Abstract
Background
We evaluated two molecular methods of HPV detection and their correlation with cytological and histological diagnosis in a large sample of Greek women.
Methods
All women with liquid-based cytology performed at a University Hospital between 2000 and 2003 were included. The Hybrid Capture 2 (HC2) kit and in house Polymerase Chain Reaction (PCR) were used for HPV DNA detection. Cervical biopsy was performed for women with ASCUS+ cytology, HPV detection, or abnormal colposcopy. Positive (PLR) and negative (NLR) likelihood ratios were calculated for cytology and HPV molecular testing for the prediction of CIN2 and greater histology.
Results
Of the 1270 women evaluated 241 (18.5%) had abnormal cytology. Cytology diagnosed high-grade squamous intraepithelial lesion (HSIL) or invasive carcinoma in 21(1.7%) cases whereas 26 (2%) women had CIN2+ or greater histology. PCR detected HPV in 397/1270 (31.3%) and HC2 in 260/1270 (20.4%) samples. Both molecular tests exhibited high reproducibility (Cohen's kappa value 0.691, 95% CI: 0.664 - 0.718). Positive likelihood ratios (PLR) of 9.4, 3.8 and 3.4 and negative likelihood ratios of 0.13, 0.21, and 0 were noted for ≥ LSIL, any positive HC2 or any positive PCR-HPV testing, for predicting CIN2+ histology, respectively. All CIN 3+ lesions harbored high risk oncogenic HPV type infections.
Conclusions
HPV infection was found in a large proportion of this population and was associated with CIN 2/3 lesions and infiltrating carcinomas. Thin prep testing and HPV detection by HC2 or PCR performed very well with regards to identifying high grade lesions in an environment with experienced examiners.
Similar content being viewed by others
Background
Large epidemiological studies performed over the last two decades have identified infection with human papilloma virus (HPV) as a necessary cause for cervical cancer[1, 2]. Furthermore, newer molecular techniques have greatly helped in identifying the association of high-risk HPV types with the development of precancerous lesions, as well as their role in cervical carcinogenesis[3, 4]. Since the vast majority of invasive cervical carcinoma cases are associated with HPV type 16 or 18 infections[5, 6], efforts have been made to develop preventive measures for infections with these high risk HPV types: two specific vaccines targeting infection with HPV types 16 and 18 have recently been introduced[7].
Specific HPV testing may not be appropriate as a primary screening due to claims that it lacks specificity[8] but it may assist in the management of women with cytological changes[9]. The association of findings from conventional cytological testing with those of newer molecular techniques is of great importance and helps to better understand the evolution of HPV infection in different epidemiological settings.
The purpose of the current study is to evaluate cytological findings from a large observational population sample in association with identification of HPV infection using newer molecular techniques, such as Hybrid Capture 2 and PCR. Furthermore, we correlate these findings with histological diagnosis.
Methods
This is a cross-sectional study performed at all specimens sent for diagnostic cytology at the Departments of Histology and Embryology, and the Department of Cytopathology of the National and Capodistrian University of Athens Medical School. Samples are addressed to these departments every day from many community and hospital-based Gynecology clinics covering the metropolitan area of Athens (roughly a population of 3.8 million people). Informed consent was obtained from all women included in this study that was performed between January 2000 and December 2003.
Cytological diagnosis
Samples for the ThinPrep Pap test were collected by means of a Brun's-like brush. The PreservCyt® vials (Cytyc Inc, Boxborough, MA, USA) containing the cell samples were addressed to the aforementioned Departments for preparation of thin-layer slides using the ThinPrep 2000 Automated Slide Processor® (Cytyc, Boxborough, MA, USA) according to the manufacturer' instructions. Cytological findings were interpreted according to the Bethesda classification system [10] and were classified as follows a) within normal limits; b) atypical squamous cells of undetermined significance (ASC-US); c) low-grade squamous intraepithelial lesion (LSIL); d) high-grade squamous intraepithelial lesion (HSIL); e) squamous cell carcinoma (SCC) or adeno-carcinoma. An experienced cytopathologist whose diagnostic experience exceeds 10 years examined all samples.
Histological diagnosis
A cervical biopsy was performed if ThinPrep testing revealed ASCUS and above cytological categories or there was positive HPV testing or there was a visible lesion upon colposcopy. Biopsy was performed by an experienced colposcopist (in practice for more than 10 years) as part of the study protocol. All women with indications consented to this procedure. The research was performed with the approval of the Institutional Review Board and in compliance with the Helsinki declaration. The three-tiered cervical intraepithelial neoplasia (CIN) grading system was used for histological diagnosis [11]. All cases were diagnosed by an experienced pathologist (more than 10 years experience). In case histology showed a CIN 2 or CIN 3 or invasive carcinoma the patient was referred for appropriate treatment.
HPV DNA detection
Two techniques were used simultaneously:
a) The commercially available Hybrid Capture 2 (HC2) kit that was used according to the manufacturer's instructions (Digene Corporation, Gaithersburg, MD). Briefly, HC2 is a signal amplification assay that uses a combination of antibody capture and chemiluminescent signal detection. An RNA probe cocktail (full genome probes) that detects 13 high-risk HPV types (16/18/31/33/35/39/45/51/52/56/58/59/68) and 5 low-risk (6/11/42/43/44) HPV types is used in the reaction with the target DNA. The RNA:DNA hybrids are captured onto a solid phase coated with universal capture antibodies specific for the RNA:DNA hybrids. Capture RNA:DNA hybrids are detected with multiple antibodies conjugated to alkaline phoshatase. The signal resulting from the chemiluminescent reaction is read and the results are interpreted. For each specimen relative light unit/cutoff values were calculated as the ratio of the specimen luminescence relative to the luminescence of the 1.0 pg/ml HPV-16 cut-off standard and reflect a semi-quantitative value of the cumulative viral burden from one or more of the examined genotypes. A relative light unit/cutoff value of ≥ 1 was considered as a positive result. Specimens testing positive for high-risk HPV types or low risk HPV types only were classified as HPV-high risk or HPV-low risk respectively, whereas specimens testing positive for both low and high risk HPV types were classified as HPV low-high risk.
b) An in house Polymerase Chain Reaction (PCR) to detect HPV DNA presence. DNA was extracted and purified from 3 ml of residual ThinPrep samples using a commercially available kit according to the manufacturer's instructions (QIAamp DNA mini kit, Tissue protocol, QIAGEN Inc, Valencia, Ca). The quantity and integrity of the DNA extracts was monitored through spectrophotometer readings and amplification of β-globin gene (endogenous control). The extracted DNA was tested for the presence of HPV DNA by PCR using the consensus (general) primers GP5+/GP6+ following a previously published protocol[12]. These primers amplify a fragment of approximately 150 base pairs (bp) from the L1 region of 22 anogenital HPV genotypes (types 6, 11, 13, 16, 18, 30-33, 35, 39, 40, 43, 45, 51, 52, 54, 55, 56, 58, 59 and 66). Positive PCR products (approximately 150 bp) visualized under UV light after electrophoresis in a 1% agarose gel in TBE buffer (0.1 mol/L Tris, 0.09 mol/L Borate, 1 mol/L EDTA) stained with ethidium bromide (0,5 μl/ml) were further analyzed using type-specific primers targeting the E6 region of the HPV genome of HPV types 6,11,16,18,31 and 33 according to a previously published protocol[13]. DNA extracted from HeLa cells infected with HPV18 was used as positive control in the type-specific HPV PCR assays. The PCR procedure was repeated twice for every sample. A representative number of PCR products from each PCR assay was sequenced (ABI PRISM® 3730XL DNA Analyzers by Lark Technologies, Inc, UK) and the PCR products were confirmed.
Statistical methods
Sensitivity, specificity, positive and negative predictive values were calculated for cytological results and molecular HPV testing against the histological diagnosis of CIN2 and above or CIN3 and above. More specifically, we calculated values considering all cytology results ≥ ASCUS as positive, or all ≥ LSIL as positive or all ≥ HSIL as positive separately. For HC2 we calculated values for any positive result and for positive high risk results (all high risk infections independent of whether they were high risk only or mixed low-high risk infections). For PCR we used either any positive result or positive results for HPV types 16, 18, 31 and 33 only. Positive and negative likelihood ratios were also calculated [14]: likelihood ratios enable the comparison of diagnostic values of tests in a way independent of the prevalence of the disease. The PLR [sensitivity/(1 - specificity)] corrects the true positive rate by the false positive rate of a test while on the other hand NLR [(1 - sensitivity)/specificity] compares the probability of a negative test in persons with disease, compared to the probability of a negative test in persons without disease [14]. Finally, a Cohen's kappa value was calculated for evaluating the agreement between the test results of HC2 and PCR methods.
Results
Cytological results
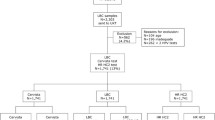
During the 4 year period 1270 women were evaluated (mean age: 34.2 ± 12.1 years old). Table 1 depicts age associations with cytological findings and HPV detection. The vast majority (n = 991, 78%) of the population examined were between 21 and 45 years old. Overall 241/1270 (19%) women were diagnosed with abnormal cytology (Table 1): more specifically, 101 (8%) women exhibited ASC-US, 119 (9.4%) were diagnosed with LSIL, 10 (0.8%) with HSIL, 10 (0.8%) with squamous cell carcinoma and 1 (0.8%) with adenocarcinoma. HSIL or infiltrating carcinoma was the cytological diagnosis in 21/1270 (1.7%) samples. Reporting rates of ASC-US/LSIL combined were 30.6% for women under 21 yrs old, 17.8% for the 22-30 yrs group, 17.7% for 30-45 yrs group, and then declined in older age groups (Table 1). HSIL+ rates increased with age, with 17/21 (81%) HSIL+ lesions diagnosed in women older than 30 years. Seven of the 11 (63.6%) invasive cancer cases were noted after 45 years of age (Table 1). Regarding HPV testing, PCR+ rates were approximately double those of ASC-US/LSIL combined for all age groups up until 65 years (Table 1).
ThinPrep diagnosis in relation to HPV DNA detection by HC2 or PCR testing is depicted in Table 2. HPV was detected by HC2 in 260/1270 (20.4%) samples and by PCR in 397/1270 (31.3%) samples. The two tests gave highly concordant results (Cohen's k value 0.691, 95% CI: 0.664 - 0.718). Discrepancies between HC2 and PCR were observed in 153 specimens. HC2 detected 8 specimens as being positive for HPV that were not detected by PCR (4 with ASCUS-all low risk HPV types; 4 with LSIL, 3 low and 1 high risk HPV types). PCR detected HPV in 145 specimens not detected by HC2, (80 with normal cytology, 45 with ASCUS, 16 with LSIL, 2 with HSIL, 1 with squamous cell carcinoma and 1 with adenocarcinoma). HC2 detected HPV in 17/21 (81%) while PCR detected HPV in 21/21 (100%) samples with HSIL+ cytology (Table 2). In the squamous cell carcinoma and adenocarcinoma categories 2/11 (18.2%) samples tested negative for HPV DNA with HC2, whereas none of these samples was negative with PCR (Table 2). In 21 cases HPV was not detected by either method: eleven cases with ASCUS and ten cases with LSIL.
HC2 detected infection with high risk or mixed low-high risk HPV types in 40/101(39.6%) of ASCUS cases. PCR detected HPV 16, 18, 31, or 33 (single or multiple infections) in 31/86 (36%) of ASCUS cases. For LSIL the corresponding figures were 77/119 (64.7%) and 63/105(60%) for HC2 and PCR respectively; for HSIL 6/10 (60%) by HC2 and 8/10 (80%) by PCR (Table 2). All cytological diagnoses (10/10) of squamous cell carcinomas tested positive for at least one of the high risk oncogenic HPV types 16, 18, 31, or 33 by PCR (Table 2).
Histological results
The comparison of ThinPrep diagnoses and histological results is presented at Table 3. In total, 426 women underwent biopsy (Table 3). From patients with ASCUS 58 out of 101 (57.4%) had CIN 1 in biopsy, whereas only 3 (3%) patients had CIN 2 or higher-grade lesions (Table 3). Only two of 119 (1.7%) patients with LSIL had CIN 2. All (100%) cases classified as HSIL with ThinPrep cytology had a biopsy diagnosis of either CIN2 or CIN 3. ThinPrep cytology correctly identified the single case with cervical adenocarcinoma as well as 10 of 11 (90.9%) subjects with a histological diagnosis of SCC (Table 3). One patient diagnosed cytologically as ASCUS had a SCC on subsequent biopsy.
Nineteen out of 26 cases (73.1%) with a biopsy diagnosis of CIN 2+ had a HC2 positive test for high risk viruses, whereas PCR detected HPV types 16, 18, 31, or 33 in 21/26 (80.8%) such cases (Table 3). For CIN 3 and invasive carcinoma, HC2 was positive in 11/15 (73.3%), whereas high risk HPV types by PCR were detected in 15/15 cases (100%) (Table 3). Seven of the 11 (63.6%) squamous cell carcinoma cases confirmed by biopsy had a single infection with either HPV type 16 or 18. The single adenocarcinoma case had an infection with HPV type 18 only. Four mixed infections (all had HPV 16, three HPV 18 or HPV 33 and two HPV 31) were diagnosed by biopsy as SCC. Sensitivities, specificities, positive and negative predictive values and positive and negative likelihood ratios for cytological and molecular HPV testing and histological diagnosis of CIN2+ or CIN3+ are provided in Table 4. Positive likelihood ratios (PLR) of 9.4, 3.8 and 3.4 were noted for ≥ LSIL, positive HC2 or positive HPV testing respectively and CIN2+ histology (Table 4). For CIN2+ histological lesion the best positive likelihood ratios (PLR) were observed with Thin prep testing exhibiting ≥ LSIL (PLR = 9.4), or ≥ HSIL (PLR = infinite), and for high risk type of virus detection either by HC2 (PLR = 5.5), or PCR (PLR = 11.4). Similar figures were observed for CIN3 (Table 4). Negative likelihood ratios of 0.13, 0.21, and 0 were noted for cytology ≥ LSIL, positive HC2 or positive HPV testing respectively and CIN2+ histology (Table 4).
Discussion
This study depicts the successful use of both HC2 and PCR in liquid based ThinPrep samples. Use of the same samples for both DNA tests was done to avoid bias. The HC2 kit assesses the presence of 18 low risk (i.e. 6,11,42,43 and 44) and high risk (i.e. 16, 18, 31, 33, 35, 39, 45, 51, 51, 56, 58, 59 and 68) HPV types [15, 16]. On the other hand, using the L1 consensus primers GP5+/GP6+ by PCR, we were able to screen for 22 HPV types. The concordance rate between the PCR and HC2 test results was high in our series. Both molecular tests exhibited high reproducibility measured by Cohen's kappa value of 0.691, similar to previous reported results [17, 18]. Although not all studies use the same PCR protocol, the concordance between HC2 and PCR testing across various studies is generally high, usually exceeding 80% [17–20]. It appears that after a certain DNA concentration cutoff point, HC2 sensitivity increases, carrying a certain risk for false positive results[17]. Newer generation kits decrease rates of false positive testing[21]. Certain cutoff levels for relative light units compared to the ones generated by the positive control samples to increase the specificity of the assay without compromising sensitivity may be used[18]. One explanation for some of the discordant results between HC2 and PCR in our study is the fact that GP5+/GP6+ primers may actually miss some cases deleted in the L1 region of the virus. In addition, some of the samples exhibiting discordance may have infection with HPV types such as HPV 42, 44 or 68 not amplified by the used primers, but still detectable by the HC2 cocktails. Moreover, HC2 probe B cocktail can react with phylogenetically related HPV types not represented in the probe such as HPV 67, CP 6108, and CP8061. Finally, the prevalence of latent infection differs across studies, depending upon demographic parameters; this may also affect the sensitivity and specificity of the two tests.
Our results confirm the higher prevalence of HPV infection in women with abnormal cytology, in concordance with most studies so far published, that have observed that the increase in HPV prevalence is related to the increasing grade of squamous intraepithelial lesions [22–26]. Rates of HPV detection in samples with normal cytology vary widely in the literature in women with normal cytology and range from 4.9% to 30.4% for the HC2 assay and from 3 to 34.3% for PCR testing [18, 19, 21, 25, 27]. This wide range is explained by the different nature of participating populations in such studies, and by technical evolutions in the diagnostic tests used[21, 28]. The higher analytical sensitivity of PCR explains the higher detection rates compared to HC2 in samples with low-grade cytological and no histological abnormalities and may be suggestive of the presence of latent HPV infection requiring a molecular test with higher analytical sensitivity.
Similar to other observations, both hybrid capture and PCR HPV DNA testing showed a strong correlation with the diagnosis of CIN lesions or squamous cell carcinoma [23, 29]. The extremely high negative predictive values of both molecular tests underlie their importance in screening for high grade lesions and are in concordance with previous publications[18]. In high grade cytological and histological lesions, HPV detection rates appeared to be a little higher with PCR than with HC2 testing; for example HC2 did not identify high risk viruses in seven instances of CIN2+ histological lesions, in which cases PCR was positive. However positive and negative likelihood ratios were similar for positive HC2 (any result) or PCR - HPV positive testing (any result) both for CIN2+ or for CIN3+ histology. Actually, HC2 had a better PLR while PCR had a better NLR in both histological categories. This means that with a positive HC2 there was a greater likelihood of disease (≥ CIN2) than with PCR, while with a negative HPV PCR there was a lesser likelihood of disease (≥ CIN2) than with HC2. Nevertheless, in essence both tests gave similar results. Likelihood ratios can be used to calculate the odds of post test probabilities if multiplied by the odds of the prevalence of the disease. For example, a patient in our study had a post test probability of disease (CIN2+) of only 5.1% (using the Bayes nomogram) if she had a negative HC2 test (according to our study she had a pre-test probability of 20.4% of having a positive test). Similarly a negative PCR for high risk types 16, 18, 31 and 33 has a post test probability of disease (CIN2+) of 1.93% (Table 4). The limitation of a population that is not systematically screened, but is consecutively enrolled for deriving this data is of course recognized. However, both molecular HPV tests did not outperform cytology that had comparable if not better results, especially if the HSIL+ cytological lesion cutoff point was used. In our study all HSIL lesions by cytology were either CIN 2 or CIN 3 by histology and all cytological diagnoses of invasive carcinomas were subsequently confirmed by histology. Possible explanations for this observation include the following: a) the experience of all doctors involved; b) the increased alertness of both the referring gynecologist and the cytopathologist examining the slides; c) the fact that the Department of Cytopathology is accredited and therefore undergoes continuous quality assessment/quality control evaluation.
Although none of the patients with normal cytology and an HPV infection had a biopsy proven lesion higher than CIN 1 in our study, these women should not be considered false-positive but as having a real risk for progression to abnormal cytological findings and cervical neoplasia [30–33]. It is known that development of precancerous lesions may shortly follow infection with HPV, despite the belief that long-term infection is a prerequisite for such an event [34]. Thus, these women should be prospectively followed by their gynecologist and submitted to cytology and other testing, as appropriate[35, 36]. The use of HPV testing has been recommended for women with ASCUS[37] and it has been shown that approximately one third of women with HSIL are subsequently identified from an initial ASCUS diagnosis [25]. These women, after positive HPV DNA testing should be referred for colposcopy[38]; our study underlines this argument, since one woman with ASCUS, had a single infection with HPV type 16 and biopsy disclosed squamous cell carcinoma. Nowadays, in the revised Bethesda 2001 classification system this case would probably be diagnosed as ASC-H. A molecular technique, more sensitive and specific than HC2 may be more appropriate for such patients.
The current observations further confirm the association of high risk HPV types with cytological detection of HSIL or invasive carcinoma but also with histologically confirmed premalignant or malignant lesions [39–41]. All cases of CIN 2, CIN 3 or carcinomas harbored single or multiple HPV high risk oncogenic type infections. In our study single infections with HPV types 16 or 18 in cases of invasive carcinoma were strong evidence for this association and support the strong rationale for preventive HPV vaccination in our population.
The rate of invasive carcinoma discovered in this study is indicative of a largely unscreened population. The reasons and possible solutions for this observation are an important public health issue and should be further investigated. Moreover the effect of the introduction of newer techniques in population screening is a matter of intense research. Despite the excellent results with cytology in this study, it is well known that screening for cytological changes may have limited sensitivity and findings are not always reproducible [8]. In a country where screening is largely opportunistic and based on self-referral results of cytological testing are expected to be much worse than the ones presented here. Moreover, molecular HPV testing should not be introduced without careful planning; results of such testing should be communicated and explained appropriately in the context of prevalence of the disease.
Conclusion
In conclusion, the present study was more of a technical nature and focused on strengths and weakness of current screening methods in correlation with histological findings. HPV rates were high in high grade lesions in accordance to published literature however HPV was also identified in a large fraction of samples with normal cytology or in samples with normal biopsies. The best approach for such samples will be guided after careful identification of the local epidemiology of the transmission and risk factors for HPV acquisition. Newer methods assessing the integration of the virus may be more appropriate in the workup of such cases. In our study in an referral center with experienced doctors, thin prep cytology and molecular HPV tests performed equally well as screening tests in all scenarios and sometimes were complementary to each other. As newer methods to detect HPV continue to evolve it is important to recognize the clinicopathological correlates of such testing. This will lead to more appropriate screening strategies in the post-vaccine era.
References
Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV: Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995, 87 (11): 796-802. 10.1093/jnci/87.11.796.
Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N: Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999, 189 (1): 12-19. 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F.
Munoz N, Castellsague X, de Gonzalez AB, Gissmann L: Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006, 24S3: S1-S10. 10.1016/j.vaccine.2006.05.115.
Clifford GM, Smith JS, Plummer M, Munoz N, Franceschi S: Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003, 88 (1): 63-73. 10.1038/sj.bjc.6600688.
Castellsague X, Diaz M, de Sanjose S, Munoz N, Herrero R, Franceschi S, Peeling RW, Ashley R, Smith JS, Snijders PJ, et al: Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: implications for screening and prevention. J Natl Cancer Inst. 2006, 98 (5): 303-315.
Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ: Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003, 348 (6): 518-527. 10.1056/NEJMoa021641.
Ames A, Gravitt P: Human papillomavirus vaccine update. Curr Infect Dis Rep. 2007, 9 (2): 151-158. 10.1007/s11908-007-0011-6.
Sahebali S, Depuydt CE, Boulet GA, Arbyn M, Moeneclaey LM, Vereecken AJ, Van Marck EA, Bogers JJ: Immunocytochemistry in liquid-based cervical cytology: analysis of clinical use following a cross-sectional study. Int J Cancer. 2006, 118 (5): 1254-1260. 10.1002/ijc.21489.
Evans MF, Adamson CS, Papillo JL, St John TL, Leiman G, Cooper K: Distribution of human papillomavirus types in ThinPrep Papanicolaou tests classified according to the Bethesda 2001 terminology and correlations with patient age and biopsy outcomes. Cancer. 2006, 106 (5): 1054-1064. 10.1002/cncr.21664.
Solomon D, Davey D, Kurman R, Moriarty A, O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T, et al: The 2001 Bethesda System: terminology for reporting results of cervical cytology. Jama. 2002, 287 (16): 2114-2119. 10.1001/jama.287.16.2114.
Wright TC, Kurman RJ, Ferenczy A: Precancerous lesions of the cervix. Blaustein's pathology of the female genital tract. Edited by: Kurman RJ. 2002, New York: Springer, 253-324. 5
de Roda Husman AM, Walboomers JM, Brule van den AJ, Meijer CJ, Snijders PJ: The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995, 76 (Pt 4): 1057-1062. 10.1099/0022-1317-76-4-1057.
Pao CC, Lin CY, Maa JS, Lai CH, Wu SY, Soong YK: Detection of human papillomaviruses in cervicovaginal cells using polymerase chain reaction. J Infect Dis. 1990, 161 (1): 113-115.
Giard RW, Hermans J: The evaluation and interpretation of cervical cytology: application of the likelihood ratio concept. Cytopathology. 1993, 4 (3): 131-137. 10.1111/j.1365-2303.1993.tb00078.x.
Clavel C, Masure M, Levert M, Putaud I, Mangeonjean C, Lorenzato M, Nazeyrollas P, Gabriel R, Quereux C, Birembaut P: Human papillomavirus detection by the hybrid capture II assay: a reliable test to select women with normal cervical smears at risk for developing cervical lesions. Diagn Mol Pathol. 2000, 9 (3): 145-150. 10.1097/00019606-200009000-00004.
Castle PE, Lorincz AT, Mielzynska-Lohnas I, Scott DR, Glass AG, Sherman ME, Schussler JE, Schiffman M: Results of human papillomavirus DNA testing with the hybrid capture 2 assay are reproducible. J Clin Microbiol. 2002, 40 (3): 1088-1090. 10.1128/JCM.40.3.1088-1090.2002.
Peyton CL, Schiffman M, Lorincz AT, Hunt WC, Mielzynska I, Bratti C, Eaton S, Hildesheim A, Morera LA, Rodriguez AC, et al: Comparison of PCR- and hybrid capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J Clin Microbiol. 1998, 36 (11): 3248-3254.
Kulmala SM, Syrjanen S, Shabalova I, Petrovichev N, Kozachenko V, Podistov J, Ivanchenko O, Zakharenko S, Nerovjna R, Kljukina L, et al: Human papillomavirus testing with the hybrid capture 2 assay and PCR as screening tools. J Clin Microbiol. 2004, 42 (6): 2470-2475. 10.1128/JCM.42.6.2470-2475.2004.
Yamazaki H, Sasagawa T, Basha W, Segawa T, Inoue M: Hybrid capture-II and LCR-E7 PCR assays for HPV typing in cervical cytologic samples. Int J Cancer. 2001, 94 (2): 222-227. 10.1002/ijc.1455.
Masumoto N, Fujii T, Ishikawa M, Mukai M, Saito M, Iwata T, Fukuchi T, Kubushiro K, Tsukazaki K, Nozawa S: Papanicolaou tests and molecular analyses using new fluid-based specimen collection technology in 3000 Japanese women. Br J Cancer. 2003, 88 (12): 1883-1888. 10.1038/sj.bjc.6601023.
Cuzick J, Beverley E, Ho L, Terry G, Sapper H, Mielzynska I, Lorincz A, Chan WK, Krausz T, Soutter P: HPV testing in primary screening of older women. Br J Cancer. 1999, 81 (3): 554-558. 10.1038/sj.bjc.6690730.
Meijer CJ, Brule van den AJ, Snijders PJ, Helmerhorst T, Kenemans P, Walboomers JM: Detection of human papillomavirus in cervical scrapes by the polymerase chain reaction in relation to cytology: possible implications for cervical cancer screening. IARC Sci Publ. 1992, 271-281. 119
Cox JT, Lorincz AT, Schiffman MH, Sherman ME, Cullen A, Kurman RJ: Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 1995, 172 (3): 946-954. 10.1016/0002-9378(95)90026-8.
Nobbenhuis MA, Walboomers JM, Helmerhorst TJ, Rozendaal L, Remmink AJ, Risse EK, Linden van der HC, Voorhorst FJ, Kenemans P, Meijer CJ: Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet. 1999, 354 (9172): 20-25. 10.1016/S0140-6736(98)12490-X.
Manos MM, Kinney WK, Hurley LB, Sherman ME, Shieh-Ngai J, Kurman RJ, Ransley JE, Fetterman BJ, Hartinger JS, McIntosh KM, et al: Identifying women with cervical neoplasia: using human papillomavirus DNA testing for equivocal Papanicolaou results. JAMA. 1999, 281 (17): 1605-1610. 10.1001/jama.281.17.1605.
Clavel C, Masure M, Bory JP, Putaud I, Mangeonjean C, Lorenzato M, Gabriel R, Quereux C, Birembaut P: Hybrid Capture II-based human papillomavirus detection, a sensitive test to detect in routine high-grade cervical lesions: a preliminary study on 1518 women. Br J Cancer. 1999, 80 (9): 1306-1311. 10.1038/sj.bjc.6690523.
Riethmuller D, Gay C, Bertrand X, Bettinger D, Schaal JP, Carbillet JP, Lassabe C, Arveux P, Seilles E, Mougin C: Genital human papillomavirus infection among women recruited for routine cervical cancer screening or for colposcopy determined by Hybrid Capture II and polymerase chain reaction. Diagn Mol Pathol. 1999, 8 (3): 157-164. 10.1097/00019606-199909000-00009.
Cuzick J, Mayrand MH, Ronco G, Snijders P, Wardle J: Chapter 10: New dimensions in cervical cancer screening. Vaccine. 2006, 24 (Suppl 3): S90-97. 10.1016/j.vaccine.2006.05.122.
Kuhn L, Denny L, Pollack A, Lorincz A, Richart RM, Wright TC: Human papillomavirus DNA testing for cervical cancer screening in low-resource settings. J Natl Cancer Inst. 2000, 92 (10): 818-825. 10.1093/jnci/92.10.818.
Hildesheim A, Schiffman MH, Gravitt PE, Glass AG, Greer CE, Zhang T, Scott DR, Rush BB, Lawler P, Sherman ME, et al: Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994, 169 (2): 235-240.
Sherman ME, Lorincz AT, Scott DR, Wacholder S, Castle PE, Glass AG, Mielzynska-Lohnas I, Rush BB, Schiffman M: Baseline cytology, human papillomavirus testing, and risk for cervical neoplasia: a 10-year cohort analysis. J Natl Cancer Inst. 2003, 95 (1): 46-52.
Kjaer SK, Brule van den AJ, Paull G, Svare EI, Sherman ME, Thomsen BL, Suntum M, Bock JE, Poll PA, Meijer CJ: Type specific persistence of high risk human papillomavirus (HPV) as indicator of high grade cervical squamous intraepithelial lesions in young women: population based prospective follow up study. BMJ. 2002, 325 (7364): 572-10.1136/bmj.325.7364.572.
Clavel C, Masure M, Bory JP, Putaud I, Mangeonjean C, Lorenzato M, Nazeyrollas P, Gabriel R, Quereux C, Birembaut P: Human papillomavirus testing in primary screening for the detection of high-grade cervical lesions: a study of 7932 women. Br J Cancer. 2001, 84 (12): 1616-1623. 10.1054/bjoc.2001.1845.
Winer RL, Kiviat NB, Hughes JP, Adam DE, Lee SK, Kuypers JM, Koutsky LA: Development and duration of human papillomavirus lesions, after initial infection. J Infect Dis. 2005, 191 (5): 731-738. 10.1086/427557.
Richart RM, Masood S, Syrjanen KJ, Vassilakos P, Kaufman RH, Meisels A, Olszewski WT, Sakamoto A, Stoler MH, Vooijs GP, et al: Human papillomavirus. International Academy of Cytology Task Force summary. Diagnostic Cytology Towards the 21st Century: An International Expert Conference and Tutorial. Acta Cytol. 1998, 42 (1): 50-58.
Lorincz AT, Richart RM: Human papillomavirus DNA testing as an adjunct to cytology in cervical screening programs. Arch Pathol Lab Med. 2003, 127 (8): 959-968.
Kaufman RH, Adam E, Icenogle J, Reeves WC: Human papillomavirus testing as triage for atypical squamous cells of undetermined significance and low-grade squamous intraepithelial lesions: sensitivity, specificity, and cost-effectiveness. Am J Obstet Gynecol. 1997, 177 (4): 930-936. 10.1016/S0002-9378(97)70296-5.
Nuovo GJ: Detection of human papillomavirus in Papanicolaou smears: correlation with pathologic findings and clinical outcome. Diagn Mol Pathol. 1998, 7 (3): 158-163. 10.1097/00019606-199806000-00006.
Ho GY, Burk RD, Klein S, Kadish AS, Chang CJ, Palan P, Basu J, Tachezy R, Lewis R, Romney S: Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995, 87 (18): 1365-1371. 10.1093/jnci/87.18.1365.
Remmink AJ, Walboomers JM, Helmerhorst TJ, Voorhorst FJ, Rozendaal L, Risse EK, Meijer CJ, Kenemans P: The presence of persistent high-risk HPV genotypes in dysplastic cervical lesions is associated with progressive disease: natural history up to 36 months. Int J Cancer. 1995, 61 (3): 306-311. 10.1002/ijc.2910610305.
zur Hausen H: Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002, 2 (5): 342-350. 10.1038/nrc798.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/10/53/prepub
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ST, AC, JG, and PK conceived and participated in the design and coordination of the study; ZV collected thin prep and histology samples; JG, AT and PK performed the thin prep evaluation; AC performed the molecular studies; JP and PK performed the pathological evaluation; ST, JP, AP and PK performed the statistical analysis and analyzed the data; ST, AC, JP and PK drafted the first draft; All authors reviewed and critically revised the first draft; All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Tsiodras, S., Georgoulakis, J., Chranioti, A. et al. Hybrid capture vs. PCR screening of cervical human papilloma virus infections. Cytological and histological associations in 1270 women. BMC Cancer 10, 53 (2010). https://doi.org/10.1186/1471-2407-10-53
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-10-53