Abstract
Background
Germ-line transformation of insects is now a widely used method for analyzing gene function and for the development of genetically modified strains suitable for pest control programs. The most widely used transposable element for the germ-line transformation of insects is piggyBac. The site of integration of the transgene can influence gene expression due to the effects of nearby transcription enhancers or silent heterochromatic regions. Position effects can be minimized by flanking a transgene with insulator elements. The scs/scs' and gypsy insulators from Drosophila melanogaster as well as the chicken β-globin HS4 insulator function in both Drosophila and mammalian cells.
Results
To minimize position effects we have created a set of piggyBac transformation vectors that contain either the scs/scs', gypsy or chicken β-globin HS4 insulators. The vectors contain either fluorescent protein or eye color marker genes and have been successfully used for germ-line transformation of Drosophila melanogaster. A set of the scs/scs' vectors contains the coral reef fluorescent protein marker genes AmCyan, ZsGreen and DsRed that have not been optimized for translation in human cells. These marker genes are controlled by a combined GMR-3xP3 enhancer/promoter that gives particularly strong expression in the eyes. This is also the first report of the use of the ZsGreen and AmCyan reef fluorescent proteins as transformation markers in insects.
Conclusion
The insulated piggyBac vectors should protect transgenes against position effects and thus facilitate fine control of gene expression in a wide spectrum of insect species. These vectors may also be used for transgenesis in other invertebrate species.
Similar content being viewed by others
Background
Transposon-mediated germ-line transformation of insects is a powerful technique for investigating gene function [1] and is also being used for the development of insect strains suitable for area-wide control programs [2, 3]. piggyBac is a type II (short inverted repeat DNA) transposon from the cabbage looper moth Trichoplusia ni [4]. Transposons related to piggyBac are found in the genomes of almost all eukaryotes [5] and piggyBac has recently been shown to be able to transpose in mammalian (human and mouse) cells as well as in the mouse germline [6]. Vectors based on the piggyBac element have been used for germ-line transformation of a wide range of insect species including coleoptera, dipteran, hymenopteran and lepidopteran species [7]. An analysis of 29,000 insertion events into the Drosophila genome found that integration was non-random but had a broader distribution than found with the P element [8]. In particular piggyBac did not show the bias for 5' regulatory regions seen with the P element. The site of integration, however, can influence transgene expression. Integration near to a tissue-specific transcription enhancer can lead to transgene expression in tissues and/or stages other than intended [9]. Additionally, integration into a heterochromatic region can lead to low levels of transgene expression [10]. These position effects are particularly problematic for regulated gene expression systems such as those based on the tetracycline-dependent transactivator (tTA) [11–13] and in instances where high levels of transgene expression are required. To be effective such systems need a low level of gene expression in the absence of inducer but high levels in its presence.
Position effects can be minimized by bracketing the transgene with insulator elements [14]. Insulators can both block the unwanted effects of a transcription enhancer and also minimize the effects of heterochromatin. In Drosophila the most widely used insulators are the specialized chromatin structures scs and scs' [15], part of the gypsy transposon [16] and the chicken β-globin 5' HS4 element [17]. These insulators appear to achieve their effects by diverse mechanisms. The chicken β-globin HS4 insulator contains multiple elements. The enhancer-blocking element contains a binding site for the CCCTC binding factor CTCF [18]. The anti-silencer function of chicken β-globin HS4 likely recruits histone-modifying enzymes as a high level of histone acetylation and methylation of histone H3 at lysine 4 are localized at the insulator [19]. It is proposed that these histone modifications counteract the spreading of a compact heterochromatic structure. The scs and scs' insulators comprise a set of divergently transcribed promoters [20]. They may also function by recruiting histone modifying enzymes. The gypsy insulator may function by establishing loop chromatin domains with the paired insulators at the base of the loop [21].
Fluorescent proteins have been widely used to identify transgenic insects [22]. Apart from DsRed, however, the reef coral fluorescent proteins have not been used in insects although they function well as transformation markers in plants [23]. Here we report the development and evaluation of piggyBac vectors containing the scs/scs', gypsy and chicken β-globin HS4 insulators, some of which carry reef coral fluorescent proteins as markers.
Results and discussion
piggyBacvectors containing the scs/scs' insulator elements
To effectively minimize position effects the transgene must be bracketed by insulator elements. In this way the insulators can both block transcription enhancers and heterochromatin spreading. We have constructed four piggyBac vectors containing the Drosophila scs and scs' insulators (Figure 1). In all of these vectors the insulators flank the marker gene. Thus both the transgene and the marker gene should be protected from position effects.
piggyBac vectors containing the scs/scs' insulator elements. The vectors contain paired scs/scs' insulators flanking the DsRed (A), AmCyan (B), ZsGreen (C) and cinnabar (cn) (D) marker genes. The location of unique restriction endonuclease cloning sites are shown. The vectors have 690 bp of 5' terminal piggyBac sequence and either 769 bp (A-C) or 907 bp (D) of 3' terminal sequence.
Three of the vectors contain reef fluorescent protein marker genes controlled by a promoter that has multiple binding sites for both the PAX6 (3xP3) and GLASS (GMR) transcription factors (Figure 2A–C). These marker genes are similar in principle to those developed previously that utilized the 3xP3 promoter [22]. However, the addition of the GMR sequence would be predicted to increase marker gene expression in the eye [24] and may be more active in some non-drosophilid species. The reef coral fluorescent proteins are available either optimized for translation in human cells or not. Codon optimization of GFP led to a significant increase in the level of GFP expression in human cells [25]. However, in some important insect pest species such as the medfly Ceratitis capitata and the Australian sheep blowfly Lucilia cuprina the protein coding genes have a very different codon bias than humans [26]. Since codon bias can significantly affect mRNA translation efficiency in insects [27], the vectors carry reef fluorescent protein genes that have not been optimized for expression in human cells. The vectors have a unique Not I site between the scs' insulator and the reef fluorescent protein marker gene for insertion of the gene of interest.
The fourth scs/scs' vector we have constructed (Figure 2D) has the D. melanogaster cinnabar (cn) eye color marker gene that encodes the enzyme kynurenine 3-monooxygenase [28]. The cn gene has been used for selection of transgenic Ae. aegypti [29] but potentially could be used in other insect species where mutations in the corresponding kynurenine 3-monooxygenase gene have been identified. The vector has a unique Pac I site downstream from the cn polyadenylation site that could be used for future gene cloning. Although all scs/scs' vectors contain only a single cloning site, both Not I and Pac I have 8 bp recognition sequences and so would be unlikely to cut most gene constructs.
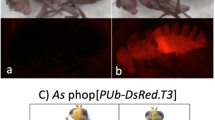
All vectors have been successfully used for germ-line transformation of D. melanogaster. Three lines were obtained with the DsRed marker, five with AmCyan, six with ZsGreen and two with the cn gene. Lines carrying the GMR-3xP3-reef fluorescent protein marker genes were easily identified as the eyes were brightly fluorescent (Figure 2). This suggests that the GMR-3xP3 promoter is a strong driver of gene expression and that the ZsGreen and AmCyan fluorescent proteins can be used as markers in insects. With the cyan filter set significant levels of ZsGreen expression were observed (Figure 2D). Thus this filter set could not be used to separately observe ZsGreen and AmCyan expression in the same tissue. The frequency of transformation (5–10% of G0 crosses) was comparable to what we obtained previously with non-insulated piggyBac vectors [23].
By visual inspection the fluorescence or color intensity appeared to be similar between lines carrying the same marker gene. A representative fly from each of the five AmCyan and six ZsGreen lines are shown in Figure 3, along with the y w parental strain. The low level of variation in marker gene expression between lines is in contrast to that previously reported for lines carrying the uninsulated 3xP3-EGFP, 3xP3-EYFP and 3xP3-ECFP marker genes [30, 31]. In a recent experiment six transformed lines were obtained with a piggyBac vector containing the uninsulated 3xP3-DsRed1 marker gene described previously [22]. These lines also show considerable line-to-line variation in fluorescence intensity (Figure 3C). These observations suggested that the scs/scs' insulators are minimizing position effects.
Comparison of fluorescence intensities of lines carrying insulated and uninsulated marker genes. A representative fly is shown from each of the lines carrying the insulated GMR-3xP3-AmCyan (A) and GMR-3xP3-ZsGreen (B) marker genes. Little variation in fluorescence intensity is seen between lines suggesting position effects had been minimized by the flanking scs and scs' insulator elements. In contrast lines carrying an uninsulated 3xP3-DsRed1 marker gene (C) show considerable variation in fluorescence intensity. Some lines are barely detectable above background fluorescence (y w) while other lines show very strong fluorescence.
To confirm these visual observations, more quantitative analyses were performed on the expression levels of the insulated AmCyan and ZsGreen lines and uninsulated DsRed1 lines shown in Figure 3. Two methods were used to estimate fluorescence intensities. Firstly, flies heterozygous for a marker gene were selected at random and examined under a stereo fluorescence microscope. Images were digitally captured and analyzed using software that can quantify color intensity of a selected region of the image (Table 1). All AmCyan, ZsGreen and DsRed1 lines were more significantly more intense than the control y w parental strain (one way analysis of variance, P < 0.05). Confirming visual observations, all AmCyan and most of the ZsGreen lines were not significantly more intense than other lines carrying the same marker gene. The two brightest ZsGreen lines (A10, A11) were significantly more intense than the three weakest lines (A7, A8M1, A9). However, the difference in color intensity between the strongest and weakest lines was less than 1.5 fold. In contrast, pairwise comparisons of the uninsulated 3xP3-DsRed1 lines found that the brightest line (R30A) was significantly more intense than all other lines as was the second brightest line (R21). Further, the brightest line had more than 20 times the color intensity of the weakest line.
In the second method, the fluorescence intensities of soluble protein extracts prepared from homogenized heads were measured using a fluorometer (Table 2). In general the results are in agreement with those obtained with the first method although there was more variability between lines, which was reflected in the statistical analyses. All AmCyan and ZsGreen lines had significantly higher fluorescence levels than the control strain (one way analysis of variance, P < 0.05). In pairwise comparisons the AmCyan lines were not significantly different from each other with the exception that the two brightest lines (A4, A5) had significantly more fluorescence than the weakest line (A2). The brightest two ZsGreen lines (A11, A8M3) had significantly higher levels of fluorescence than the other four ZsGreen lines. Other pairwise comparisons found little significant differences except that the A9 and A10 lines had significantly higher fluorescence levels than the A7 and A8M1 lines. While a statistically significant difference was found in some pairwise comparisons it is important to note that at most the fluorescence intensities differ by no more than two fold. While insulators do protect against position effects, it would be expected that the expression levels of insulated transgenes would not be identical between lines [13, 32]. In contrast the fluorescence levels of the uninsulated 3xP3-DsRed1 lines differ by as much as 7 fold (Table 2). In pairwise comparisons, the brighter lines (R30A, R21) had significantly more fluorescence than all the other lines carrying the same marker gene.
piggyBac vectors containing the gypsy or β-globinHS4 insulators
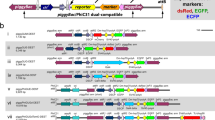
The β-globin 5' HS4 insulator sequences were introduced into 3xP3-EYFP and 3xP3-ECFP marked piggyBac constructs. The resulting vectors, pBac{3xP3-EYFP>>af>>} and pBac{3xP3-ECFP>>af>>} were used to insulate transgene constructs of the tTA/TRE binary expression system when introducing conditional embryonic lethality into Drosophila melanogaster [13]. For completeness, maps of these vectors are shown in Figure 4. The transgenic lines carrying the insulated TRE-responder constructs seemed to work more reliably and to mediate higher gene expression than their non-insulated counterparts [13], which indicates the functionality of the insulator elements in protecting the embedded transgenes from position effects.
piggyBac vectors containing the β- globin 5' HS4 and gypsy insulator elements. The vectors contain either the ECFP (A), EYFP (B) or DsRed1 (C) marker genes. The β-globin 5' HS4 (A, B) and gypsy (C) insulator elements do not bracket the marker genes but do flank the unique AscI and FseI cloning sites. The vectors have 685 bp of 5' terminal piggyBac sequence and 1051 bp of 3' terminal sequence.
The chicken β-globin 5' HS4 insulator sequences are relatively long (two times 2.4 kb) and seem to work efficiently. However, the tandem repetition of the element on either side of the insulated region causes problems of recombination when growing these plasmid constructs in bacteria. These constructs can only be grown in recombinase-negative bacteria, like STBL2 (Life Technologies, Rockville, MD). To avoid this complication in generating insulated constructs, another smaller insulator from the gypsy element was chosen and introduced into a 3xP3-DsRed marked piggyBac construct [22] resulting in the transformation vector pBac{3xP3-DsRed>af>} (Figure 4C). This vector has been successfully used in several germ-line transformation experiments in D. melanogaster. More than thirty lines have been generated at an efficiency comparable to that reported previously for piggyBac vectors carrying 3xP3-fluorescent protein marker genes [31].
The gypsy and chicken β-globin 5' HS4 vectors contain single AscI and FseI endonuclease restriction sites, which allow for a described two step cloning procedure using the shuttle vector pSLfa1180fa [31]. In contrast to the scs/scs' vectors, the gypsy or chicken β-globin 5' HS4 vectors were constructed to insulate only the transgene of interest without insulating the transformation marker gene. In this arrangement, the transgene is also insulated from the cis-regulatory sequences driving marker gene expression.
Conclusion
We have described insulated piggyBac vectors carrying different fluorescent protein and eye color markers that have been successfully used for germ-line transformation in D. melanogaster. The use of the reef coral fluorescent proteins AmCyan and ZsGreen is the first report that these proteins can serve as effective transformation markers in insects. It is likely that all of the insulated vectors will provide some protection against position effects in most insect species. The chicken β-globin HS4 insulator is of vertebrate origin yet effectively blocks position effects in D. melanogaster [17]. Although the gypsy and scs insulators are from D. melanogaster they have both been shown to be effective at protecting against heterochromatin silencing in a cultured human cell line [33]. Thus the vectors described herein should facilitate fine control of gene expression in a broad range of insect species. Also, these insulated vectors may be modified by changing the respective promoter to use them in mammalian systems.
Methods
Recombinant DNA
pB [SCS-GMR-3xP3-DsRed-SCS']: The piggyBac plasmid p3E1.2 (provided by M. Fraser) was first modified by the removal of the SalI and EcoRI sites. This modified 3E1.2 was then digested with PstI and BglII, blunt ended with T4 DNA polymerase, and the scs-scs' cassette from pELBA6 (provided by Paul Schedl) inserted to create pB [SCS-SCS']. The plasmid pB [SCS-SCS'] was digested with SalI and a XhoI-ScaI fragment containing the GMR enhancer [24] inserted to create pB [SCS-GMR-SCS']. The 3xP3 fragment from pHer [3xP3-EGFPaf] [31] was inserted into pDsRed (BD Biosciences Clontech) to create p3xP3DsRed. Subsequently, a fragment containing the polyadenylation site from the D. melanogaster hsp70 gene was cloned into the NotI site of p3xP3DsRed to create p3xP3DsRedpolyA. Finally, the 3xP3DsRedpolyA fragment was cloned into pB [SCS-GMR-SCS'] to create pB [SCS-GMR-3xP3-DsRed-SCS'].
pB [SCS-GMR-3xP3-AmCyan-SCS'] and pB [SCS-GMR-3xP3-ZsGreen-SCS']: The plasmid pBCpolyA was created by cloning the hsp70 3' flanking (polyA) sequence as a StuI-PstI fragment into the EcoRV-PstI sites of pBCKS+ (Stratagene). Fragments containing the AmCyan and ZsGreen genes (BD Biosciences Clontech) were then cloned into the PspOMI-Asp718 sites of pBCpolyA as NotI-Asp718 fragments to create pBCAmCyanpolyA and pBCZsGreenpolyA respectively. pB [SCS-GMR-3xP3-DsRed-SCS'] was then digested with NotI-Asp718, the DsRedHsp70polyA gene cassette removed and substituted with the NotI-Asp718 fragments containing either the AmCyanpolyA or the ZsGreenpolyA sequences to create pB [SCS-GMR-3xP3-AmCyan-SCS'] and pB [SCS-GMR-3xP3-ZsGreen-SCS'] respectively.
pB [SCS-cn-SCS']: A 7.2 kb Asp718/SphI fragment containing the D. melanogaster cn gene [28] was treated with T4 DNA polymerase to make blunt ends then inserted into the EcoRV site of pElba6 (kindly provided by Paul Schedl), which contains the scs and scs' insulators. The resulting plasmid was digested with NotI and Bsp120I and ligated with NotI digested pB [Ccw] (provided by A. Handler) essentially replacing the C. capitata w gene with the scs-cn-scs' cassette.
The construction of the β-globin 5' HS4 insulator vectors, pBac{3xP3-EYFP>>af>>} and pBac{3xP3-ECFP>>af>>}, is described in [13]. pBac{3xP3-DsRed>af>} was constructed by introducing a 400 bp PCR fragment, amplified with primers NH_FgyFse (5'-CCGGCCGGCCTTGAATTCGAGCTCGGTACC-3') and NH_RgyNgo (5'-CCCGCCGGCAATTGATCGGCTAAATGG-3') from pBS-gypsyHIII (provided by V. Pirrotta) and cut with NgoMIV, into FseI-cut pBac{3xP3-DsRed>af}, which itself was constructed by introducing a 400 bp PCR fragment, amplified with primers NH_FgyBss (5'-CCCGCGCGCTTGAATTCGAGCTCGGTACC-3') and NH_RgyAsc (5'-CCGGCGCGCCAATTGATCGGCTAAATGG-3') from pBS-gypsyHIII and cut with BssHII, into AscI-cut pBac{3xP3-DsRedaf} [22]. During the ligations in the different steps, the AscI site is restored at the 3'-end of the first gypsy element and the FseI site is restored at the 5'-end of the second gypsy element, respectively. In this construct, ">" symbolizes the insulator region of the gypsy retrotransposon.
Transformation of D. melanogasterand detection of fluorescent proteins
Germ-line transformation of D. melanogaster was carried out essentially as previously described [34]. The recipient strains used were y w and cn bw for piggyBac vectors containing the fluorescent protein and cn marker genes respectively. Transformants carrying the GMR-3xP3-reef fluorescent proteins marker genes were identified by florescence using an Olympus SZX12-RFL3 dissecting microscope with the appropriate excitation/barrier filter sets (EX 420–460 nm, EM 480–520 nm for AmCyan; EX460-90 nm, EM510-550 nm for ZsGreen; EX520-550 nm, EM580+ nm for DsRed). Images were captured using an Olympus DP70 digital camera. To compare expression levels of fluorescent proteins, single flies were selected at random from each line, anaesthetized then examined together. Thus identical camera settings were used for flies from each line. The intensity of fluorescence in the eyes was measured using the analySIS software package (Soft Imaging System GmbH) from an arbitrary selection of eye ommatidia using appropriate filter sets for the fluorescent protein marker. The DP-70 camera settings were exposure time 500 microseconds for AmCyan and ZsGreen lines and 2500 microseconds for the uninsulated DsRed1 lines. Further there was no color offset and sharpness and contrast were at maximum. These settings were used to avoid pixel saturation, while still detecting the weakest lines. Flies heterozygous for the marker gene were obtained by back crossing to the parental y w strain. The amount of fluorescence in the eyes of the transgenic lines was also quantified using a fluorometer. Heads of ten flies heterozygous for the fluorescent marker were dissected, snap frozen in liquid nitrogen and ground in 100 μL RIPA buffer (50 mM Tris HCl, pH 8.5; 300 mM NaCl; 1× protease inhibitors [Roche]), kept on ice for 30 minutes and centrifuged for 10 minutes at 14,000 rpm at 4°C using an Eppendorf 5417R microcentrifuge and the supernatant collected. 15–60 μL aliquots were taken and fluorescence detected using a Fluorostar Galaxy fluorometer (BMG Technologies) at the appropriate wavelengths (AmCyan: EX450 nm, EM490 nm/10 nm; ZsGreen EX450 nm, EM500 nm/12 nm; DsRed1, EX450 nm, EM590 nm/20 nm). For all extracts there was a linear response between volume of extract and level of fluorescence. Three extractions were performed for each line. The amount of protein in the supernatant was estimated using the Bradford assay at 595 nm. Statistical analyses were performed using the SigmaStat for Windows v3.11 software package. The Holm-Sidak method was used for the one way analysis of variance calculation.
References
Wimmer EA: Innovations: applications of insect transgenesis. Nature Reviews Genetics. 2003, 4 (3): 225-232. 10.1038/nrg1021.
Robinson AS, Franz G, Atkinson PW: Insect transgenesis and its potential role in agriculture and human health. Insect Biochemistry & Molecular Biology. 2004, 34 (2): 113-120. 10.1016/j.ibmb.2003.10.004.
Scott MJ, Heinrich JC, Li X: Progress towards the development of a transgenic strain of the Australian sheep blowfly (Lucilia cuprina) suitable for a male-only sterile release program. Insect Biochem Mol Biol. 2004, 34 (2): 185-192. 10.1016/j.ibmb.2003.11.001.
Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ: Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989, 172 (1): 156-169. 10.1016/0042-6822(89)90117-7.
Sarkar A, Sim C, Hong YS, Hogan JR, Fraser MJ, Robertson HM, Collins FH: Molecular evolutionary analysis of the widespread piggyBac transposon family and related "domesticated" sequences. Mol Genet Genomics. 2003, 270 (2): 173-180. 10.1007/s00438-003-0909-0.
Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T: Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005, 122 (3): 473-483. 10.1016/j.cell.2005.07.013.
Handler AM: Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochemistry & Molecular Biology. 2002, 32 (10): 1211-1220. 10.1016/S0965-1748(02)00084-X.
Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J: A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004, 36 (3): 283-287. 10.1038/ng1314.
O'Kane CJ, Gehring WJ: Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987, 84 (24): 9123-9127. 10.1073/pnas.84.24.9123.
Wallrath LL, Elgin SC: Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995, 9 (10): 1263-1277.
Heinrich JC, Scott MJ: A repressible female-specific lethal genetic system for making transgenic insect strains suitable for a sterile-release program. Proc Natl Acad Sci U S A. 2000, 97 (15): 8229-8232. 10.1073/pnas.140142697.
Thomas DD, Donnelly CA, Wood RJ, Alphey LS: Insect population control using a dominant, repressible, lethal genetic system. Science. 2000, 287 (5462): 2474-2476. 10.1126/science.287.5462.2474.
Horn C, Wimmer EA: A transgene-based, embryo-specific lethality system for insect pest management. Nature Biotechnology. 2003, 21 (1): 64-70. 10.1038/nbt769.
Kuhn EJ, Geyer PK: Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol. 2003, 15 (3): 259-265. 10.1016/S0955-0674(03)00039-5.
Kellum R, Schedl P: A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991, 64 (5): 941-950. 10.1016/0092-8674(91)90318-S.
Geyer PK, Corces VG: DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992, 6 (10): 1865-1873.
Chung JH, Whiteley M, Felsenfeld G: A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993, 74 (3): 505-514. 10.1016/0092-8674(93)80052-G.
Bell AC, West AG, Felsenfeld G: The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999, 98 (3): 387-396. 10.1016/S0092-8674(00)81967-4.
West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G: Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Molecular Cell. 2004, 16 (3): 453-463. 10.1016/j.molcel.2004.10.005.
Kuhn EJ, Hart CM, Geyer PK: Studies of the role of the Drosophila scs and scs' insulators in defining boundaries of a chromosome puff. Molecular & Cellular Biology. 2004, 24 (4): 1470-1480. 10.1128/MCB.24.4.1470-1480.2004.
Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P: Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001, 291 (5503): 495-498. 10.1126/science.291.5503.495.
Horn C, Schmid BG, Pogoda FS, Wimmer EA: Fluorescent transformation markers for insect transgenesis. Insect Biochemistry & Molecular Biology. 2002, 32 (10): 1221-1235. 10.1016/S0965-1748(02)00085-1.
Wenck A, Pugieux C, Turner M, Dunn M, Stacy C, Tiozzo A, Dunder E, van Grinsven E, Khan R, Sigareva M, Wang WC, Reed J, Drayton P, Oliver D, Trafford H, Legris G, Rushton H, Tayab S, Launis K, Chang YF, Chen DF, Melchers L: Reef-coral proteins as visual, non-destructive reporters for plant transformation. Plant Cell Rep. 2003, 22 (4): 244-251. 10.1007/s00299-003-0690-x.
Hay BA, Wolff T, Rubin GM: Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994, 120 (8): 2121-2129.
Haas J, Park EC, Seed B: Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996, 6 (3): 315-324. 10.1016/S0960-9822(02)00482-7.
Nakamura Y, Gojobori T, Ikemura T: Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000, 28 (1): 292-10.1093/nar/28.1.292.
Carlini DB, Stephan W: In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics. 2003, 163 (1): 239-243.
Warren WD, Palmer S, Howells AJ: Molecular characterization of the cinnabar region of Drosophila melanogaster: identification of the cinnabar transcription unit. Genetica. 1996, 98 (3): 249-262. 10.1007/BF00057589.
Coates CJ, Jasinskiene N, Miyashiro L, James AA: Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 1998, 95 (7): 3748-3751. 10.1073/pnas.95.7.3748.
Horn C, Jaunich B, Wimmer EA: Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev Genes Evol. 2000, 210 (12): 623-629. 10.1007/s004270000111.
Horn C, Wimmer EA: A versatile vector set for animal transgenesis. Dev Genes Evol. 2000, 210 (12): 630-637. 10.1007/s004270000110.
Fitzsimons HL, Henry RA, Scott MJ: Development of an insulated reporter system to search for cis-acting DNA sequences required for dosage compensation in Drosophila. Genetica. 1999, 105 (3): 215-226. 10.1023/A:1003801402153.
van der Vlag J, den Blaauwen JL, Sewalt RG, van Driel R, Otte AP: Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J Biol Chem. 2000, 275 (1): 697-704. 10.1074/jbc.275.1.697.
Li X, Heinrich JC, Scott MJ: piggyBac-mediated transposition in Drosophila melanogaster: an evaluation of the use of constitutive promoters to control transposase gene expression. Insect Molecular Biology. 2001, 10: 447-455. 10.1046/j.0962-1075 .2001.00283.x.
Acknowledgements
Plasmid DNAs were kindly provided by Paul Schedl, Malcolm Fraser, Bruce Hay, Al Handler, Bill Warren, Vincenzo Pirrotta. We thank Peter Farley, Ranjita Sengupta and Shrabani Saha for help with fluorometric assays, protein quantification assays and statistical analyses respectively. This work was supported by a grant from Meat and Wool Innovation (formerly WoolPro) to MJS and the Robert Bosch Foundation providing a junior research group to EAW. AS is supported by a postdoctoral fellowship from Massey University, CH was a fellow of the Fonds der Chemischen Industrie and EAW was an EMBO Young Investigator.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
AS, constructed the scs/scs' AmCyan and ZsGreen vectors, performed assays for quantifying fluorescence intensities, assisted with transgenesis and drafting of manuscript.
AA, constructed the scs/scs' DsRed vectors and assisted with transgenesis.
EJB, performed transgenesis experiments with AmCyan and ZsGreen vectors.
JCH, constructed scs/scs' vector with cinnabar marker gene
XL, performed transgenesis experiments with scs/scs' vectors containing DsRed and cn marker genes.
CH, constructed gypsy and β-globin insulated vectors and performed transgenesis experiments with these vectors.
EAW, supervised development of gypsy and β-globin insulated vectors and assisted with drafting of manuscript.
MJS, designed the scs/scs' vectors, assisted with screening for transgenics, drafted the manuscript and supervised Massey personnel.
Abhimanyu Sarkar, Asela Atapattu contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Sarkar, A., Atapattu, A., Belikoff, E.J. et al. Insulated piggyBac vectors for insect transgenesis. BMC Biotechnol 6, 27 (2006). https://doi.org/10.1186/1472-6750-6-27
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1472-6750-6-27