Abstract
Background
GLUT10 (gene symbol SLC2A10) is a facilitative glucose transporter within the type 2 diabetes (T2DM)-linked region on chromosome 20q12-13.1. Therefore, we evaluated GLUT10 as a positional candidate gene for T2DM in Caucasian Americans.
Methods
Twenty SNPs including 4 coding, 10 intronic and 6 5' and 3' to the coding sequence were genotyped across a 100 kb region containing the SLC2A10 gene in DNAs from 300 T2DM cases and 310 controls using the Sequenom MassArray Genotyping System. Allelic association was evaluated, and linkage disequilibrium (LD) and haplotype structure of SLC2A10 were also determined to assess whether any specific haplotypes were associated with T2DM.
Results
Of these variants, fifteen had heterozygosities greater than 0.80 and were analyzed further for association with T2DM. No evidence of significant association was observed for any variant with T2DM (all P ≥ 0.05), including Ala206Thr (rs2235491) which was previously reported to be associated with fasting insulin. Linkage disequilibrium analysis suggests that the SLC2A10 gene is contained in a single haplotype block of 14 kb. Haplotype association analysis with T2DM did not reveal any significant differences between haplotype frequencies in T2DM cases and controls.
Conclusion
From our findings, we can conclude that sequence variants in or near GLUT10 are unlikely to contribute significantly to T2DM in Caucasian Americans.
Similar content being viewed by others
Background
Multiple genetic studies have been carried out that link human chromosome 20q13.1-13.2 to type 2 diabetes (T2DM) [1–5]. This linkage evidence has led investigators to search for T2DM susceptibility genes in this genomic region. Our laboratory has carried out analysis of specific genes [6–8] and developed high resolution physical maps of the region [9–11]. In an association analysis of genetic markers Price et al. [12] identified three regions of T2DM susceptibility. Among the genes mapped to the linkage disequilibrium regions, a novel facilitative glucose transporter (GLUT) was identified and designated GLUT10 (gene symbol SLC2A10) [6, 13]. The gene spans 28 kb of genomic sequence, is split into 5 exons and 4 introns [6, 13] and is expressed mainly in heart, liver, lung, skeletal muscle, pancreas, placenta, thyroid, and adipose tissue [6, 13, 14]. In Xenopus oocytes, human GLUT10 exhibited a high affinity, saturable 2-deoxy-D-glucose transport activity [6].
Glucose transport plays a central role in metabolism. Defects in glucose transport have been implicated in the reduced insulin sensitivity and in the increased insulin resistance observed in type 2 diabetes [14, 15]. The genes coding for the classical GLUT1-5 proteins have been extensively analyzed for mutations contributing to type 2 diabetes, but no common causative mutation has been identified [16, 17]. Both the function and genomic location of the novel SLC2A10 gene are consistent with a role in T2DM. In addition, Andersen et al. recently observed evidence for association of an Ala206Thr polymorphism in SLC2A10 with lower fasting insulin levels although they observed no evidence for association with T2DM [18]. We have carried out a systematic genetic evaluation of SLC2A10 to assess association with T2DM.
Methods
Subjects
The individual samples evaluated in this study consisted of a collection of 300 unrelated Caucasian T2DM patients with end-stage renal disease (ESRD), with a corresponding collection of 310 randomly-ascertained unrelated Caucasian subjects without known diabetes. Both cases and controls were recruited simultaneously. This group will be referred to as "T2DM-ESRD"; their ascertainment and recruitment have been previously described in detail [19]. The T2DM-ESRD subjects have a mean age at diagnosis of diabetes of 46.5 ± 12.8 years, mean BMI at recruitment of 28.5 ± 7.0, and mean maximum reported BMI of 36.1 ± 8.3, mean duration of diabetes > 15 years, and mean HbA1c of 8.6%.
SNP selection and genotyping
Twenty SNPs used in this study were selected from the dbSNP public database (rs1004571, rs6012006, rs4810542, rs4810544, rs2425897, rs4428069, rs2425902, rs6094438, rs2235491, rs2076293, rs2425906, rs2425907, rs6094440, rs998422, rs2425911, rs3091904, rs6018008, rs707507, rs1059217, rs6090547). SNPs with frequency information were preferentially selected. The majority of the SNPs had minor allele frequencies greater than 0.2.
Genotyping was performed on a Sequenom MassArray Genotyping System using methods previously described [19]. Discordance between blind duplicate samples included in the genotyping was <0.9% and the call rate for each assay was set at ≥ 90%. Average call rate was 98.1% (range 95.6% to 100%).
Statistical analysis
Pearson's test of homogeneity of proportions was applied to analyze allele frequency differences between diabetic and nondiabetic subjects SNPs were tested for Hardy-Weinberg equilibrium and pairwise linkage disequilibrium statistics D' calculated using the SNP-Analysis software package [20].
HAPLO.SCORE [21] was used to test for association of haplotypes within the case-control populations. HAPLO.SCORE uses posterior population-based frequency weighting of haplogenotypes that have prior consistency with observed heterozygous diplotypes. Variance estimates are inflated to account for uncertainty in specific haplotype assignment. We performed a global test of association between GLUT10 haplotypes and T2DM status, followed by individual tests of haplotype risk, in a collapsed 2 × 2 analysis. The skip.haplo option was set to 0.01. DANDELION [22], another expectation-maximization (EM) algorithm-based program for haplotype analyses was used to calculate haplotype frequencies in specific subject groups. A P < 0.05 was considered statistically significant.
Results
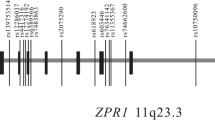
To examine the association of SLC2A10 variants with T2DM, 20 SNPs spanning the genomic region were evaluated. The average SNP density was 1 SNP/5.2 kb with inter-SNP distances ranging from 0.078 kb to 26.856 kb. The location of the SNPs relative to the exon-intron structure of SLC2A10 is shown in Figure 1. The 20 SNPs were genotyped in a collection of Caucasian T2DM-ESRD cases (n = 300) and controls (n = 310) and single SNP association analysis was performed on those SNPs with minor allele frequencies greater than 0.20. Five of the SNPs are located in coding regions (rs6094438, rs2235491, rs6018008, rs707507, rs1059217), but only rs1059217 had a minor allele frequency greater than 0.20. The coding variant rs2235491 (Ala206Thr) had a low minor allele frequency of 0.04, but was included since it has been previously described as associated with reduced fasting insulin [18]. The remaining coding variants, rs6094438, rs6018008, rs707507, are so rare that they are very poorly informative with minor allele frequencies ≤ 0.01. In addition two SNPs in the noncoding regions, rs4428069 and rs609440, were also found to be nonpolymorphic.
Genomic map of the SLC2A10 gene with the locations of the 20 genotyped SNPs. The shaded regions are exons, numbered 1–5. The ruler along the bottom represents the relative location and spacing of SNPs in kilobases within the 40 kb region containing SLC2A10. Note that this does not have a uniform scale.
In the control population, rs2076293 (P = <0.0001) and rs6090547 (P = 0.00043) did not conform to Hardy-Weinberg equilibrium. The allelic association analysis is summarized in Table 1, which shows the SNP identifier, the relevant alleles for each SNP, frequencies in cases and controls, and P-values. This analysis revealed moderate evidence of association between SNP rs2076293 and T2DM (P = 0.025). In addition, genotypic association analysis showed moderate evidence of association of this variant with T2DM under an additive model (P = 0.039). However, these results should be viewed with caution since the proportions of the genotypes for the variant are significantly out of Hardy-Weinberg equilibrium in the control population.
Linkage disequilibrium (LD) and haplotype structure of SLC2A10 were determined to assess whether any specific haplotypes were associated with T2DM. Inter-SNP D' statistics were calculated for the gene region as shown in Figure 2 using LD Viewer (unpublished). The upper portion of the matrix displays the calculated inter-SNP D' values, where a D' ≥ 0.70 is defined as high LD. The lower portion of the matrix shows the calculated P-values for each D' value, where a P-value ≤ 0.05 is significant (e.g. the calculated D' value is significant). Using these criteria (D' ≥ 0.70, P ≤ 0.05), SLC2A10 SNPs appear to be one LD block covering a 14 kb region which begins with variant rs2425902 in intron 1 and ends beyond the 3'-UTR with variant rs1059217.
Marker-to-marker D' plot for the SLC2A10 SNPs. Inter-SNP D' values are graphically represented using LD Viewer which generates a color coded plot of the pair-wise statistics in the upper portion of the matrix, and a color coded plot of the calculated P-value for each LD measurement in the bottom portion of the matrix.
Haplotype frequencies were estimated and association analyses performed using the programs Dandelion [22] and HAPLO.SCORE [21]. Shown in Table 2 are the results from the analyses evaluating 7 SNP haplotypes within the single major LD block of the SLC2A10 gene. Three common haplotypes are estimated and no significant evidence of haplotype association was observed with T2DM. It is important to note that although the variants rs2235491 and rs2235491 were within the LD block, they were removed from the analysis due to their low minor allele frequencies (rs2235491) or due to departure from Hardy-Weinberg equilibrium (rs2235491).
Discussion
SNPs spanning the SLC2A10 genomic sequence have been genotyped in a Caucasian American T2DM case-control population. Most of the coding SNPs validated in this gene region were so rare in this study sample that they were uninformative for our analysis. The remaining SNPs did not show significant evidence for association to T2DM. This included SNP rs2235491 (Ala206Thr) which was previously reported to be associated with fasting insulin (but without evidence of associaton with T2DM) [18]. Linkage disequilibrium analysis suggests that there is one LD block covering a 14 kb region beginning with variant rs2425902 in intron 1 and ending with variant rs1059217 in the 3'UTR region. Haplotype analysis based on the LD structure did not reveal evidence for association with T2DM. However, the study is not powered (with the criterion MAF > 0.1) or designed to address less common SNPs or haplotypes. We have carried out a detailed power analysis for this case-control study group elsewhere (Bento et al, submitted). Briefly, assuming a T2DM disease prevalence of 10% and modest multiplicative genotype risk ratios of 1.5, the power to detect association at a nominal SNP significance level of 5% is 74.7% under an additive trend test with risk allele frequency of 10%, and >95% for risk allele frequency 20%.
Conclusion
From these observations, we can conclude that variants in or near GLUT10 do not contribute substantially to T2DM in this sample of Caucasian Americans. This does not exclude the possibility that evidence for association could be observed in a larger case-control study or a study with cases ascertained in a different manner. However, another study has confirmed these negative association results. When examining SNPs in SLC2A10 using a Finnish population, no significant evidence for T2DM association with any SNP was observed [23]. In addition, this same study and another performed in a Danish population could not confirm the published association of Ala206Thr and fasting insulin [18, 24]. It is noteworthy that we have observed evidence for association to T2DM with the nearby PTPN1 gene, and to a lesser extent, with the HNF4A gene which is also located within 20q12-13.1.
Abbreviations
- SNP- single nucleotide polymorphism:
-
T2DM- Type 2 diabetes, LD- linkage disequilibrium, ESRD- end stage renal disease, BMI- body mass index, GLUT10- glucose transporter 10, SLC2A10- (gene name for GLUT10) solute carrier family 2 member 10, PTPN1- protein tyrosine phosphatase non-receptor type 1, HNF4A- hepatocyte nuclear factor 4-alpha
References
Bowden DW, Sale M, Howard TD, Qadri A, Spray BJ, Rothschild CB, Akots G, Rich SS, Freedman BI: Linkage of genetic markers on human chromosomes 20 and 12 to NIDDM in Caucasian sib pairs with a history of diabetic nephropathy. Diabetes. 1997, 46 (5): 882-886.
Ji L, Malecki M, Warram JH, Yang Y, Rich SS, Krolewski AS: New susceptibility locus for NIDDM is localized to human chromosome 20q. Diabetes. 1997, 46 (5): 876-881.
Klupa T, Malecki MT, Pezzolesi M, Ji L, Curtis S, Langefeld CD, Rich SS, Warram JH, Krolewski AS: Further evidence for a susceptibility locus for type 2 diabetes on chromosome 20q13.1-q13.2. Diabetes. 2000, 49 (12): 2212-2216.
Permutt MA, Wasson JC, Suarez BK, Lin J, Thomas J, Meyer J, Lewitzky S, Rennich JS, Parker A, DuPrat L, Maruti S, Chayen S, Glaser B: A genome scan for type 2 diabetes susceptibility loci in a genetically isolated population. Diabetes. 2001, 50 (3): 681-685.
Zouali H, Hani EH, Philippi A, Vionnet N, Beckmann JS, Demenais F, Froguel P: A susceptibility locus for early-onset non-insulin dependent (type 2) diabetes mellitus maps to chromosome 20q, proximal to the phosphoenolpyruvate carboxykinase gene. Hum Mol Genet. 1997, 6 (9): 1401-1408. 10.1093/hmg/6.9.1401.
Dawson PA, Mychaleckyj JC, Fossey SC, Mihic SJ, Craddock AL, Bowden DW: Sequence and functional analysis of GLUT10: a glucose transporter in the Type 2 diabetes-linked region of chromosome 20q12-13.1. Mol Genet Metab. 2001, 74 (1-2): 186-199. 10.1006/mgme.2001.3212.
Fossey SC, Kuroda S, Price JA, Pendleton JK, Freedman BI, Bowden DW: Identification and characterization of PRKCBP1, a candidate RACK-like protein. Mamm Genome. 2000, 11 (10): 919-925. 10.1007/s003350010174.
Price JA, Fossey SC, Sale MM, Brewer CS, Freedman BI, Wuerth JP, Bowden DW: Analysis of the HNF4 alpha gene in Caucasian type II diabetic nephropathic patients. Diabetologia. 2000, 43 (3): 364-372. 10.1007/s001250050055.
Fossey SCMJCPJKSJRBJTHSRSSFBIBDW: A high resolution 6.0 Mb transcript map of the Type 2 diabetes susceptibility region on human chromosome 20. Genomics. 2000, 76: 45-57. 10.1006/geno.2001.6584.
Price JA, Brewer CS, Howard TD, Fossey SC, Sale MM, Ji L, Krolewski AS, Bowden DW: A physical map of the 20q12-q13.1 region associated with type 2 diabetes. Genomics. 1999, 62 (2): 208-215. 10.1006/geno.1999.6007.
Bento JLMJCRBHPFBIBDW: A SNP map of human chromosome 20q12-13.1. American Journal of Human Genetics. 2003, 73 supplemental: 486-
Price JABCSHTDFSCRSSFBIWJPBDW: Construction of a physical map of chromosome 20q12-13.1 and linkage disequilibrium analysis in diabetic nephropathy patients. American Journal of Human Genetics. 1997, 58 supplemental: A241-
McVie-Wylie AJ, Lamson DR, Chen YT: Molecular cloning of a novel member of the GLUT family of transporters, SLC2a10 (GLUT10), localized on chromosome 20q13.1: a candidate gene for NIDDM susceptibility. Genomics. 2001, 72 (1): 113-117. 10.1006/geno.2000.6457.
Wood IS, Hunter L, Trayhurn P: Expression of Class III facilitative glucose transporter genes (GLUT-10 and GLUT-12) in mouse and human adipose tissues. Biochem Biophys Res Commun. 2003, 308 (1): 43-49. 10.1016/S0006-291X(03)01322-6.
Matthaei S, Stumvoll M, Kellerer M, Haring HU: Pathophysiology and pharmacological treatment of insulin resistance. Endocr Rev. 2000, 21 (6): 585-618. 10.1210/er.21.6.585.
Rothman DL, Magnusson I, Cline G, Gerard D, Kahn CR, Shulman RG, Shulman GI: Decreased muscle glucose transport/phosphorylation is an early defect in the pathogenesis of non-insulin-dependent diabetes mellitus. Proc Natl Acad Sci U S A. 1995, 92 (4): 983-987.
Cline GW, Petersen KF, Krssak M, Shen J, Hundal RS, Trajanoski Z, Inzucchi S, Dresner A, Rothman DL, Shulman GI: Impaired glucose transport as a cause of decreased insulin-stimulated muscle glycogen synthesis in type 2 diabetes. N Engl J Med. 1999, 341 (4): 240-246. 10.1056/NEJM199907223410404.
Andersen G, Rose CS, Hamid YH, Drivsholm T, Borch-Johnsen K, Hansen T, Pedersen O: Genetic variation of the GLUT10 glucose transporter (SLC2A10) and relationships to type 2 diabetes and intermediary traits. Diabetes. 2003, 52 (9): 2445-2448.
Bento JL, Palmer ND, Mychaleckyj JC, Lange LA, Langefeld CD, Rich SS, Freedman BI, Bowden DW: Association of protein tyrosine phosphatase 1B gene polymorphisms with type 2 diabetes. Diabetes. 2004, 53 (11): 3007-3012.
Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA: Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002, 70 (2): 425-434. 10.1086/338688.
Green LELEMLCD: Power comparison of phase-known versus phase-unknown haplotype analyses for case-control designs. Am J Hum Genet. 2001, 69: 1948a-
Mohlke KL, Skol AD, Scott LJ, Valle TT, Bergman RN, Tuomilehto J, Boehnke M, Collins FS: Evaluation of SLC2A10 (GLUT10) as a candidate gene for type 2 diabetes and related traits in Finns. Mol Genet Metab. 2005, 85 (4): 323-327. 10.1016/j.ymgme.2005.04.011.
Rose CS, Andersen G, Hamid YH, Glumer C, Drivsholm T, Borch-Johnsen K, Jorgensen T, Pedersen O, Hansen T: Studies of relationships between the GLUT10 Ala206Thr polymorphism and impaired insulin secretion. Diabet Med. 2005, 22 (7): 946-949. 10.1111/j.1464-5491.2005.01547.x.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/6/42/prepub
Acknowledgements
This work was supported by NIH grants R01 DK56289 to DWB.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
Dr. Bento carried out the majority of the genotyping and data analysis. The data analysis plan was designed and supervised by Drs. Mychaleckyj and Rich. Dr. Hirakawa carried out the initial laboratory studies identifying and genotyping SNPs in the coding sequence of GLUT10. Dr. Freedman oversaw recruitment, diagnosis, and clinical characterization of the subjects used in this study. Drs. Bowden and Segade conceived the overall study design and supervised the performance of the study. All authors participated in writing and editing the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bento, J.L., Bowden, D.W., Mychaleckyj, J.C. et al. Genetic analysis of the GLUT10 glucose transporter (SLC2A10) polymorphisms in Caucasian American type 2 diabetes. BMC Med Genet 6, 42 (2005). https://doi.org/10.1186/1471-2350-6-42
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-6-42