huguet F, Vernant JP, Daniel F, Béné MC, Ifrah N, Thomas X, Dombret H, Macintyre E: NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a group for research on adult acute lymphoblastic leukemia (GRAALL) study. Blood. 2009, 17: 3918-24.
Mansour MR, Duke V, Foroni L, Patel B, Allen CG, Ancliff PJ, Gale RE, Linch DC: Notch1 mutations are secondary events in some patients with T-Cell acute lymphoblastic leukemia. Clin Cancer Res. 2007, 13: 6964-9. 10.1158/1078-0432.CCR-07-1474.
Erbilgin Y, Sayitoglu M, Hatirnaz O, Dogru O, Akcay A, Tuysuz G, Celkan T, Aydogan G, Salcioglu Z, Timur C, Yuksel-Soycan L, Ure U, Anak S, Agaoglu L, Devecioglu O, Yildiz I, Ozbek U: Prognostic significance of NOTCH1 and FBXW7 mutations in pediatric T-ALL. Dis Markers. 2010, 28: 353-60.
Mansur MB, Ford AM, van Delft FW, Gonzalez D, Emerenciano M, Maia RC, Greaves M, Pombo-de-Oliveira MS: Occurrence of identical NOTCH1 mutation in non-twinned sisters with T-cell acute lymphoblastic leukemia. Leukemia. 2011, 25: 1368-70. 10.1038/leu.2011.96.
Bhanushali AA, Babu S, Thangapandi VR, Pillai R, Chheda P, Das BR: Mutations in the HD and PEST domain of Notch-1 receptor in T-cell acute lymphoblastic leukemia: report of novel mutations from Indian population. Oncol Res. 2010, 19: 99-104. 10.3727/096504010X12864748215007.
Zhu YM, Zhao WL, Fu JF, Shi JY, Pan Q, Hu J, Gao XD, Chen B, Li JM, Xiong SM, Gu LJ, Tang JY, Liang H, Jiang H, Xue YQ, Shen ZX, Chen Z, Chen SJ: Notch1 mutations in T-cell acute lymphoblastic leukemia: prognostic significance and implication in multifactorial leukemogenesis. Clin Cancer Res. 2006, 12: 3043-9. 10.1158/1078-0432.CCR-05-2832.
Li Y, Chen SH, Yang L, Chen S, Lin C, Wang L, Lu Y, Geng S, Du X, Schmidt CA: Change in expression pattern of TCR-CD3 complex in patients with multiple myeloma. Hematology. 2011, 16: 143-50. 10.1179/102453311X12953015767491.
Zheng H, Chen Y, Chen S, Niu Y, Yang L, Li B, Lu Y, Geng S, Du X, Li Y: Expression and distribution of PPP2R5 gene in leukemia. J Hematol Oncol. 2011, 4: 21-10.1186/1756-8722-4-21.
Chen S, Zha X, Yang L, Li B, Zhong L, Li Y: Deficiency of CD3gamma, delta, epsilon and zeta expression in T-cells from AML patients. Hematology. 2011, 16: 31-6. 10.1179/102453311X12902908411832.
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001, 25: 402-8. 10.1006/meth.2001.1262.
Ferrando AA: The role of Notch1 signaling in T-ALL. Hematology Am Soc Hematol Educ Program. 2009, 353-61.
Kox C, Zimmermann M, Stanulla M, Leible S, Schrappe M, Ludwig WD, Koehler R, Tolle G, Bandapalli OR, Breit S, Muckenthaler MU, Kulozik AE: The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 2010, 24: 2005-13. 10.1038/leu.2010.203.
Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC: Leukemia-associated mutations within the Notch1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006, 12: 4642-51.
Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, Happich M, Muckenthaler MU, Kulozik AE: Activating Notch1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006, 108: 1151-17. 10.1182/blood-2005-12-4956.
Acknowledgements
This work was supported by Grants from National Natural Science Foundation of China (no. 30871091), the Fundamental Research Funds for the Central Universities (No. 21610603) and Science and Technology Innovation Key Project of Guangdong Higher Education Institutes (kjcxzd1013).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YQL contributed to concept development and study design. CLL and SHC performed PCR and sequencing, HTZ, LJY and YBZ performed the real-time PCR. CYW, LJY, BL and HT were responsible for collection of clinical data. YQL and CLL and HTZ coordinated the study and helped drafting the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lin, C., Zheng, H., Wang, C. et al. Mutations increased overexpression of Notch 1 in T-cell acute lymphoblastic leukemia. Cancer Cell Int 12, 13 (2012). https://doi.org/10.1186/1475-2867-12-13
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2867-12-13