Abstract
Background
Chromosomal Comparative Genomic Hybridization (CGH) has been applied to all stages of cervical carcinoma progression, defining a specific pattern of chromosomal imbalances in this tumor. However, given its limited spatial resolution, chromosomal CGH has offered only general information regarding the possible genetic targets of DNA copy number changes.
Methods
In order to further define specific DNA copy number changes in cervical cancer, we analyzed 20 cervical samples (3 pre-malignant lesions, 10 invasive tumors, and 7 cell lines), using the GenoSensor microarray CGH system to define particular genetic targets that suffer copy number changes.
Results
The most common DNA gains detected by array CGH in the invasive samples were located at the RBP1-RBP2 (3q21-q22) genes, the sub-telomeric clone C84C11/T3 (5ptel), D5S23 (5p15.2) and the DAB2 gene (5p13) in 58.8% of the samples. The most common losses were found at the FHIT gene (3p14.2) in 47% of the samples, followed by deletions at D8S504 (8p23.3), CTDP1-SHGC- 145820 (18qtel), KIT (4q11-q12), D1S427-FAF1 (1p32.3), D9S325 (9qtel), EIF4E (eukaryotic translation initiation factor 4E, 4q24), RB1 (13q14), and DXS7132 (Xq12) present in 5/17 (29.4%) of the samples.
Conclusion
Our results confirm the presence of a specific pattern of chromosomal imbalances in cervical carcinoma and define specific targets that are suffering DNA copy number changes in this neoplasm.
Similar content being viewed by others
Background
Uterine cervix carcinoma (UCC) represents the second cause of death among the female population worldwide. The fact that more than 99% of all the cervical invasive tumors are positive for infection with high risk human papillomavirus (HPV) suggests that this is one of the most important factors for the development of this neoplasm [1, 2]. These viruses can induce cellular transformation by several mechanisms; the viral oncoproteins E6 and E7 can interact with cellular proteins involved in important cellular functions, such as tumor suppression, apoptosis, cell cycle control, genomic instability, transcriptional regulation and immune evasion [3].
The induction of genomic instability by HPV seems to be particularly important for the establishment and development of an invasive tumor [4, 5] since this increased genomic plasticity would generate cellular clones with enhanced transforming and invasive potential [6].
Metaphase comparative genomic hybridization (mCGH) has been applied to study different stages of this tumor [4, 7–19], detecting specific patterns of chromosomal imbalances that arises very early during the development of cervical carcinoma, suggesting that the gain of chromosome 3q is one of the most important genetic alteration that defines the transition from a pre-malignant lesion to an invasive carcinoma [4]. Some of these imbalances have been related to specific clinical behaviors, such as the presence of lymph node metastases [9]. However, given the spatial resolution of mCGH [20], little is known about the identity of specific genes that might be the targets of regional chromosomal imbalances. Matrix-based CGH or array CGH overcomes this problem increasing the sensitivity for the detection of DNA copy number changes at specific loci, through the use of well defined genomic DNA fragments whose mapping location is known, arrayed onto a solid surface [21–23], thereby achieving a resolution of copy number imbalances up to the single gene level.
In order to refine the patterns of chromosomal imbalances present in cervical carcinoma, and trying to identify specific genes that might be targets of copy number changes in this tumor, we applied microarray CGH on 20 uterine cervix-derived samples (three pre-malignant lesions, 10 invasive tumors and seven UCC derived cell lines) to detect DNA copy number changes at the single gene level.
Methods
Cervical tissues
All described procedures have been evaluated and approved by the local committee of ethics of the Mexican Institute of Social Security (IMSS), and all samples were taken after informed consent from the patients. The pre-malignant lesions and the invasive tumors were collected by colposcopy-directed biopsies at the Gynecology Department of the Hospital General de México, Mexico City. The biopsies were divided in three sections. The central part was used for genomic DNA extraction using the Wizard Genomic kit (Promega, Madison, WI, USA), and the extremes were fixed with 70% ethanol overnight and paraffin embedded. Hematoxilin-eosin stained sections from these biopsies were analyzed in order to confirm the presence of at least 70% tumoral cells in the samples.
Cell lines
The cell lines included in this study were: CasKi, SiHa, both positive for HPV16, and HeLa (HPV18) The CaLo and ViBo cell lines were established from stage IIB invasive tumors, while INBL and RoVa from a stage IVA tumor. These cells are HPV18 positive and were established from tumor explants at the laboratory of Cell differentiation and Cancer of the National University of Mexico [24]. The chromosomal CGH profiles of CaLo, ViBo, INBL and RoVa have been published recently [19].
HPV detection and typing
HPV detection was carried out by PCR using the consensus primers MY09 and MY11 for the L1 region of the viral genome. After a 5 min. denaturation at 94°C, 100 ng of DNA were subjected to 35 amplification cycles with the following parameters: 94°C for 1 min., 55°C for 2 min. and 73°C for 3 min., with a final extension step of 7 min. at 72°C. The amplicon was labeled using the Big Dye sequencing kit and sequenced on an ABI371 sequencer (Applied Biosystems, Foster City, CA, USA). BLAST http://www.ncbi.nlm.nih.gov/BLAST/ sequence comparison was used in order to define the viral type.
Microarray CGH
Microarray CGH was performed using the GenoSensor Array 300 system, following the manufacturer's instructions (ABBOT-Vysis, Downers Grove, IL, USA). Each array contains 861 spots, representing 287 chromosomal regions that are commonly altered in human cancer, such as telomeres, regions involved in microdeletions, oncogenes, and tumor suppressor genes. Briefly, 100 ng of genomic DNA were labeled by a random primer reaction during two hours. Tumor DNA was labeled with Cy3 and the normal female reference DNA with Cy5. After the labeling reaction, the probes were digested with DNAse at 15°C for one hr., followed by two ethanol-purifications; finally the probe size was checked by gel electrophoresis. The hybridization mixture consisted of 2.5 μl of each of the differentially labeled DNAs plus 25 μl of hybridization buffer provided in the kit. This mixture was denatured at 80°C for 10 min. at 80°C, followed by incubation at 37°C for one hr. Five μl of this probe were applied onto the spotted area of the array under a coverslip and hybridized in a humid chamber containing 50% formamide (FA)/2XSSC at 37°C for 72 hrs. After hybridization, the arrays were washed 3X in 50%FA/2XSSC at 40°C for 10 min/wash, followed by four 5 min. washes in 1XSSC at room temperature. Finally, the arrays were briefly rinsed in distilled water, mounted and counterstained in the dark for 45 min. with DAPI (4,6-diamino-2-phenylindole).
Image capture and analysis
Array analysis was performed immediately after counterstaining using the GenoSensor scanner and software. This system generates a "genomic analysis report", indicating which chromosome regions in the array are involved in copy number changes, as well as a spreadsheet containing the data generated by a single experiment. In order to compare all the experiments, a database was created using the normalized, bias corrected, tumor/normal ratio value of each experiment [see additional file 1]. Since each spot in the array is present in triplicates, the median of the three spots of each probe in the array was calculated and its log2 transformed value was used for further analysis. A fluorescence ratio >1.25 (log2 = 0.32) was considered as a DNA gain, while DNA losses were scored when the ratio was <0.75 (log2 = -0.41). A ratio >2 (log2 = 1) was considered as a high copy number amplification.
Results
HPV detection and typing
One of the premalignant lesions was positive for HPV16 infection; one for HPV31 and the other for HPV58. In the invasive tumors, seven were positive for HPV16 and in three cases, we were not able to detect HPV sequences with the oligonucleotides we used for PCR amplification. As expected, CasKi and SiHa were positive for HPV16, while HeLa, INBL, CaLo, ViVo and RoVa were positive for HPV18.
Microarray comparative genomic hybridization
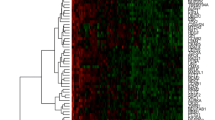
All our samples, except one pre-malignant lesion, presented alterations, ranking from 1/287 (alterations/total targets in the array) in a pre-malignant lesion to 175/287alterations in the cell line RoVa. We found almost twice the number of DNA gains than DNA losses (571 vs. 298) and the average number of copy number alterations (ANCA=total number of alterations in the sample collective/total number of cases) was 43.45 per case.
One of the pre-malignant lesions did not show any alteration, while amplifications at MSH2-KCNK12 (2p22.3-2p22.1), TCL1A (14q32.1) and TOP1 (20q12) were found in a second pre-malignant lesion and DMBT1 (10q25.3), ERBB2 (17q12), and 4qTEL11 (4qtel) amplification was found in the third sample from this group.
In the invasive tumors and the cell lines, the most common amplifications (58.8% of the samples) were found at the clones RBP1-RBP2 (retinol binding protein 1 and 2, 3q21-q22), present as a high copy number amplification (HCNA) in two samples; DAB2 (disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila), 5p13; C84C11/T3 (5ptel) and D5S23 (5p15.2), followed by gains of Tp63 (3q27-q29, 2 HCNA); EGFR (Epidermal growth factor receptor, 7p12.3-p12.1, 4 HCNA) and D5S2064 (5p15.2), in 52.9% of the invasive samples and amplification of INS (Insulin, 11ptel) in 47% of the samples.
The most common deletion was found at the clone corresponding to the FHIT (Fragile histidine triad) gene (3p14.2), present in 47% of the invasive samples, followed by deletions at D8S504 (8p23.3), CTDP1-SHGC- 145820 (18qtel), KIT (4q11-q12), D1S427-FAF1 (1p32.3), D9S325 (9qtel), EIF4E (eukaryotic translation initiation factor 4E, 4q24), RB1 (13q14), and DXS7132 (Xq12) present in 29.4% of the samples. A histogram of the DNA copy number alterations detected in the tumor samples analyzed by array CGH is presented in figure 1. The results of these experiments can be accessed through the Progenetix CGH database http://www.progenetix.com[25].
Discussion
Previous studies using chromosomal CGH have delimited a specific pattern of chromosomal imbalances in cervical carcinoma. However, there is little knowledge regarding the identity of particular genes that might be the targets for these copy number changes, making microarray CGH an attractive method in order to define these particular gene targets.
There was concordance between the alterations detected by microarray CGH and the pattern of chromosomal alterations already described by chromosomal CGH. Although the array that we used did not cover the entire genome, we were able to detect alterations at particular genes and genetic markers that might be related to the transformation process in the cervical epithelium.
It is important to notice that the limited number of pre-malignant lesions analyzed did not allowed us to detect any particular region that might be related with this stage of the disease. However, an interesting candidate gene amplified in one pre-malignant sample and in 5 invasive tumors was MSH2-KCNK12 (2p22.3-2p22.1). This gene is the human homolog of the E. coli mismatch repair gene mutS, and has been found mutated in hereditary nonpolyposis colon cancer. Higher MSH2 expression has been described in cervical intraepithelial neoplasias and invasive cervical carcinomas than in non-neoplastic cervical lesions. An altered expression of this gene has also been proposed as an important event during cervical carcinogenesis [26, 27]. Interestingly, the invasive samples showing MSH2 amplification presented with a high number of alterations (>40), suggesting a possible connection between increased copy number of this gene and chromosomal instability in invasive cervical carcinomas.
One of the most important genetic events during cervical carcinoma progression is the gain of 3q. This alteration has been detected in early stages of cervical transformation and in cooperation with other imbalances, seems to play an important role in tumor development. Microarray analysis identified among the most prevalent alterations in cervical tumors and cell lines the amplification of the RBP1- RBP2 (Retinol binding protein 1 and 2, 58.8%) and Tp63 (52.9% of the samples) genes, located at 3q21-q22 and 3q27-q29, respectively.
Tp63 is a homolog of the p53 tumor suppressor gene. Its protein is inactivated by the E6 HPV oncoprotein and plays a primordial role in the development of squamo-stratified epithelia. Tp63 is highly expressed in the basal stratum of these epithelia with diminished expression in the differentiated strata, suggesting that the presence of this protein preserves the self-renewal capacity of the epithelial stem cells after an asymmetric division, in which one of the daughter cell must conserve its epithelial stem-cell properties and the other daughter cell is committed to the differentiation process [28]. This protein has been detected in human cervical tissues in the basal and parabasal layers of the ectocervical squamous epithelium, and it is not present in the differentiated layers. In premalignant lesions and invasive squamous tumors, a strong p63 expression has been described [29, 30]. The presence of this protein has also been associated with poor survival and locoregional failure after radiation and chemotherapy [31]. Expression of the epidermal growth factor receptor (EGFR, 7p12.3-p12.1), which was found amplified in 52.9% of the invasive tumors that we analyzed, was found to be a prognostic predictor of extrapelvic failure after treatment, and the expression of both molecules was found to be a very good risk factor measurement in patients with stage IIB squamous cell carcinoma of the uterine cervix, who had received radiotherapy and concurrent chemotherapy [31].
DAB2 on 5p13 was amplified in 58.8% of the invasive cases. The DAB2 gene has been identified as a potent tumor suppressor gene in prostate and ovarian carcinoma [32], and loss of expression of this gene has been associated with the transition of ovarian epithelial cells to premalignant states [33]. DAB2 has been implicated in cell positioning control and seems to mediate the requirement for basement membrane attachment of epithelial cells [34]. To our knowledge, there are no available reports analyzing the expression of this gene in the uterine cervix or in cervical carcinoma. The amplification of this gene seems contrary to its putative role as a tumor suppressor gene. A possible explanation for this observation might be the loss of one allele followed by the amplification of the remaining chromosome. However, since array CGH does not offer any type of information regarding the parental origin of the amplified chromosome, this situation can not be confirmed.
Detection of the TERC gene amplification has been recently proposed as a potential marker for the evaluation of cervical carcinoma progression [35]; however, we detected amplification of the clone representing this gene in less than 10% of the samples analyzed by CGH arrays. FISH analysis, as described by Heselmayer et al., detected a higher prevalence of nuclei with a diploid pattern than those with a tetraploid pattern, even in the high grade lesions. Furthermore, the percentage of nuclei with more than 2 copies of 3q, including the tetraploid cells ranged between 3.3 to 50% of the CIN3 (cervical intraepithelial neoplasia grade 3). Dellas et al., [9] used in situ hybridization to analyze the prevalence of 3q amplifications in cervical cancer tissue arrays, detecting low level amplifications in most of the tumors studied. These results suggest that these low copy number gains might not be adequately detected by chromosome or even array CGH, due to the contamination with normal cells and/or the presence of a high number of diploid or tetraploid cells in the sample.
Regarding DNA losses, the FHIT (fragile histidine triad, 3p14.2) gene suffered losses in 47% of the cases. Aberrant expression of this gene has been well documented in cervical carcinoma and has been related to lymph node metastasis, parametrial invasion, and vaginal involvement in invasive tumors [36]. An association between FHIT gene abnormalities and infection with particular HPV types has been suggested, since 87% of the cases with absent FHIT expression were positive for HPV16 infection [37]. Furthermore, abnormal expression of this gene has been found in significantly younger patients than those with normal expression, suggesting that abnormalities in the regulation of this gene might be accelerating carcinogenesis in cooperation with HPV [37]. These observations might be related to the preferential integration of HPV into fragile sites, particularly FRA3B, where FHIT is located [38].
Conclusion
In conclusion, microarray CGH allowed the detection of particular genes located in regions with common DNA copy number changes in cervical carcinoma. Further studies using CGH arrays with a higher resolution and the possibility to combine LOH with copy number changes, might be useful for the detection of gene specific targets that are relevant for the genesis and progression of cervical carcinoma.
Abbreviations
- CGH:
-
Comparative Genomic Hybridization, UCC: Uterine cervix carcinoma, HPV: Human papilloma virus, DAPI: 4,6-diamino-2-phenylindole.
References
Walboomers J, Jacobs M, Manos M, Bosch X, Kummer A, Shah K, Snijders P, Peto J, Meijer C, Muñoz N: Human Papilloma Virus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999, 189: 12-19. 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F.
Bosch X, Muñoz N: The viral etiology of cervical cancer. Virus Res. 2002, 89: 183-190. 10.1016/S0168-1702(02)00187-9.
Münger K, Howley P: Human papillomavirus immortalization and transforming functions. Virus Res. 2002, 89: 213-228. 10.1016/S0168-1702(02)00190-9.
Ried T, Hesselmeyer K, Blegen H, Schröck E, Auer G: Genomic changes defining the genesis, progression and malignancy in solid human tumors: a phenotype/genotype correlation. Genes Chrom Cancer. 1999, 25: 195-204. 10.1002/(SICI)1098-2264(199907)25:3<195::AID-GCC1>3.0.CO;2-8.
Pihan G, Wallace J, Zhou Y, Doxsey S: Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003, 63: 1398-1404.
Cahill DP, Kinzler KW, Vogelstein B, Lengauer C: Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999, 9: M57-M60. 10.1016/S0962-8924(99)01661-X.
Heselmeyer K, Schröck E, Du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T: Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996, 93: 479-484. 10.1073/pnas.93.1.479.
Heselmeyer K, Macville M, Schröck E, Blegen H, Hellström A, Shah K, Auer G, Ried T: Advanced stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chrom Cancer. 1997, 19: 233-240. 10.1002/(SICI)1098-2264(199708)19:4<233::AID-GCC5>3.0.CO;2-Y.
Dellas A, Torhorst J, Jiang F, Proffit J, Schultheiss E, Holzgreve W, Sauter G, Mihatsch M, Moch H: Prognostic value of genomic alterations in invasive cervical squamous cell carcinoma stage IB detected by comparative genomic hybridization. Cancer Res. 1999, 59: 3475-3479.
Kirchhoff M, Rose H, Petersen B, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm S, Philip J: Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chrom Cancer. 1999, 24: 144-150. 10.1002/(SICI)1098-2264(199902)24:2<144::AID-GCC7>3.0.CO;2-9.
Allen D, White D, Hutchins A, Scurry J, Tabrizi S, Garland S, Armes J: Progressive genetic aberrations detected by comparative genomic hybridization in squamous cell carvical cancer. Br J Cancer. 2000, 83: 1659-1663. 10.1054/bjoc.2000.1509.
Kirchhoff M, Rose H, Petersen B, Maahr J, Gerdes T, Philip J, Lundsteen C: Comparative genomic hybridization reveals non-random chromosomal aberrations in early preinvasive cervical lessions. Cancer Genet Cytogenet. 2001, 129: 47-51. 10.1016/S0165-4608(01)00424-1.
Umayahara K, Numa F, Suehiro Y, Sakata A, Nawata S, Ogata H, Suminami Y, Sakamoto M, Sasaki K, Kato H: Comparative Genomic Hybridization detects genetic alterations during early stages of cervical cancer progression. Genes Chrom Cancer. 2002, 33: 98-102. 10.1002/gcc.1215.
Harris CP, Lu XY, Narayan G, Singh B, Murty VV, Rao PH: Comprehensive molecular cytogenetic characterization of cervical cancer cell lines. Genes Chrom Cancer. 2003, 36: 233-241. 10.1002/gcc.10158.
Narayan G, Pulido HA, Koul S, Lu XY, Harris CP, Yeh YA, Vargas H, Posso H, Terry MB, Gissmann L, Schneider A, Mansukhani M, et al: Genetic analysis identifies putative tumor supressor sites at 2q35-q36.1 and 2q36.3-q37.1 involved in cervical cancer progression. Oncogene. 2003, 22: 3489-3499. 10.1038/sj.onc.1206432.
Rao PH, Arias-Pulido H, Lu XY, Harris CP, Vargas H, Zhang FF, Narayan G, Schneider A, Terry MB, Murty VV: Chromosomal amplifications, 3q gain and deletions of 2q33-q37 are the frequent genetic changes in cervical carcinoma. BMC Cancer. 2004, 4: 5-10.1186/1471-2407-4-5.
Solinas-Toldo S, Dürst M, Lichter P: Specific chromosomal imbalances in human papillomavirus transfected cells during progression toward immortality. Proc Natl Acad Sci USA. 1997, 94: 3854-3859. 10.1073/pnas.94.8.3854.
Hidalgo A, Schewe C, Petersen S, Salcedo M, Gariglio P, Schlüns K, Dietel M, Petersen I: Human papilloma virus status and chromosomal imbalances in primary cervical carcinomas and tumor cell lines. Eur J Cancer. 2000, 36: 542-548. 10.1016/S0959-8049(99)00323-8.
Hidalgo A, Monroy A, Arana R, Taja L, Vázquez G, Salcedo M: Chromosomal imbalances in four new uterine cervix carcinoma derived cell lines. BMC Cancer. 2003, 3 (1): 8-10.1186/1471-2407-3-8.
Lichter P, Joos S, Bentz M, Lampel S: Comparative genomic hybridization: uses and limitations. Semin Hematol. 2000, 37: 348-357. 10.1053/shem.2000.16594.
Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P: Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chrom Cancer. 1997, 20: 399-407. 10.1002/(SICI)1098-2264(199712)20:4<399::AID-GCC12>3.0.CO;2-I.
Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, et al: High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet. 1998, 20: 207-211. 10.1038/2524.
Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO: Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet. 1999, 23: 41-46. 10.1038/14385.
Caceres-Cortes JR, Alvarado-Moreno JA, Waga K, Rangel-Corona R, Monroy-Garcia A, Rocha-Zavaleta L, Urdiales-Ramos J, Weiss-Steider B, Haman A, Hugo P, Brousseau R, Hoang T: Implication of tyrosine kinase receptor and steel factor in cell density-dependent growth in cervical cancer and leukemias. Cancer Res. 2001, 61: 6281-6289.
Baudis M, Cleary ML: Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001, 17: 1228-1229. 10.1093/bioinformatics/17.12.1228.
Giarnieri E, Mancini R, Pisani T, Alderisio M, Vecchione A: Msh2, Mlh1, Fhit, p53, Bcl-2, and Bax expression in invasive and in situ squamous cell carcinoma of the uterine cervix. Clin Cancer Res. 2000, 6: 3600-3606.
Kwasniewska A, Gozdzicka-Jozefiak A, Postawski K, Miturski R: Evaluation of DNA mismatch repair system in cervical dysplasias and invasive carcinomas related to HPV infection. Eur J Gynaecol Oncol. 2002, 23: 231-235.
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F: p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999, 398: 714-718. 10.1038/19539.
Quade BJ, Yang A, Wang Y, Sun D, Park J, Sheets EE, Cviko A, Federschneider JM, Peters R, McKeon FD, Crum CP: Expression of the p53 homologue p63 in early cervical neoplasia. Gynecol Oncol. 2001, 80: 24-29. 10.1006/gyno.2000.5953.
Wang T, Chen B, Yang Y, Chen H, Wang Y, Cviko A, Quade B, Sun D, Yang A, Mckeon F, Crum C: Histologic and immunophenotypic classification of cervical carcinomas by expression of the p53 homologue p63: A study of 250 cases. Hum Pathol. 2001, 32: 479-486. 10.1053/hupa.2001.24324.
Cho NH, Kim YB, Park TK, Kim GE, Park K, Song KJ: P63 and EGFR as prognostic predictors in stage IIB radiation-treated cervical squamous cell carcinoma. Gynecol Oncol. 2003, 91: 346-353. 10.1016/S0090-8258(03)00504-3.
Chen H, Toyooka S, Gazdar AF, Hsieh JT: Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J Biol Chem. 2003, 278: 3121-3130. 10.1074/jbc.M208230200.
Yang DH, Smith ER, Cohen C, Wu H, Patriotis C, Godwin AK, Hamilton TC, Xu XX: Molecular events associated with dysplastic morphologic transformation and initiation of ovarian tumorigenicity. Cancer. 2002, 94: 2380-2392. 10.1002/cncr.10497.
Sheng Z, Sun W, Smith E, Cohen C, Sheng Z, Xu XX: Restoration of positioning control following Disabled-2 expression in ovarian and breast tumor cells. Oncogene. 2000, 19: 4847-4854. 10.1038/sj.onc.1203853.
Huang LW, Chao SL, Chen TJ: Reduced Fhit expression in cervical carcinoma: correlation with tumor progression and poor prognosis. Gynecol Oncol. 2003, 90: 331-337. 10.1016/S0090-8258(03)00318-4.
Butler D, Collins C, Mabruk M, Leader MB, Kay EW: Loss of Fhit expression as a potential marker of malignant progression in preinvasive squamous cervical cancer. Gynecol Oncol. 2002, 86: 144-149. 10.1006/gyno.2002.6712.
Takizawa S, Nakagawa S, Nakagawa K, Yasugi T, Fujii T, Kugu K, Yano T, Yoshikawa H, Taketani Y: Abnormal Fhit expression is an independent poor prognostic factor for cervical cancer. Br J Cancer. 2003, 88: 1213-1216. 10.1038/sj.bjc.6600892.
Thorland EC, Myers SL, Gostout BS, Smith DI: Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003, 22: 1225-1237. 10.1038/sj.onc.1206170.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/5/77/prepub
Acknowledgements
This work was partially funded through the 7114 and 34686 grants from the Mexican Council of Science and technology (CONACyT) and the Mexican Institute for Social Security (IMSS-FOFOI FP- 2001–2003). AH, GVO, CP, RL were recipients of scholarships from the CONACyT, IMSS and DGEP-UNAM. We would like to thank Abbott-Vysis for providing the CGH array system for this analysis. This work was submitted in partial fulfillment of the requirements for the Ph.D. degree of HA at the Ph.D. in Biomedical Sciences, National University of Mexico.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they do not have any competing interests.
Authors' contributions
AH: Performed the microarray CGH experiments, data analysis and paper writing; MB: Help with data submission to the Progenetix database, data analysis; IP: provided training for the experiments, CGH data analysis; PP: tissue processing; GV: HPV typing; DH: sample collection; JG: provided access to the samples; ML: access to samples, histopathological analysis; RL: sample collection; CP: DNA extraction; JG: Help with data analysis; KV: DNA extraction; BA: HPV typing; MS: project coordinator.
Electronic supplementary material
12885_2005_265_MOESM1_ESM.txt
Additional File 1: Raw data from the CGH microarray experiments. This is text file containing the raw data from the microarrays, the first column denotes the identification of the clone, column 2 represents the cytogenetic position of the clone, sample data begins in column 3. (TXT 114 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hidalgo, A., Baudis, M., Petersen, I. et al. Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer 5, 77 (2005). https://doi.org/10.1186/1471-2407-5-77
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-5-77