Abstract
Background
Identification and characterization of the prostate stem cell is important for understanding normal prostate development and carcinogenesis. The flow cytometry-based side population (SP) technique has been developed to isolate putative adult stem cells in several human tissue types including the prostate. This phenotype is mainly mediated by the ATP-binding cassette membrane transporter ABCG2.
Methods
Immunolocalization of ABCG2 was performed on normal prostate tissue obtained from radical prostatectomies. Normal human prostate SP cells and ABCG2+ cells were isolated and gene expression was determined with DNA array analysis and RT-PCR. Endothelial cells were removed by pre-sorting with CD31.
Results
ABCG2 positive cells were localized to the prostate basal epithelium and endothelium. ABCG2+ cells in the basal epithelium constituted less than 1% of the total basal cell population. SP cells constituted 0.5–3% of the total epithelial fraction. The SP transcriptome was essentially the same as ABCG2+ and both populations expressed genes indicative of a stem cell phenotype, however, the cells also expressed many genes in common with endothelial cells.
Conclusion
These results provide gene expression profiles for the prostate SP and ABCG2+ cells that will be critical for studying normal development and carcinogenesis, in particular as related to the cancer stem cell concept.
Similar content being viewed by others
Background
Experimental evidence suggests that prostatic epithelial stem cells exist and are likely localized to the basal epithelium [1]. Basal, luminal secretory and a small population of neuroendocrine cells constitute the epithelial component of prostatic acini. Basal and luminal cells may belong to two functional cell types descended from a common stem cell type. We are interested in identifying and isolating this prostatic stem cell. Studies to date suggest that stem cells from diverse tissue sources may contain a common set of gene transcripts, which are required for maintenance of the stem cell phenotype [2]. Considerable research efforts have been directed towards discovery of markers associated with the putative prostate stem cell, including the side population (SP) phenotype [3], integrin α2β1 (CD49b/CD29) [4, 5] and PROM1 (CD133) [6]. Identification and characterization of a stem/progenitor cell population is important to our understanding of not only normal prostate development but also the cancer process, particularly in regard to cancer stem cells [7]. This knowledge may lead to the development of effective cancer treatment strategies such as differentiation and cell-based therapy.
The ATP-binding cassette membrane transporter ABCG2 (BCRP/Bcrp1) functions as an energy-dependent efflux pump, and was first identified in the breast cancer cell line MCF-7 [8]. ABCG2 is highly expressed in human endothelial cells and plays an important role in the blood-brain barrier [9–11], but it is rarely expressed in most other differentiated cell types [12]. Expression of ABCG2 is associated with multi-drug resistance; more significantly, ABCG2 is the molecular determinant for the SP phenotype and has been postulated as a universal stem cell marker [13]. Goodell et al. discovered a small and distinct SP of whole bone marrow cells based on their capacity to efflux the fluorescent dye Hoechst 33342 [14]. Remarkably, although this SP comprised ~0.1% of total bone marrow cells, it accounted for virtually all of the hematopoietic stem cell (HSC) activity as demonstrated by bone marrow repopulation assays [15]. Subsequent studies of ABCG2-null mice have attributed this dye efflux to expression of ABCG2 [13]. Since the initial discovery of the hematopoietic SP, an analogous population has been detected in embryonic stem cells, the liver, heart, and many other organs including the prostate [3, 13, 16, 17]. Collectively, these studies provide evidence that the SP phenotype, and therefore ABCG2 expression, may represent a feature shared by stem cells of different tissue origins. However, other recent studies have found no direct correlation between SP cells and ABCG2 expression [18]. Both SP and known stem/progenitor cells express other ABC transporters including ABCB1 (MDR-1), ABCC1 and ABCA2, suggesting that these latter molecules may also be involved in determining the SP phenotype [19–21].
ABCG2 expression in the prostate has been reported in both the epithelium [22] and endothelium [23]. The SP of the prostate has been previously isolated and characterized as integrin α2+ and containing a subpopulation of quiescent (~12%) cells [3]. Immunohistochemical analysis of both normal and cancerous ABCG2+ cells shows that this subset also lacks the androgen receptor (AR) protein, and it has been proposed that ABCG2-mediated efflux of androgen is a mechanism for maintenance of the prostate stem cell phenotype [24].
In the cancer stem cell model, tumors are thought to contain phenotypically diverse populations of cancer cells, but only a minority of these cells (10–35%) possess the ability to form new tumors [7]. It is postulated that these cancer "stem" cells drive tumor growth and expansion, and are resistant to therapy. For breast cancer tumors, it was found that as few as 100 tumorigenic (CD44+/CD24-/low) cells could form new tumors that contained both the tumorigenic and non-tumorigenic cell types [25]. These cancer cells, like stem cells, can self-renew as well as "differentiate" into other cancer cell types to produce tumor heterogeneity.
The goal of this study was to determine if the SP of normal human prostate and ABCG2+ cells are similar populations that express putative stem cell markers. Furthermore, we sought to more precisely define the patterns of ABCG2 expression in prostate tissue, determine if the non-endothelial ABCG2 expressing cells could be isolated, and finally to characterize those cells by comparing the transcriptome from sorted SP cells with that of cells isolated using an antibody (5D3) that recognizes an external epitope of human ABCG2 [26]. This transcriptome data can be used for future comparison with candidate cancer stem cells isolated from prostate tumors.
Methods
Cell lines and prostate tissue
The breast cancer cell line MCF-7 and prostate cancer line C4-2 were obtained from American Type Culture Collection (Manassas, VA) and cultured in RPMI-1640 (Cambrex BioScience, Walkersville, MD) media supplemented with 10% heat-inactivated fetal bovine serum (FBS). The tissue samples that were used in this study consisted of cancer-free samples obtained from 30 radical prostatectomies. All tissue samples were obtained under approval by the University of Washington Institutional Review Board. Samples of cancer-free prostate parenchyma were collected following a standard protocol. To minimize RNA degradation, upon receipt of a radical specimen, 3-mm thick transverse sections were made of the prostate after inking the exterior surface (the surgical margin). Between 2 and 10 g of tissue from the anterior aspect of the prostate (transition zone) were excised. A frozen section of a block of tissue corresponding to that non-frozen tissue from which the cells were excised was taken and histologically assessed to confirm that the specimen was free of cancer. The sample was minced and digested by overnight incubation at room temperature in an aqueous solution of 0.2% collagenase type I (Invitrogen, Carlsbad, CA) in RPMI-1640 media supplemented with 5% FBS and 10-8 M dihydrotestosterone on a magnetic stirrer. The resultant cell suspension was filtered with a 70-μm Falcon cell strainer to remove non-digested tissue, diluted with an equal volume of Hanks balanced salt solution (HBSS), and aspirated with an 18-gauge needle. The resultant predominantly single cell preparation was partitioned into stromal and epithelial fractions on a discontinuous Percoll density gradient (Amersham Pharmacia, Piscataway, NJ) as described previously [27, 28]. High purity is obtained by this method [27]. Cells banding at a density of ρ = 1.07 were collected as the epithelial cell fraction and used for cell sorting. Because stromal cells (ρ = 1.035) are less dense they cannot sediment into the epithelial density band and the epithelial fraction is therefore relatively free of stromal cells.
Immunohistochemistry
Blocks of unfixed prostate tissue, typically from the peripheral zone where cancer arises, were harvested at the time of surgery. Serial 5-μm sections were prepared from randomly selected frozen blocks, fixed in cold acetone, and processed for immunohistochemistry. Immunohistochemistry was performed as described previously, using a three-step indirect avidin-biotin-peroxidase procedure [29]. The primary antibodies used were mouse monoclonal anti-ABCG2 (BXP-34, Chemicon, Temecula, CA) diluted 1:100 in phosphate-buffered saline (PBS), and CD138 (MI15, BD-PharMingen, San Diego, CA) diluted 1:100 in PBS. Antigen was localized using biotinylated anti-mouse IgG (BA-2000, Vector Labs, Burlingame, CA) as the secondary antibody, and diaminobenzidine tetrahydrochloride was the chromogen. The sections were counterstained in hematoxylin. Immunostained sections were imaged with an Olympus BX41 microscope (Olympus, Melville, NY) equipped with a MircoFire digital camera (Optronics, Goleta, CA). Percentage of ABCG2+/CD138+ basal cells was obtained by manual counting of 5 representative CD138+ glands (approximately 1 in 20 glands in a field contained ABCG2-positive basal epithelial cells). Composite images were constructed with Photoshop CS (Adobe Systems, San Jose, CA).
Hoechst 33342 labeling and SP sorting
The epithelial cell fraction enriched by Percoll gradient was resuspended at a concentration of 106 cells/ml in HBSS, supplemented with 10% FBS, 20 mM N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid (HEPES), pH 7.4, and 1% D-glucose. Hoechst 33342 (Molecular Probes, Eugene, OR) was added to a concentration of 2–5 μg/ml. Cells were incubated at 37°C for 90 min and then placed on ice until analysis. Fluorescence-activated cell sorting (FACS) was done using a high-speed cell sorter (InFlux, Cytopeia, Seattle, WA) following the protocol of Goodell et al. [14]. Fluorescence from the Hoechst-stained cells was excited by 100–150 mW UV from an Innova 305c argon ion laser (Coherent, Santa Clara, CA). Fluorescence was monitored at two emission bands from 400 – 480 nm (Hoechst blue) and from 690 – 800 nm (Hoechst red). Epithelial cells were sorted by magnetic cell sorting (MACS, Miltenyi Biotech, Auburn, CA) for CD44+ (basal) cells prior to SP sort. In MACS sorting, cells were resuspended in 0.1% bovine serum albumin (BSA)-HBSS and labeled with phycoerythrin (PE)-conjugated CD44 antibody (BD-PharMingen). After the primary antibody, the cells were incubated with anti-PE microbeads (Miltenyi Biotec). The cell suspension was filtered and then sorted by using the "DOUBLE POSITIVE SORT" program of an AutoMACS system (Miltenyi Biotec). Both the positive and negative fractions were analyzed by FACS to assess sorting efficiency using a FACSCalibur flow cytometer (Becton Dickenson, Mountainview, CA). The selected epithelial cells were then sorted for SP. The sorted SP cells were lysed in RLT Buffer (Qiagen, Germantown, MD) and RNA was isolated using RNeasy kits (Qiagen). The RNA quality was monitored by using Agilent 2100 Bioanalyzer with RNA Nano Labchip (Agilent Technologies, Palo Alto, CA). Some degradation was evident, likely due to the SP sorting process.
MACS isolation of ABCG2+ cells
The epithelial cell fraction was resuspended in 0.1% BSA-HBSS, and labeled with anti-CD31-PE followed by anti-PE microbeads. This step was intended to first remove ABCG2+/CD31+ endothelial cells. The CD31-negative fraction was then incubated with anti-ABCG2-PE. The reaction was stopped by the addition of 1 ml 0.1% BSA-HBSS and centrifugation. The labeled cells were resuspended and 15 μl paramagnetic microbead-conjugated anti-PE antibody was added for 15 min. Afterwards, the positive and negative cells were separated with an AutoMACS using the "DOUBLE POSITIVE SORT" program. Aliquots of the positive and negative cell fractions were analyzed by FACS to assess sort efficiency. The ABCG2-positive cells were pelleted by centrifugation and resuspended in RLT for storage at -80°C. RNA was isolated and the RNA quality was monitored. Only RNA samples that were of sufficient concentration and showed no degradation were used for array hybridization.
Affymetrix expression profiling
Five separate biological replicates of the sorted cell population were assayed to produce a dataset using the Human Genome U133 Plus 2.0 GeneChips (Affymetrix, Santa Clara, CA). The U-133 Plus 2.0 array contains probesets representing 54,675 genes, splice variants, and ESTs. In addition to the SP and ABCG2+ cells, transcriptomes of the following prostate cell types have been previously determined: CD104+ (basal epithelial) cells, CD26+ (luminal epithelial) cells, CD49a+ (stromal) cells, and CD31+ (endothelial) cells [30]. The GeneChips were prepared, hybridized, and scanned according to the protocols provided by Affymetrix. Briefly, 200 ng of RNA was reverse transcribed with poly (dT) primer containing a T7 promoter, and the cDNA was made double-stranded. In vitro transcription was performed to produce unlabeled cRNA. Next, first-strand cDNA was produced with random primer. cDNA was made double-stranded with poly (dT) primer/T7 promoter. Finally, in vitro transcription was performed with biotinylated ribonucleotides. The biotin-labeled cRNA was hybridized to the GeneChips. The chips were washed and stained with streptavidin-PE using an Affymetrix FS-450 fluidics station. Data was collected with an Affymetrix GeneChip Scanner 3000.
Data analysis
Comparative analysis between the SP, ABCG2+ and the other cell-type datasets was used to identify genes specific to stem cells. CEL files produced by GeneChip Operating Software (GCOS, Affymetrix) were loaded into GeneSpring 7.2 (Agilent) via the robust multiple array average (RMA) preprocessor. The GeneSpring RMA preprocessor uses the same analysis algorithm for Affymetrix array data as the open source Bioconductor project. GeneSpring's replicate error model was used for our analysis. Genes in the dataset with an average raw fluorescence signal <50 were considered to be undetected by the experiment. Statistical significance of differential expression between the various cell sorts was determined by 1-way ANOVA at p < 0.05.
Gene expression validations
Reverse transcriptase-polymerase chain reaction (RT-PCR) was used to validate expression. For cell lines and tissue specimens, 1 μg RNA was reverse transcribed with superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) at 50 C for 50 mins followed by 10 min at 70 C. Gene-specific primers for PCR (Table 1) were designed to produce amplicons from 100–650 bp in size. PCR was carried out at 95 C 30 s, 55 C 30 s, 72 C 1 min for 35 cycles. PCR products were resolved on 2% agarose gels. The ribosomal gene RPL13a served as the internal reference for each sample. Quantitative real-time PCR was performed by using SYBR Green I on ABI 7900HT (Applied Biosystems, Foster City, CA). Up to 1 μg of RNA from CD31+ and ABCG2+ (5D3+) cells were used for cDNA synthesis. The cDNA was then used as template for real-time PCR with gene specific primers. The quantitative PCR (qPCR) assay was run in a 96-well Optical Reaction Plate (Applied Biosystems) with 20 μl final volume per well. Each sample was run in triplicate for each gene. The reaction contained 3–5 μl template cDNA, 10 μl SYBR Green PCR Master Mix (Applied Biosystems) and 10 pmol each of forward and reverse primer. Gene-specific primers for qPCR were designed using Primer Express™ 1.5 (Applied Biosystems) (Table 2). Primer Sets were initially validated for equal amplification efficiencies of the genes. qPCR was carried out at 95 C 30 s, 55 C 30 s, 72 C 1 min for 40 cycles with RPL13a as the internal reference for each sample. The comparative Ct method was used to assess relative mRNA levels between two samples using CD31+ cells as a calibrator basis for comparison.
Results
Immunolocalization of ABCG2+ epithelial cells to the basal epithelium
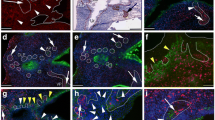
Fig. 1 shows the result of prostate immunostaining of ABCG2 and the prostate basal epithelial marker CD138 (syndecan 1) [29]. The ABCG2 antibody recognizes an internal epitope of the human ABCG2 protein [31]. All basal cells of a particular acinus were labeled by CD138 but only about 3% of these cells were also positive for ABCG2. Most glands had very few or no ABCG2+ cells. Furthermore, distribution of the ABCG2-positive cells within the basal epithelium consisted of both single cells and cell clusters. Three cell types could be distinguished: ABCG2+/CD138+ basal cells, ABCG2-/CD138+ basal cells, and ABCG2+/CD138- endothelial cells of vessels, which are known to express ABCG2 [11].
ABCG2 + cells in the prostate epithelium. Serial sections of normal human prostate were stained for ABCG2 (A – C) or CD138 (D – F). A subpopulation of cells in the basal epithelium is ABCG2+. Endothelial cells of capillaries (black arrow) are also ABCG2+ but are CD138- (red arrow). All basal cells of a particular small gland were ABCG2+/CD138+ (B). Original magnification is 100×, magnification for C and F is 200×.
Prostate epithelial SP
Hoechst 33342 labeling and analysis of human prostate epithelial cells by FACS detected a distinct SP as shown in Fig. 2. The prostate SP constituted 0.5–3% of the total epithelial cell fraction. Light scattering analysis of the SP cells (Fig. 2A) indicated that they were smaller and less granular than the non-SP cells, which is characteristic of stem cells [32, 33]. The Hoechst profile was plotted with BLUE on the y-axis and RED on the x-axis (Fig. 2B). The SP sorting protocol was also used on two ABCG2+ cancer cell lines (MCF-7 and C4-2) as positive controls (Fig. 2C and 2D respectively).
Prostate epithelial side population. Prostate cells prepared from tissue specimens were analyzed for Hoechst 33342 dye efflux. The cytograms show (A) forward and side light scatter of the SP and non-SP populations as defined by (B) the dye efflux characteristics of the cells. Cytograms show the dye efflux characteristics of (C) the prostate cancer cell line C4-2 and (D) the breast cancer cell line MCF-7 used as positive controls.
Prostate epithelial ABCG2+
CD31 labeling and sorting of prostate epithelial cells by MACS resulted in removal of 99.94% CD31-positive cells (Fig 3). The CD31 negative fraction was subsequently sorted using ABCG2 (Fig. 3).
Prostate SP, ABCG2+, and endothelial cell transcriptomes
Analysis of the transcriptomes of prostate SP and ABCG2+ cells revealed that the SP cell population was not significantly different from the ABCG2+ population. Fig. 4 shows the expression overlap of SP, ABCG2+, and endothelial cell transcriptomes in a Venn diagram. The SP transcriptome contained 5,011 probesets while the ABCG2+ transcriptome contained 23,973 probesets, and the endothelial transcriptome contained 21,145 probesets. Of the probesets detected in the SP transcriptome 23 were not detected in the ABCG2+ or endothelial datasets. However, the low number of probesets present in the SP data suggests that RNA quality (perhaps due to Hoechst toxicity) may have significantly affected the number of genes detected. Overall, the ABCG2+ transcriptome contained 3,704 probesets not detected in the SP or endothelial data. In addition to ABCG2, all three populations contained other ABC transporters including ABCE1, ABCB1, ABCG1 and ABCE1. The expression of stem cell markers detected by the arrays for each population is listed in Table 3. Of these, NR6A1 and NES were detected in the SP, ABCG2+ and CD31+ cell populations; and BMP-4, POU5F1, KIT and VM were detected in the ABCG2+ and CD31+ populations. Differentiation associated genes keratin 18 (K18), prostate acid phosphatase (PAP), and prostate stem cell antigen (PSCA) were downregulated in both SP and ABCG2+ cell populations compared to the prostate basal epithelial (CD104+) and luminal epithelial (CD26+) cell transcriptomes.
Molecular signature of stem cells in prostate SP cells
SP and non-SP cells were analyzed by RT-PCR. Analysis of the sorted SP cells showed that they were enriched in ABCG2 expression in comparison to the non-SP, and were AR- (Fig. 5A). In addition to ABCG2, the SP cells were shown to differentially express the following genes: (a) nestin (NES), a marker first identified in neural stem cells [34], and subsequently in other stem cell types, with expression higher in SP than non-SP; (b) telomerase (TERT), a known marker of stem cells, again higher in SP than non-SP; and (c) BMI-1, a transcription repressor specific for the induction of telomerase in epithelial cells and the extension of proliferative lifespan, and for the maintenance of hematopoietic stem cells [35], with expression confined mainly to SP. RT-PCR analysis of CD31+ endothelial cells also showed expression of these genes (Fig. 5B). The sorted SP cells likely still contained some CD31+ (ABCG2+) endothelial cells as indicated by RT-PCR (Fig. 5A). Additionally, qPCR was performed on CD31+ and ABCG2+ cells. Increased levels of expression for TERT, CD133, BMI-1, NES and ABCG2 were evident in the ABCG2+ cells vs CD31+ was verified (Fig. 6). These results suggest that the SP, ABCG2+ and CD31+ cells each have some intrinsic expression of genes characteristic of stem cells, but that the ABCG2+ sorting results in a greater enrichment of these genes than in CD31+ cells.
Gene expression analysis of prostate SP cells. (A): Expression of various stem cell genes in human prostate SP (SP+) vs. non-SP (SP-), by reverse transcription polymerase chain reaction (RT-PCR). The ribosomal protein-encoding gene RPL13a served as the reaction control. Nestin, telomerase, Bmi-1 are reported in the literature to be stem cell genes. CD133 has been reported as a prostate stem cell gene. Androgen receptor (AR) is known to be expressed by differentiated cells. The SP+ and SP-cells were also CD31+, indicating possible endothelial contamination. (B): Prostate endothelial cells (CD31+) also expressed these genes.
Discussion
Immunohistochemical analysis of ABCG2 expression identified two distinct positive populations in the prostate: an endothelial population and a nonendothelial basal epithelial population. Identification of nonendothelial expression was accomplished by comparing serial sections stained with the basal cell marker CD138 to ABCG2 staining patterns. Nonendothelial ABCG2 cells were identified as CD138+/ABCG2+. Nonendothelial staining for ABCG2 was observed in small clusters of cells in the basal epithelium of a few prostate glands. Endothelial CD138-/ABCG2+ staining was observed in capillaries in the stroma, or at periglandular location.
Using a protocol established for mouse bone marrow SP analysis, we isolated an SP in normal human prostate tissue obtained from radical prostatectomies. The forward and side scatter plot revealed a smaller and less granular population of cells corresponding to the SP defined by the Hoechst RED/BLUE profile. Transcriptome analysis of the prostate SP was compared to previously sorted prostate cell transcriptomes [30]. In contrast to the CD antibody-sorted cell types, far fewer genes were detected in SP sorted cells. RNA integrity is critical for determining gene expression, and suboptimal RNA quality in the SP sorted cells was the probable cause of this decrease in genes detected. Hoechst 33342 is a DNA intercalating agent that results in significant cellular toxicity [36], and may have caused RNA degradation in cells isolated in this fashion. Several studies have addressed the potential toxicity of Hoechst 33342 [36, 37], however, very little research has looked at the effects of the SP technique on the quality of RNA isolated from these cells. To mitigate the effects of Hoechst toxicity on RNA degradation, the monoclonal antibody, clone 5D3, was used to generate a transcriptome for the ABCG2+ prostate epithelial cells. MACS was chosen as the separation method because the procedure is much faster than FACS (minutes vs. hours).
Transcriptome analysis of the SP and ABCG2+ cells from the prostate included the endothelial (CD31+) population [30] since these cells also express ABCG2. As expected, all three populations (prostate SP, ABCG2+ and CD31+) expressed higher levels of ABCG2 than the other prostate cell types: luminal, stromal, basal, and epithelial (data not shown). Additionally, all three populations contained other ABC transporters including ABCE1, ABCB1, ABCG1 and ABCE1 [see Additional file 1]. The ABCG2+ (5D3) transcriptome dataset was found to contain many genes present in endothelial cells, although it did contain 232 unique probesets with a raw fluorescence signal >100 [see Additional file 2]. The sorting scheme used to isolate ABCG2+ cells by presorting with CD31-PE might have been inefficient in removing all CD31+ cells. However, MACS sorting of ABCG2+ cells did result in a more complete transcriptome than the SP sorting method [see Additional file 3], as evidenced by the nearly 5-fold increase in number of genes identified. RT-PCR analysis revealed that SP cells expressed higher levels of the putative stem cell markers NES, TERT, and BMI-1, in addition to ABCG2 compared to non-SP cells. In the same analysis, other markers, such as the endothelial marker CD31 [29] and the prostate stem cell marker CD133 [6] were expressed at similar levels by both SP and non-SP cells. CD31+ cells also expressed NES, BMI-1, TERT and CD133. qPCR analysis of ABCG2+ cells verified that the ABCG2+ sorting resulted in enrichment for these genes as compared to CD31+ cells. Based on these transcriptome and RT-PCR analyses, the distinction between the prostate endothelial cell and the putative prostate stem cell isolated based on SP expression is not entirely clear. The ABCG2+ population shares many endothelial genes as well, however, based on transcriptome and qPCR analyses, it seems to be a more distinct population than the SP cells. Because of their expression of ABCG2, endothelial cells are a major potential contaminant of stem cells isolated by methods targeting this marker. Alternative sorting schemes to isolate purer ABCG2+/CD31- populations are currently being explored in our lab.
Our study was not designed to provide a functional assay for these putative stem cells. The sorting methods utilized resulted in very small numbers of cells that were insufficient for in vitro studies. Additionally, the fact that these cells express ABCG2, which is hypothesized to efficiently pump out differentiation inducing molecules [24], may render them non-responsive to differentiation signaling. This transcriptome data has, however, provided a wealth of targets for further studies aimed at identifying the mechanisms that govern the process of self-renewal and development of prostate stem cells.
Conclusion
Our data show that ABCG2 efflux of Hoechst 33342 or antibodies to ABCG2 could be used in isolating a subset of prostate cells that may include the prostate stem cells. The prostate cells isolated by SP and antibodies for ABCG2 have gene expression characteristics consistent with the endothelial cell phenotype and a number of stem cell markers. Because of potential RNA degradation caused by the SP process, it may be that antibody sorting based on ABCG2 expression is a better method of isolation for the putative prostate stem cell. Future studies are needed to determine more conclusively whether SP or ABCG2+ results in organ-specific stem cell enrichment in the prostate, or whether other putative markers such as integrin α2β1 (CD49b/CD29) [4, 5] and PROM1 (CD133) [6] are better suited for prostate stem cell isolation.
References
Bonkhoff H, Remberger K: Differentiation pathways and histogenetic aspects of normal and abnormal prostatic growth: a stem cell model. Prostate. 1996, 28 (2): 98-106. 10.1002/(SICI)1097-0045(199602)28:2<98::AID-PROS4>3.0.CO;2-J.
Seruya M, Shah A, Pedrotty D, du Laney T, Melgiri R, McKee JA, Young HE, Niklason LE: Clonal population of adult stem cells: life span and differentiation potential. Cell Transplant. 2004, 13 (2): 93-101.
Bhatt RI, Brown MD, Hart CA, Gilmore P, Ramani VA, George NJ, Clarke NW: Novel method for the isolation and characterisation of the putative prostatic stem cell. Cytometry A. 2003, 54 (2): 89-99. 10.1002/cyto.a.10058.
Collins AT, Habib FK, Maitland NJ, Neal DE: Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001, 114 (Pt 21): 3865-3872.
Hudson DL, O'Hare M, Watt FM, Masters JR: Proliferative heterogeneity in the human prostate: evidence for epithelial stem cells. Lab Invest. 2000, 80 (8): 1243-1250.
Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT: CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004, 117 (Pt 16): 3539-3545. 10.1242/jcs.01222.
Reya T, Morrison SJ, Clarke MF, Weissman IL: Stem cells, cancer, and cancer stem cells. Nature. 2001, 414 (6859): 105-111. 10.1038/35102167.
Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD: A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci U S A. 1998, 95 (26): 15665-15670. 10.1073/pnas.95.26.15665.
Aronica E, Gorter JA, Redeker S, van Vliet EA, Ramkema M, Scheffer GL, Scheper RJ, van der Valk P, Leenstra S, Baayen JC, Spliet WG, Troost D: Localization of breast cancer resistance protein (BCRP) in microvessel endothelium of human control and epileptic brain. Epilepsia. 2005, 46 (6): 849-857. 10.1111/j.1528-1167.2005.66604.x.
Kusuhara H, Sugiyama Y: Active efflux across the blood-brain barrier: role of the solute carrier family. NeuroRx. 2005, 2 (1): 73-85. 10.1602/neurorx.2.1.73.
Zhang W, Mojsilovic-Petrovic J, Andrade MF, Zhang H, Ball M, Stanimirovic DB: The expression and functional characterization of ABCG2 in brain endothelial cells and vessels. Faseb J. 2003, 17 (14): 2085-2087.
Schinkel AH, Jonker JW: Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003, 55 (1): 3-29. 10.1016/S0169-409X(02)00169-2.
Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP: The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001, 7 (9): 1028-1034. 10.1038/nm0901-1028.
Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC: Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996, 183 (4): 1797-1806. 10.1084/jem.183.4.1797.
Goodell MA, Rosenzweig M, Kim H, Marks DF, DeMaria M, Paradis G, Grupp SA, Sieff CA, Mulligan RC, Johnson RP: Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat Med. 1997, 3 (12): 1337-1345. 10.1038/nm1297-1337.
Shimano K, Satake M, Okaya A, Kitanaka J, Kitanaka N, Takemura M, Sakagami M, Terada N, Tsujimura T: Hepatic oval cells have the side population phenotype defined by expression of ATP-binding cassette transporter ABCG2/BCRP1. Am J Pathol. 2003, 163 (1): 3-9.
Martin CM, Meeson AP, Robertson SM, Hawke TJ, Richardson JA, Bates S, Goetsch SC, Gallardo TD, Garry DJ: Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004, 265 (1): 262-275. 10.1016/j.ydbio.2003.09.028.
Naylor CS, Jaworska E, Branson K, Embleton MJ, Chopra R: Side population/ABCG2-positive cells represent a heterogeneous group of haemopoietic cells: implications for the use of adult stem cells in transplantation and plasticity protocols. Bone Marrow Transplant. 2005, 35 (4): 353-360. 10.1038/sj.bmt.1704762.
Lechner A, Leech CA, Abraham EJ, Nolan AL, Habener JF: Nestin-positive progenitor cells derived from adult human pancreatic islets of Langerhans contain side population (SP) cells defined by expression of the ABCG2 (BCRP1) ATP-binding cassette transporter. Biochem Biophys Res Commun. 2002, 293 (2): 670-674. 10.1016/S0006-291X(02)00275-9.
Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK: A distinct "side population" of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004, 101 (39): 14228-14233. 10.1073/pnas.0400067101.
Benchaouir R, Rameau P, Decraene C, Dreyfus P, Israeli D, Pietu G, Danos O, Garcia L: Evidence for a resident subset of cells with SP phenotype in the C2C12 myogenic line: a tool to explore muscle stem cell biology. Exp Cell Res. 2004, 294 (1): 254-268. 10.1016/j.yexcr.2003.11.005.
Fetsch PA, Abati A, Litman T, Morisaki K, Honjo Y, Mittal K, Bates SE: Localization of the ABCG2 mitoxantrone resistance-associated protein in normal tissues. Cancer Lett. 2006, 235 (1): 84-92. 10.1016/j.canlet.2005.04.024.
Maliepaard M, Scheffer GL, Faneyte IF, van Gastelen MA, Pijnenborg AC, Schinkel AH, van De Vijver MJ, Scheper RJ, Schellens JH: Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res. 2001, 61 (8): 3458-3464.
Huss WJ, Gray DR, Greenberg NM, Mohler JL, Smith GJ: Breast cancer resistance protein-mediated efflux of androgen in putative benign and malignant prostate stem cells. Cancer Res. 2005, 65 (15): 6640-6650. 10.1158/0008-5472.CAN-04-2548.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF: Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003, 100 (7): 3983-3988. 10.1073/pnas.0530291100.
Zhou S, Morris JJ, Barnes Y, Lan L, Schuetz JD, Sorrentino BP: Bcrp1 gene expression is required for normal numbers of side population stem cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. Proc Natl Acad Sci U S A. 2002, 99 (19): 12339-12344. 10.1073/pnas.192276999.
Liu AY, True LD, LaTray L, Nelson PS, Ellis WJ, Vessella RL, Lange PH, Hood L, van den Engh G: Cell-cell interaction in prostate gene regulation and cytodifferentiation. Proc Natl Acad Sci U S A. 1997, 94 (20): 10705-10710. 10.1073/pnas.94.20.10705.
Kassen A, Sutkowski DM, Ahn H, Sensibar JA, Kozlowski JM, Lee C: Stromal cells of the human prostate: initial isolation and characterization. Prostate. 1996, 28 (2): 89-97. 10.1002/(SICI)1097-0045(199602)28:2<89::AID-PROS3>3.0.CO;2-I.
Liu AY, True LD: Characterization of prostate cell types by CD cell surface molecules. Am J Pathol. 2002, 160 (1): 37-43.
Oudes AJ, Campbell DS, Sorensen CM, Walashek LS, True LD, Liu AY: Transcriptomes of human prostate cells. BMC Genomics. 2006, 7 (1): 92-10.1186/1471-2164-7-92.
Diestra JE, Scheffer GL, Catala I, Maliepaard M, Schellens JH, Scheper RJ, Germa-Lluch JR, Izquierdo MA: Frequent expression of the multi-drug resistance-associated protein BCRP/MXR/ABCP/ABCG2 in human tumours detected by the BXP-21 monoclonal antibody in paraffin-embedded material. J Pathol. 2002, 198 (2): 213-219. 10.1002/path.1203.
Chen Z, de Paiva CS, Luo L, Kretzer FL, Pflugfelder SC, Li DQ: Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004, 22 (3): 355-366. 10.1634/stemcells.22-3-355.
Barrandon Y, Green H: Cell size as a determinant of the clone-forming ability of human keratinocytes. Proc Natl Acad Sci U S A. 1985, 82 (16): 5390-5394. 10.1073/pnas.82.16.5390.
Lobo MV, Arenas MI, Alonso FJ, Gomez G, Bazan E, Paino CL, Fernandez E, Fraile B, Paniagua R, Moyano A, Caso E: Nestin, a neuroectodermal stem cell marker molecule, is expressed in Leydig cells of the human testis and in some specific cell types from human testicular tumours. Cell Tissue Res. 2004, 316 (3): 369-376. 10.1007/s00441-003-0848-4.
Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF: Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003, 423 (6937): 302-305. 10.1038/nature01587.
Uchida N, Fujisaki T, Eaves AC, Eaves CJ: Transplantable hematopoietic stem cells in human fetal liver have a CD34(+) side population (SP)phenotype. J Clin Invest. 2001, 108 (7): 1071-1077. 10.1172/JCI200113297.
Zhang X, Chen J, Davis B, Kiechle F: Hoechst 33342 induces apoptosis in HL-60 cells and inhibits topoisomerase I in vivo. Arch Pathol Lab Med. 1999, 123 (10): 921-927.
The National Institutes of Health resource for stem cell research. [http://stemcells.nih.gov/info/scireport/appendixE.asp]
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2490/7/6/prepub
Acknowledgements
We thank Lillian Eichner, Melanie Leong and Christina P. Shadle for assistance with cell culture. We also thank Bruz Marzolf and Pamela Troisch and the microarray facility at the Institute for Systems Biology. This work was supported by a grant, DK63630, from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
LEP: study design, experiments, and drafting of the manuscript. AJO: gene expression array and data analysis. TWP: side population sorting. YAG: qPCR and data analysis. LSW: flow cytometry. LDT: tissue acquisition, immunohistochemistry and pathology. AYL: study conception, study design and coordination. All authors have read and approved the final manuscript.
Electronic supplementary material
12894_2006_92_MOESM1_ESM.pdf
Additional file 1: ABC transporters in SP, ABCG2+ and endothelial cell transcriptomes. Transcriptome analysis of prostate SP, ABCG2+ and endothelial cells detected 19 probesets for ABC genes with a raw fluorescence signal > 100. (PDF 22 KB)
12894_2006_92_MOESM2_ESM.xls
Additional file 2: ABCG2 unique genes. Transcriptome analysis of prostate ABCG2+ detected 1291 probesets not detected in the SP or endothelial data with a raw fluorescence signal > 50, 232 probesets with signal > 100. (XLS 276 KB)
12894_2006_92_MOESM3_ESM.pdf
Additional file 3: SP unique genes. Transcriptome analysis of prostate SP detected 17 probesets not detected in the ABCG2+ or endothelial data with a raw fluorescence signal > 50, 8 probesets with signal > 100. (PDF 21 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Pascal, L.E., Oudes, A.J., Petersen, T.W. et al. Molecular and cellular characterization of ABCG2 in the prostate. BMC Urol 7, 6 (2007). https://doi.org/10.1186/1471-2490-7-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2490-7-6