Abstract
Background
Congenital forms of hearing impairment can be caused by mutations in the estrogen related receptor beta (ESRRB) gene. Our initial linkage studies suggested the ESRRB locus is linked to high caries experience in humans.
Methods
We tested for association between the ESRRB locus and dental caries in 1,731 subjects, if ESRRB was expressed in whole saliva, if ESRRB was associated with the microhardness of the dental enamel, and if ESRRB was expressed during enamel development of mice.
Results
Two families with recessive ESRRB mutations and DFNB35 hearing impairment showed more extensive dental destruction by caries. Expression levels of ESRRB in whole saliva samples showed differences depending on sex and dental caries experience.
Conclusions
The common etiology of dental caries and hearing impairment provides a venue to assist in the identification of individuals at risk to either condition and provides options for the development of new caries prevention strategies, if the associated ESRRB genetic variants are correlated with efficacy.
Similar content being viewed by others
Background
Dental caries is a major public health problem and is estimated to affect 60 to 90 percent of school children as well as a vast number of adults [1]. Also, data from across the world show that children with hearing disorders suffer from poor oral health [2–12]. Congenital forms of hearing impairment can be caused by mutations in the estrogen related receptor beta (ESRRB) gene [13–16]. ESRRB is located in the 14q24.3 locus, which was linked to dental caries through a genome-wide linkage scan [17]. This is not the first time that hearing loss is associated with alterations of dental structures. Distinct mutations of the dentin sialophosphoprotein gene (DSPP), a gene involved in the initial mineralization of the dentin matrix, are responsible for the clinical manifestations of dentinogenesis imperfecta 1 with or without autosomal dominant progressive high frequency sensorineural hearing loss (DFNA39) [18]. In addition, a case control study of 572 college age musicians showed that ESRRB nonsynomous SNP rs61742642 (P386S) was associated with bilateral notches in their ears and thus suffered from hearing loss due to acoustic overload [19].
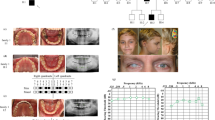
Estrogen has a role in the preservation of hearing in aging human adults, and ESRRB binds to estrogen-responsive elements of downstream transcription targets of estrogen signaling [20]. Alterations in transcription, mediated by the glucocorticoid receptor (GR), can contribute to the phenotype of hearing loss related to ESRRB mutations because GR, like ESRRB, is widely expressed during and after maturation of the mouse and rat cochlea. In addition, ESRRB might repress transcriptional activity mediated by GR [13, 20, 21]. Here we investigated whether loss of function of ESRRB, which in humans leads to hearing impairment, also leads to increased dental caries experience. We used multiple experiments to attempt to gain a thorough idea of how ESRRB plays a role in dental decay. Figure 1 provides a visual representation of how the work was conceptualized and developed.
Methods
Subjects studied are summarized in Table 1. Overall, we studied 1,731 individuals, including two consanguineous families (Figure 2) carrying ESRRB mutations associated with congenital hearing loss. Subjects had ancestry in the Philippines, Turkey, Brazil, Argentina, and the Czech Republic. These populations have been described previously [13, 16, 17, 22–26]. The two consanguineous families are from Turkey and the Czech Republic and have been described previously as well [13, 16]. The study was approved by the Institutional Review Board of the University of Pittsburgh, appropriate oversight committees of human participation in research in the Philippines, Turkey, Brazil, Argentina, and the Czech Republic, and written informed consent was obtained from each person included in the study. Whole blood was collected for genetic analysis in the cases of the Philippines and the two consanguineous families from Turkey and the Czech Republic. The remaining study groups had DNA purified from whole saliva. Dental caries data was recorded by the use of the Decayed-Missing due to caries-Filled Teeth index (DMFT). Detailed descriptions of the assessments for each population are in the Additional file 1.
Dental caries and tooth loss information was collected for two consanguineous families (one from Turkey [13] segregating a seven base pair duplication mutation and one from the Czech Republic [16] segregating the missense mutation R291L). Families reported on the status of their teeth based on what they were told by their dentist. The detailed reports are presented in Additional file 1: Table S1. Based on these reports, dental caries status was defined as high or low caries experience. These families have recessive hearing impairment due to mutations in ESRRB. It is important to note that the family from Turkey comes from a region of low socioeconomic status with limited access to dental care, whereas the family from the Czech Republic resides in a metropolitan area with better access to dental care. Dental data were collected by phone interview (E.K. in Turkey and D.S.B. in the Czech Republic). Of the 17 family members contacted by E.K., 15 provided information regarding their dental caries experience. D.S.B. was able to obtain information from four family members. Dental caries experience between ESRRB mutation carriers and non-carriers was compared using the Fisher’s exact test.
To study 14q24.3, single nucleotide polymorphisms (SNPs) were selected using data from the International HapMap project on Caucasians and Chinese (http://www.hapmap.org), which were viewed using Haploview [27]. Twenty-five single nucleotide polymorphisms (SNPs) were selected in 14q24.3 for fine-mapping and are listed in Additional file 1: Table S2 based on pairwise linkage disequilibrium and gene structure data. The Graphical Overview of Linkage Disequilibrium (GOLD) software was used to calculate pairwise linkage disequilibrium between the SNPs and help interpret data [28].
Genotyping was performed using Taqman chemistry end-point analysis. Association between the chosen SNPs and recorded dental caries experience was tested with the transmission disequilibrium test (TDT) implemented in the Family-Based Association Test (FBAT) statistical program [29]. Bonferroni correction was implemented to correct for multiple comparisons and significance was set at 0.002 (0.05/25). Eight SNPs that showed a trend for association with dental caries experience in the Filipino dataset were studied in the additional four population datasets. These eight SNPs were all present within or flanking the ESRRB gene. Data were analyzed using the PLINK software [30]. In order to derive a summary statistic for association with the eight SNPs across populations, a random-effects meta-analysis model was used to estimate the odds ratio for the presence of the associated allele determined by the fine-mapping of the Filipino families. Before pooling the data, we estimated Cochran’s Q statistic, which indicates the degree of heterogeneity. There was no significant evidence of heterogeneity overall (Q = 7.0, p = 0.429). A random-effects model was used because it includes variance components both within and between studies. Moreover, because the random-effects model generally yields a wider confidence interval than a fixed-effects model, the random-effects model is more conservative [31].
The less common allele of rs17074565 in 13q31.1 was associated with dental caries and was predicted to disrupt a binding site of GR [32]. Lower expression levels of GR in whole saliva are also associated with high dental caries experience [32]. In the Filipino sample, we tested if the eight ESRRB SNPs that showed a trend for association with dental caries also interacted with the SNP rs17074565. We observed the transmission of alleles from parents heterozygous for both the rs17074565 SNP and the ESRRB SNPs to estimate if specific allele combinations were transmitted more often than expected.
All of the exons and exon-intron boundaries of ESRRB were sequenced and compared with the reference sequence transcript ENST00000505752 obtained from Ensembl Genome Browser (http://useast.ensembl.org/index.html). Ninety-three samples from the Turkish cohort were used (62 caries samples and 31 caries free control samples). Primers are listed in Additional file 1: Table S3.
Total RNA isolated from a subset of 94 subjects from the Argentinean population described above was used to test if ESRRB expression can be detected in whole saliva. Subsequent cDNA synthesis from 100 ng of total RNA was accomplished by using High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Primers specific for the three ESRRB isoforms [13] were tested (ESRRB short, long, and Delta10 isoforms listed in Additional file 1: Table S4); GAPDH was our endogenous control. Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems). Quantification of ESRRB expression levels compared to GAPDH was performed by 2-DeltaDeltaCT method [33]. Real-time PCR amplification was performed with an initial denaturation at 95°C for five minutes, 60 cycles at 95°C for 45 seconds, 55°C for 45 seconds, and finally 72°C for 90 seconds in a 7900HT Real-time PCR machine. Real-time results were confirmed by western blotting analysis. ESRRB expression levels were analyzed based on the presence of zero, one, or two copies of lesser common alleles, sex, and dental caries experience. Non-parametric tests were used in all comparisons.
Total RNA from the submaxillary salivary gland epidermoid carcinoma cell line HTB-41™ (American Type Culture Collection) was isolated and studied. cDNA synthesis and real-time PCR conditions used were described above. GAPDH was used as the endogenous control. Amplification of cDNA was performed with SYBR Green PCR Master Mix (Applied Biosystems).
Enamel samples from extracted premolar teeth from 100 orthodontic adolescent patients (63 with high dental caries experience and 37 with low caries experience, Table 1) from Istanbul University were used in enamel microhardness testing (Figure 3). The enamel samples came from premolars and were used to test the association between genetic variation in ESRRB and enamel microhardness at baseline, after simulating artificial caries, and after fluoride treatment. The goal was to test the hypothesis that ESRRB influences dental caries by generating a more susceptible enamel surface to acidic dissolution. The Ethics Committee of Istanbul University approved this study, and informed consent from all participating patients was obtained. Subjects age ranged from ten to 32 years (mean age of 17.2 years; 38 males and 62 females). The mean DMFT ranged from zero to 17 (mean DMFT 5.2; 63 with high caries experience and 37 with low caries experience). Tooth samples were cleaned of any remnants and stored in a 10% formalin solution (pH = 7.0) at room temperature until the initial polishing. The crowns of each tooth were separated from the roots and then separated again buccolingually and mesiodistally. The five surfaces studied were occlusal, mesial, buccal, distal, and lingual. The surfaces were sanded for one minute, at a force of 1 lbf, while moving at a speed of 20 rpm on paper of 320, 400, and 600 grit, and then polished for seven minutes at a force of 1 lbf at a speed of 25 rpm in 6 μm, 1 μm, and 0.25 μm diamond suspension. Sample baseline microhardness was tested using a microhardness tester (IndentaMet 1100, Buehler Ltd.) with a knoop diamond. Five indentations under a load of 25 grams for five seconds were made. Next, artificial caries was simulated by immersing the samples in 24 mL of demineralizing solution (1.3 mmol/L Ca, 0.78 mmol/L P, 0.05 mol/L acetate buffer, 0.03 μg F/mL, pH:5.0) at 37°C for 16 hours. Microhardness was again measured by five indentations created just below the initial ones. These indentations were then exposed for ten minutes to a fluoride solution, created from toothpaste containing sodium fluoride (1,400 ppm fluoride), to determine if microhardness for the artificial caries lesions would be brought back to baseline levels after fluoride exposure. Again, surface microhardness was measured with five more indentations below the previous ten.
Enamel microhardness study design. Image above A depicts the five tooth surfaces upon which enamel microhardness was tested. Image above B is a close-up of the occlusal surface. Image above C shows the appearance of an enamel specimen ready to be tested. Image below D shows the testing unit. Microhardness was tested at baseline, after artificial caries creation, and after exposure to a fluoridated solution (panels above E, F, and G. are schematic representations of the assessments).
The results of the microhardness testing were compared to the genotyping of DNA extracted from saliva from each of the 100 patients. Results were analyzed using the PLINK software package28. Mean microhardness at baseline, after artificial caries lesion creation, and after fluoride application was calculated. Subjects were divided into two comparison groups: above and below the means. We made comparisons by surface, as well as by using the mean enamel microhardness of all five surfaces combined. A p-value of 0.0004 was considered statistically significant to accommodate for multiple comparisons (Bonferroni correction: 0.05/144).
Expression of Esrrb during enamel development was determined by immunohistochemical analysis of sections of mouse mandibular molars at postnatal day four (secretory stage) and postnatal day eleven (maturation stage).
Results
Of the 25 SNPs used for fine-mapping the region 14q24.3, eight SNPs within or flanking ESRRB were found to be over-transmitted in a sample population from the Philippines (Figure 4). Linkage disequilibrium was assessed for these markers and is presented graphically as Additional file 1: Figure S1. Studies of additional populations also indicated associations (details in Additional file 1: Tables S5 through S8). Populations are summarized in Table 1. Figure 5 shows the odds ratios for the association of rs1676303 in samples from the Philippines, Turkey, Brazil, and Argentina. We also show odds ratios when the Brazilian samples, which had lower dental caries experience, were excluded from the meta-analysis (meta-analysis of the additional SNPs can be found in Additional file 1: Figures S2 through S8).
Odds ratios for over-representation of the T allele (rs1676303) in individuals with high dental caries. A. Meta-analysis with the five studied groups suggest an association between ESRRB rs1676303 and dental caries. B. When the two study groups with less dental caries are removed from analysis, the association between ESRRB rs1676303 and dental caries becomes stronger.
Two families with DFNB35 hearing loss and ESRRB mutations were contacted for this study, one from Turkey [13] and the other from the Czech Republic [16]. Of the 17 Turkish DFNB35 family members with a recessive seven base pair duplication in exon 8 of ESRRB (c.1018_1024dupGAGTTTG) and hearing impairment [13] contacted by phone interview, 15 provided information regarding their dental caries experience (Figure 2). The ten members of the family who are carriers for the ESRRB mutation (six homozygous, four heterozygous) have severe dental caries, with many if not all teeth affected by caries. Three of the five individuals without ESRRB mutations were caries free, and two of the five had low caries experience (Fisher’s exact test, p = 0.02). The DFNB35 family from the Czech Republic has a recessive missense mutation (R291L) in ESRRB and hearing impairment. Both the mother of the affected child and the mother’s father had high dental caries experience. The affected four-year-old child is caries-free in his primary dentition, and his father is apparently affected by periodontal disease. It was not possible to define the father’s dental caries status.
We also found statistical evidence of an interaction between ESRRB SNPs and a SNP predicted to disrupt a GR binding site in families with high dental caries experience (Table 2).
From the sequencing of ESRRB exons and exon-intron boundaries, SNPs rs10132091, rs61742642, rs3813545, rs3829784, rs45533334, rs35544003, rs2361292, and rs55835922 were found in our samples. There is no evidence indicating these SNPs are disease-causing variants. No mutations causing hearing impairment were found. Individuals with dental caries have an over-representation of the T allele of rs55835922 (74% versus 54%; p = 0.01). The SNP rs61742642 is a missense mutation (P386S), but its frequency was just slightly elevated in cases with dental caries (13% versus 9.5%). SNP rs35544003 is a synonymous change not thought to have any detrimental effect. Detailed sequencing results are listed in Additional file 1: Table S9.
Only expression of the short ESRRB isoform listed in Additional file 1: Table S4 was detected, both by real time PCR and western blot analyses (Additional file 1: Figure S9). Additionally, expression of the short ESRRB isoform was detected in the submaxillary salivary gland epidermoid carcinoma cell line HTB-41™. The data from the real time PCR experiments indicate that adult females express ESRRB in whole saliva in higher levels than men (p = 0.01). Furthermore, a statistical association was found between ESRRB expression in whole saliva of children and rs745011 allele distribution (p = 0.04). In a dominant model, statistical association was found between ESRRB expression and rs10132091 genotypes (p = 0.03), between low dental caries experience and rs10132091 genotypes (p = 0.05), and between low ESRRB expression in whole saliva of adults and rs6574293 genotypes (p = 0.04). In a recessive model, statistical association was found between low ESRRB expression and rs2860216 genotypes (p = 0.02).
Through experiments that tested the effects of acid dissolution of the enamel surface, we reasoned that ESRRB variants contributes to formation of an enamel structure that is more susceptible to the acidic effects involved in the initiation of dental caries. Distribution of alleles of SNP rs4903419 was different between subjects with harder and softer enamel at baseline under a recessive model (The G allele was associated with harder enamel, p = 0.0007). Also, the distribution of alleles of SNP rs6574293 was different when enamel microhardness at the distal surface was compared after the creation of an artificial caries lesion and after fluoride application (The A allele was associated with harder enamel after fluoride application, p = 0.0006). Complete results are summarized in Additional file 1: Table S10.
Immunohistochemical stain with rabbit polyclonal antibody, demonstrated that Esrrb is expressed by mouse ameloblasts, the cells that deposit tooth enamel, during the secretory stage of amelogenesis in mice (postnatal day four, Additional file 1: Figure S10), but not during the later maturation stage of dental development, such as postnatal day eleven.
Discussion
Mice that are Esrrb-deficient, or which have a conditional knockout of the Esrrb gene, exhibit head-tossing, head-bobbing, and running in circles caused by inner-ear defects [34, 35]. In humans, the ESRRB autosomal recessive hearing impairment indicates that ESRRB is essential for inner-ear development [13–16]. We showed that SNPs in the ESRRB locus are also associated with dental caries experience in multiple populations, but particular populations are less influenced by factors that protect against the disease. Results were clearer when we removed the “healthier” groups from the pooled analysis (Figure 5), leaving the ones with limited access to dental care, similar cultural and social behaviors, and sub-optimal exposure to fluoridated drinking water. When we evaluated dental caries in two families with previously described hearing impairment and ESRRB mutations, the evidence clearly showed that the severity of dental destruction was much more apparent in mutation carriers. Upon testing the enamel of human teeth in regards to genetic variation in ESRRB, we found evidence that “softer” enamel is associated with ESRRB SNPs.
ESRRB is suggested to repress transcriptional activity mediated by GR, and these two proteins are widely expressed during and after maturation of the mouse and rat cochlea [13]. We previously showed that a SNP in 13q31.1 (rs17074565) was associated with dental caries and potentially disrupts the GR binding site [32]. We found statistical evidence that rs17074565 and ESRRB SNPs are over-transmitted together in families with high dental caries experience.
Both clinical and archeological evidence suggest that women have higher levels of dental caries [36–43], although these differences are not evident when studies are performed in individuals with similar socioeconomic levels and environments [44–46]. The differences are suggested to be the consequence of sex disparities and bias related to the risk factors modulating dental caries [47]. On the other hand, men appear more commonly to have faster hearing deterioration, in part due to the types of occupations that favor males [48]. This evidence is promising in the sense that ESRRB detection in whole saliva can be explored not only in regards to risks of dental caries, but also related to risks of hearing loss related to aging or occupational hazard (i.e., dentists [49]). The rationale for this suggestion comes from the hypothesis that ESRRB could cause congenital forms of hearing impairment as well as increased susceptibility to the acquired forms of hearing loss. A similar phenomenon happens with diabetes. Data on susceptibility genes and familial clustering for Type 1 and Type 2 Diabetes in humans, mice, and rats suggest the possibility of shared genetic susceptibility to both Type 1 and Type 2 Diabetes in humans [50, 51].
We have demonstrated that informed candidate-gene selection aids in identifying specific variants with a role in complex traits that may be otherwise missed by genome-wide association studies [52–55]. The association of dental caries and hearing impairment provides a venue to assist in the identification of individuals at risk to either condition and provides options for the development of new strategies of prevention for both caries and hearing loss, if the associated ESRRB genetic variants are correlated with efficacy.
Conclusions
ESRRB, a gene when mutated causes a form of hearing impairment, also contributes to dental decay likely by influencing the formation of an enamel surface more susceptible to demineralization under acidic conditions.
Web resources
The URLs for presented data are as follows:
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP.
Ensemble Genome Browser: http://www.ensembl.org/index.html.
HapMap Project: http://hapmap.ncbi.nlm.nih.gov.
PLINK: http://pngu.mgh.harvard.edu/~purcell/plink/.
Primer3: http://biotools.umassmed.edu/bioapps/primer3_www.cgi.
UCSC Genome Bioinformatics: http://genomebrowser.ucsc.edu.
References
World Health Organisation: The World Oral Health Report 2003. 2003, http://www.who.int/oral_health/publications/report03/en/,
Semczuk B: Studies on the role of the state of dentition in the physiopathology of the auditory organ. IV. Studies on the effect of dentition on the etiology of acoustic trauma. Ann Univ Mariae Curie Sklodowska Med. 1967, 22: 173-178.
Dahl T: Dental findings in children with hearing disorders. Zahn Mund Kieferheilkd Zentralbl. 1975, 63: 24-31.
Mpozinakou M, Papaphragkou K, Galanopoulou E, Giamerelos G, Pantazopoulos P: Management of dental problems in the deaf child. Stomatologia (Athenai). 1977, 34: 45-49.
Masierowska A, Fetkowska-Mielnik K: Oral hygiene and dental status in children with visual and auditory function impairment in 2 educational institutions in Lublin. Czas Stomatol. 1986, 39: 311-317.
Parlato M, Alessandrella G: Survey of dental caries incidence in a group of deaf children. Arch Stomatol (Napoli). 1990, 31: 81-83.
Gupta DP, Chowdhury R, Sarkar S: Prevalence of dental caries in handicapped children of Calcutta. J Indian Soc Pedod Prev Dent. 1993, 11: 23-27.
Vanderley ATVO: University of Pernanbuco School of Dentistry Master’s thesis. Behavior and social factors and its relation with dental caries prevalence on institutionalized special children (with auditive and visual deficiency), in the city of Recife – PE. 2001, 132-
Al-Qahtani Z, Wyne AH: Caries experience and oral hygiene status of blind, deaf and mentally retarded female children in Riyadh, Saudi Arabia. Odontostomatol Trop. 2004, 27: 37-40.
Jain M, Mathur A, Kumar S, Dagli RJ, Duraiswamy P, Kulkarni S: Dentition status and treatment needs among children with impaired hearing attending a special school for the deaf and mute in Udaipur, India. J Oral Sci. 2008, 50: 161-165.
Wei H, Wang YL, Cong XN, Tang WQ, Wei PM: Survey and analysis of dental caries in students at a deaf-mute high school. Res Dev Disabil. 2012, 33: 1279-1286.
Nqcobo CB, Yengopal V, Rudolph MJ, Thekiso M, Joosab Z: Dental caries prevalence in children attending special needs schools in Johannesburgh, Gauteng Province, South Africa. SADJ. 2012, 67: 308-313.
Collin RW, Kalay E, Tariq M, Peters T, van der Zwaag B, Venselaar H, Oostrik J, Lee K, Ahmed ZM, Caylan R, Li Y, Spierenburg HA, Eyupoglu E, Heister A, Riazuddin S, Bahat E, Ansar M, Arslan S, Wollnik B, Brunner HG, Cremers CW, Karaguzel A, Ahmad W, Cremers FP, Vriend G, Friedman TB, Riazuddin S, Leal SM, Kremer H: Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am J Hum Genet. 2008, 82: 125-138.
Ben Saïd M, Ayedi L, Mnejja M, Hakim B, Khalfallah A, Charfeddine I, Khifagi C, Turki K, Ayadi H, Benzina Z, Ghorbel A, Castillo ID, Masmoudi S, Aifa MH: A novel missense mutation in the ESRRB gene causes DFNB35 hearing loss in a Tunisian family. Eur J Med Genet. 2011, 54: e535-e541.
Lee K, Khan S, Ansar M, Santos-Cortez RL, Ahmad W, Leal SM: A novel ESRRB deletion is a rare cause of autosomal recessive nonsyndromic hearing impairment among Pakistani families. Genet Res Int. 2011, 2011: 368915-
Brožková DS, Laštůvková J, Machalová E, Lisoňová J, Trková M, Seeman P: DFNB35 due to a novel mutation in the ESRRB gene in a Czech consanguineous family. Int J Pediatr Otorhinolaryngol. 2012, 76: 1681-1684.
Vieira AR, Marazita ML, Goldstein-McHenry T: Genome-wide scan finds suggestive caries loci. J Dent Res. 2008, 87: 435-439.
Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X: Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001, 27: 201-204.
Henrich VC, Phillips SL, Richter SJ, Teglas S, Morehouse R: Abstracts of the Annual Meeting of the American Society of Human Genetics. A polymorphism in human estrogen-related receptor beta (ESRRβ) is associated with early indications of hearing loss from acoustic overload in young adult musicians. 2012, http://www.ashg.org/2012meeting/abstracts/fulltext/f120122450.htm,
Hultcrantz M, Simonoska R, Stenberg AE: Estrogen and hearing: a summary of recent investigations. Acta Otolaryngol. 2006, 126: 10-14.
Trapp T, Holsboer F: Heterodimerization between mineralocorticoid and glucocorticoid receptors increases the functional diversity of corticosteroid action. Trends Pharmacol Sci. 1996, 17: 145-149.
Patir A, Seymen F, Yildirim M, Deeley K, Cooper ME, Marazita ML, Vieira AR: Enamel formation genes are associated with high caries experience in Turkish children. Caries Res. 2008, 42: 394-400.
Shimizu T, Ho B, Deeley K, Briseño-Ruiz J, Faraco IM, Schupack BI, Brancher JA, Pecharki GD, Küchler EC, Tannure PN, Lips A, Vieira TC, Patir A, Yildirim M, Poletta FA, Mereb JC, Resick JM, Brandon CA, Orioli IM, Castilla EE, Marazita ML, Seymen F, Costa MC, Granjeiro JM, Trevilatto PC, Vieira AR: Enamel formation genes influence enamel microhardness before and after cariogenic challenge. PLoS One. 2002, 7: 45022-
Shimizu T, Deeley K, Briseño-Ruiz J, Faraco IM, Poletta FA, Brancher JA, Pecharki GD, Küchler EC, Tannure PN, Lips A, Vieira TC, Patir A, Yildirim M, Mereb JC, Resick JM, Brandon CA, Cooper ME, Seymen F, Costa MC, Granjeiro JM, Trevilatto PC, Orioli IM, Castilla EE, Marazita ML, Vieira AR: Fine-Mapping of 5q12.1-13.3 unveils new genetic contributors to caries. Caries Res. 2013, 47: 273-283.
Briseño-Ruiz J, Shimizu T, Deeley K, Dizak PM, Ruff TD, Faraco IM, Poletta FA, Brancher JA, Pecharki GD, Küchler EC, Tannure PN, Lips A, Vieira TC, Patir A, Koruyucu M, Mereb JC, Resick JM, Brandon CA, Letra A, Silva RM, Cooper ME, Seymen F, Costa MC, Granjeiro JM, Trevilatto PC, Orioli IM, Castilla EE, Marazita ML, Vieira AR: Role of TRAV locus in low caries experience. Hum Genet. 2013, 132: 1015-1025.
Tannure PN, Küchler EC, Lips A, Costa MC, Luiz RR, Granjeiro JM, Vieira AR: Genetic variation in MMP20 contributes to higher caries experience. J Dent. 2012, 40: 381-386.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265.
Abecasis GR, Cookson WO: GOLD-graphical overview of linkage disequilibrium. Bioinformatics. 2000, 16: 182-183.
Horvath S, Xu X, Laird NM: The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet. 2001, 9: 301-306.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, Bakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007, 81: 559-575.
Berlin JA, Laird NM, Sacks HS, Chalmers TC: A comparison of statistical methods for combining event rates from clinical trials. Stat Med. 1989, 8: 141-151.
Küchler EC, Deeley K, Ho B, Linkowski S, Meyer C, Noel J, Kouzbari MZ, Bezamat M, Granjeiro JM, Antunes LS, Abreu FV, Costa MC, Tannure PN, Seymen F, Koruyucu M, Patir A, Mereb JC, Poletta FA, Castilla EE, Orioli IM, Marazita ML, Vieira AR: Genetic mapping of high caries experience on human chromosome 13. BMC Med Genet. 2013, 14: 116-
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001, 25: 402-408.
Mitsunaga K, Araki K, Mizusaki H, Morohashi K, Haruna K, Nakagata N, Giguère V, Yamamura K, Abe K: Loss of PGC-specific expression of the orphan nuclear receptor ERR-beta results in reduction of germ cell number in mouse embryos. Mech Dev. 2004, 121: 237-246.
Chen J, Nathans J: Estrogen-related receptor beta/NR3B2 controls epithelial cell fate and endolymph production by the stria vascularis. Dev Cell. 2007, 13: 325-337.
National Research Council: A Survey of the Literature of Dental Caries. 1952, Washington D.C.: National Academy of Sciences, 567-
Koch G, Martinsson T: Socio-odontologic investigation of school children with high and low caries frequency. I. Socioeconomic background. Odontol Revy. 1970, 21: 207-228.
Varenne B, Petersen PE, Ouattara S: Oral health status of children and adults in urban and rural areas of Burkina Faso, Africa. Int Dent J. 2004, 54: 83-89.
Madléna M, Hermann P, Jáhn M, Fejérdy P: Caries prevalence and tooth loss in Hungarian adult population: results of a national survey. BMC Public Health. 2008, 8: 364-
Demirci M, Tuncer S, Yuceokur AA: Prevalence of caries on individual tooth surfaces and its distribution by age and gender in university clinic patients. Eur J Dent. 2010, 4: 270-279.
Costa SM, Vasconcelos M, Haddad JP, Abreu MH: The severity of dental caries in adults aged 35 to 44 years residing in the metropolitan area of a large city in Brazil: a cross-sectional study. BMC Oral Health. 2012, 12: 25-
Maru AM, Narendran S: Epidemiology of dental caries among adults in a rural area in India. J Contemp Dent Pract. 2012, 2012 (13): 382-388.
Lukacs JR, Largaespada LL: Explaining sex differences in dental caries prevalence: saliva, hormones, and “life-history” etiologies. Am J Hum Biol. 2006, 18: 540-555.
Potgieter M, Morse EH, Relenbach FM, Dall R: The food habits and dental status of some Connecticut children. J Dent Res. 1956, 35: 638-644.
Zita AC, McDonald RE, Andrews AL: Dietary habits and dental caries experience in 200 children. J Dent Res. 1959, 38: 860-865.
Clancy KL, Bibby BG, Goldberg HJ, Ripa LW, Barenie J: Snack foods and caries in adolescence. J Dent Res. 1977, 56: 568-573.
Martinez-Mier EA, Zandona AF: The impact of gender on caries prevalence and risk assessment. Dent Clin North Am. 2013, 57: 301-315.
Linssen AM, van Boxtel MP, Joore MA, Anteunis LJ: Predictors of hearing acuity: Cross-sectional and longitudinal analysis. J Gerontol A Biol Sci Med Sci. 2013, [Epub ahead of print]
Gurbuz MK, Catli T, Cingi C, Yaz A, Bal C: Occupational safety threats among dental personnel and related risk factors. J Craniofac Surg. 2013, 24: e599-e602.
Eftychi C, Howson JM, Barratt BJ, Vella A, Payne F, Smyth DJ, Twells RC, Walker NM, Rance HE, Tuomilehto-Wolf E, Tuomilehto J, Undlien DE, Rønningen KS, Guja C, Ionescu-Tîirgovişte C, Savage DA, Todd JA: Analysis of the type 2 diabetes-associated single nucleotide polymorphisms in the genes IRS1, KCNJ11, and PPARG2 in type 1 diabetes. Diabetes. 2004, 53: 870-873.
Ikegami H, Fujisawa T, Ogihara T: Mouse models of type 1 and type 2 diabetes derived from the same closed colony: genetic susceptibility shared between two types of diabetes. ILAR J. 2004, 45: 268-277.
Shaffer JR, Wang X, Feingold E, Lee M, Begum F, Weeks DE, Cuenco KT, Barmada MM, Wendell SK, Crosslin DR, Laurie CC, Doheny KF, Pugh EW, Zhang Q, Feenstra B, Geller F, Boyd HA, Zhang H, Melbye M, Murray JC, Weyant RJ, Crout R, McNeil DW, Levy SM, Slayton RL, Willing MC, Broffitt B, Vieira AR, Marazita ML: Genome-wide association scan for childhood caries implicates novel genes. J Dent Res. 2011, 90: 1457-1462.
Wang X, Shaffer JR, Zeng Z, Begum F, Vieira AR, Noel J, Anjomshoaa I, Cuenco KT, Lee MK, Beck J, Boerwinkle E, Cornelis MC, Hu FB, Crosslin DR, Laurie CC, Nelson SC, Doheny KF, Pugh EW, Polk DE, Weyant RJ, Crout R, McNeil DW, Weeks DE, Feingold E, Marazita ML: Genome-wide association scan of dental caries in the permanent dentition. BMC Oral Health. 2012, 12: 57-
Shaffer JR, Feingold E, Wang X, Lee M, Cuenco KT, Weeks DE, Weyant RJ, Crout R, McNeil DW, Marazita ML: GWAS of dental caries patterns in the permanent dentition. J Dent Res. 2013, 92: 38-44.
Zeng Z, Shaffer JR, Wang X, Feingold E, Weeks DE, Lee M, Cuenco KT, Wendell SK, Weyant RJ, Crout R, McNeil DW, Marazita ML: Genome-wide association studies of pit-and-fissure- and smooth-surface caries in permanent dentition. J Dent Res. 2013, 92: 432-437.
Rozen S, Skaletsky H: Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000, 132: 365-386.
Dausset J: Le centre d'etude du polymorphisme humain. Presse Med. 1986, 13: 1801-
Jindal A, McMeans M, Narayanan S, Rose EK, Jain S, Marazita ML, Menezes R, Letra A, Carvalho FM, Brandon CA, Resick JM, Mereb JC, Poletta FA, Lopez-Camelo JS, Castilla EE, Orioli IM, Vieira AR: Women are more susceptible to caries but individuals born with clefts are not. Int J Dent. 2011, 2011: 454532-
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/15/81/prepub
Acknowledgements
We would like to thank the individuals that participated in this study for their support. We would also like to thank Eric Davis and Maggie Saludis for their contributions to the enamel microhardness portion of the study, Paula Noh for helping to optimize primers for sequencing, and Sarah Vinski for administrative support. This research is supported by NIH grants R01-DE018914 and R01-DE016148, and Czech Republic grant IGA MHCZ NT14348.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors have no competing interests to declare.
Authors’ contributions
MLW and ARV wrote the manuscript. All authors critically revised and approved the version submitted for publication. MLW, H-YH, EK, DSB, TS, MB, KD, ECC, JF, TDR, VMT, RCS, MH, JB-R, SKR, HO, and TJ generated and analyzed data and helped interpret results. JMG, LSA, LAA, FVA, MCC, and PNT collected clinical and biological patient data at the Brazilian site. JMG. ECK, and MCC helped design the component of this study developed in Brazil. EK, MK, AP, and MB collected clinical and biological patient data at the Turkish site. FS and EK helped design the component of this study developed in Turkey. FAP, JCM, IMO, MLM, and ECC helped design the component of this study developed in Argentina. FAP, ARV, and JB-R collected clinical and biological patient data at the Argentinean sites. ARV designed and provided oversight on all components of this project. All authors read and approved the final manuscript.
Electronic supplementary material
12881_2014_1227_MOESM1_ESM.docx
Additional file 1: Figure S1: Linkage Disequilibrium (D’) of ESRRB SNPs Showing Association with Dental Caries. Figure S2. Results of Meta-Analyses (rs745011). Figure S3. Results of Meta-Analyses (rs1077430). Figure S4. Results of Meta-Analyses (rs2860216). Figure S5. Results of Meta-Analyses (rs4903399). Figure S6. Results of Meta-Analyses (rs4903419). Figure S7. Results of Meta-Analyses (rs6574293). Figure S8. Results of Meta-Analyses (rs10132091). Figure S9. Western Blot of ESRRB in Whole Saliva from 10 Healthy Female Subjects. Figure S10. Immunohistochemical analysis showing Esrrb expression in secretory stage ameloblasts. Table S1. Dental Caries Status of the Two Families Segregating ESRRB Mutations and DFNB35. Table S2. SNPs Studied and Summary of Results of Fine-Mapping of the Filipino Population. Table S3. Primer Sets for Sequencing ESRRB exons and exon-intron boundaries. used for polymerase chain reaction (PCR) amplification were designed using Primer3 software [56] and supplied by Integrated DNA Technologies (Integrated DNA Technologies, Inc.). The samples were sent to Functional Biosciences, Inc. for purification and sequencing. The sequences were then verified against the reference sequence transcript and the sequences from two unrelated CEPH (Foundation Jean Dausset-Centre d’Etude du Polymorphisme Humain) DNA samples [57] using Sequencher 5.1 software (Gene Codes Corporation). Table S4. Primer Sets and Detailed Methods for RT-PCR Experiments. Table S5. Results of Association Studies in the Turkish Children. Table S6. Results of Association Studies in the Brazilian (Nova Friburgo) Children. Table S7. Results of Association Studies in the Brazilian (Rio de Janeiro) Children. Table S8. Results of Association Studies in the Argentinean Population. Table S9. Summary of Sequencing Results and Case-Control Comparisons. Table S10. Workflow and Results of Enamel Microhardness Testing [1, 17, 20, 24, 55, 56, 58]. (DOCX 1 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Weber, M.L., Hsin, HY., Kalay, E. et al. Role of estrogen related receptor beta (ESRRB) in DFN35B hearing impairment and dental decay. BMC Med Genet 15, 81 (2014). https://doi.org/10.1186/1471-2350-15-81
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-15-81