Abstract
Background
Recent genome wide association studies (GWAS) and previous positional linkage studies have identified more than 50 single nucleotide polymorphisms (SNPs) associated with obesity, mostly in Europeans. We aimed to assess the contribution of some of these SNPs to obesity risk and to the variation of related metabolic traits, in Mexican children.
Methods
The association of six European obesity-related SNPs in or near FTO, NPC1, ENPP1, NEGR1, GNPDA2 and MC4R genes with risk of obesity was tested in 1,463 school-aged Mexican children (N cases = 514; N controls = 949). We also assessed effects of these SNPs on the variation of body mass index (BMI), fasting serum insulin levels, fasting plasma glucose levels, total cholesterol and triglyceride levels, in a subset of 1,171 nonobese Mexican children.
Results
We found a significant effect of GNPDA2 rs10938397 on risk of obesity (odds ratio [OR] = 1.30; P = 1.34 × 10-3). Furthermore, we found nominal associations between obesity risk or BMI variation and the following SNPs: ENPP1 rs7754561, MC4R rs17782313 and NEGR1 rs2815752. Importantly, the at-risk alleles of both MC4R rs17782313 and NPC1 rs1805081 showed significant effect on increased fasting glucose levels (β = 0.36 mmol/L; P = 1.47 × 10-3) and decreased fasting serum insulin levels (β = −0.10 μU/mL; P = 1.21 × 10-3), respectively.
Conclusion
Our present results suggest that some obesity-associated SNPs previously reported in Europeans also associate with risk of obesity, or metabolic quantitative traits, in Mexican children. Importantly, we found new associations between MC4R and fasting glucose levels, and between NPC1 and fasting insulin levels.
Similar content being viewed by others
Background
Obesity and associated comorbidities (such as cardiovascular diseases, type 2 diabetes, musculoskeletal disorders and some cancers) represent a major public health problem that, in recent years, has reached epidemic proportions. In Mexico, obesity is the most common nutritional disorder of children, and the prevalence of this disease has increased alarmingly over the past decade: recent data from ENSA 2006 (‘Encuesta Nacional de Salud’, i.e. The National Health Census) reported an increase in overweight and obesity from 18% in 1999 to 26% in 2006, in young Mexican children (between 5 and 11 years old) [1]. Overweight in childhood or adolescence is a risk factor for overall mortality in adulthood [2].
The prevalence of obesity varies across populations and it is noteworthy that the Mexican population has been disproportionally affected. Although environmental changes (linked to the ‘westernization’ of ways of life) can explain the increase in prevalence of obesity on a global level, individual variation of body mass index (BMI) persists in the same environment, and the heritability of this trait is very high. Indeed, heritability estimates of BMI range between 40% and 70% according to studies [3]. The high genetic susceptibility to obesity (and both insulin resistance and type 2 diabetes) in the Mexican population may be ascribed to the American Indian heritage [4].
To date, genome wide association studies (GWAS) and linkage studies, mostly performed in European adult populations, have identified more than 50 loci associated with obesity or BMI [5–10]. However, replication attempts have yielded inconsistent outcomes [11–13]. In particular, replication in other populations is not obvious, and several factors (such as ethnic differences in linkage disequilibrium patterns, ethnic-specific associations, gene × environment interactions) may puzzle the picture. Furthermore, very few studies have been performed in children presenting with severe obesity in non-European populations.
In the present study, we aimed to perform a follow-up replication study including sixEuropean obesity-associated genetic variants, in Mexican children.
Methods
Study participants
In the present study, we analyzed 1,685 children (aged 6 to 12 years) of Mexican origin, from five different states of Mexico (San Luis Potosí, Queretaro, Tijuana, Guanajuato and Mexico city), who were randomly selected and invited to participate in a cross-sectional study between 2007 and 2011 from public and private schools. Child assent was obtained and parents provided written informed consent. We collected data from the children and parent or legal guardian per child by direct questioning.
Participants were scheduled for clinical laboratory evaluation following a 12 h overnight fasting. Blood samples were drawn to assess levels of fasting glucose, fasting insulin, total cholesterol and triglycerides levels. Biochemical variables were measured using an ILab 350 Clinical Chemistry System (Instrumentation Laboratory IL). Weight was measured with a digital scale (Seca) and height was measured with a portable stadiometer (Seca 225). BMI was calculated and classified according to the ‘Centers for Disease Control and Prevention 2000’ (CDC 2000) reference [14]. CDC 2000 growth charts are based on 5 U.S nationally representative surveys conducted between 1963 and 1994, in which Mexican American children were included [14]. According to those growth charts, for ages 2 to 20 years, overweight was defined as a BMI-for-age between the 85th and 95th percentiles, while obesity was defined when BMI-for-age was higher than the 95th percentile [14, 15]. Insulin resistance was defined as: homeostasis model assessment of insulin resistance (HOMA-IR = [(Fasting glucose (mg/dL)) (Fasting insulin (μU/mL))]/405) ≥ 3.4 (that is the 90th percentile of HOMA-IR in a population of healthy Mexican children [16, 17]).
The study was approved by the local ethics committees (‘Instituto Mexicano del Seguro Social’, i.e. the Mexican national health service; reference number: 2011-000-079) and is in compliance with the Helsinki Declaration.
SNPs Selection and genotyping
To achieve a power of at least 50%, with an odds ratio of 1.25, we only selected SNPs with minor allele frequencies ≥ 15% in the Mexican population according to the HapMap database. These SNPs were identified by GWAS or meta-analyses of GWAS in European populations [6, 7, 9, 10]. Furthermore, we selected a SNP that was significantly associated with childhood obesity in a French population, according to a linkage association study [8]. The expected statistical power per SNP (estimated by Quanto software) is reported in Additional file 1: Table S1.
Genomic DNA was isolated from peripheral blood white cells using a QIAamp DNA kit (Qiagen), purity and integrity was verified by 260/280 nm measurements and by electrophoresis in 0.8% agarose gels, stained with ethidium bromide.
All DNA samples were genotyped for the MC4R rs17782313, NEGR1 rs2815752, ENPP1 rs7754561, NPC1 rs1805081, GNPDA2 rs10938397 and FTO rs1421085 using TaqMan assays on an ABI7900 system, following the manufacturer’s protocol (Applied Biosystems). The genotype success rate was at least 98%, and no deviation (P ≥ 0.01) from Hardy-Weinberg equilibrium was observed in our population. Thirty duplicate quality control samples were included and were genotyped with 100% concordance.
Statistical analysis
The effect of SNPs on obesity status was assessed using a logistic regression model adjusted for age, gender and Mexican state (1: San Luis Potosí, 2: Queretaro, 3: Tijuana, 4: Guanajuato and 5: Mexico city), under an additive model. We also analyzed the effect of SNPs on several metabolic quantitative traits (BMI, fasting serum insulin, fasting plasma glucose, total cholesterol and triglycerides) using linear regressions under an additive model adjusted for age, gender and Mexican state. Data for fasting serum insulin and triglycerides were logarithmically transformed before statistical analysis. By applying Bonferroni correction, a significant p-value has been considered when below 1.4 × 10-3 (0.05/36) and a p-value between 0.05 and 1.4 × 10-3 has been considered as nominally significant. All statistical analyses were performed using the SPSS software (version 14.0).
Results
From an initial sample of 1,685 children (between 6 and 12 years old), we extracted 949 lean and 514 obese children (see clinical characteristics in Table 1) for the case–control study, and 1,171 nonobese children (with BMI < 95th percentile) for the study of metabolic quantitative traits including BMI, fasting serum insulin, fasting plasma glucose, total cholesterol and triglycerides. As expected, obese children exhibited higher values of insulin and cholesterol levels (P ≤ 0.001) compared to lean children. Insulin resistance was found in 45% of obese children.
In both studies, we genotyped six SNPs that are known to be associated with risk of obesity in European populations: rs17782313 near MC4R[6]; rs2815752 near NEGR1[7]; rs7754561 near ENPP1[8]; rs1805081 in exon 6 of NPC1[9]; rs10938397 near GNPDA2[7] and rs1421085 in intron 1 of FTO[10].
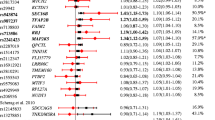
We identified a significant contribution of GNPAD2 rs10938397 to risk of obesity (G risk allele [in European populations]; odds ratio[95% confidence interval] [OR] = 1.30[1.11;1.53]; P = 1.34 × 10-3; Table 2). Furthermore, we found nominal association between risk of obesity and both ENPP1 rs7754561 (G risk allele; OR = 0.84[0.72;0.97]; P = 0.020; Table 2) and MC4R rs17782313 (C risk allele; OR = 1.40[1.06;1.83]; P = 0.016; Table 2). This last MC4R SNP also showed a nominal effect on BMI (C risk allele; β[standard error] = 0.41[0.16] kg/m2; P = 0.012; Table 3). Moreover, we identified a nominal association between NEGR1 rs2815752 and BMI (A risk allele; β = 0.24[0.10] kg/m2; P = 0.019; Table 3). Of note, we did not find any significant association between risk of obesity (or BMI variation) and either NPC1 rs2815752 or FTO rs1421085 (Tables 2 and Table 3).
In our study of quantitative traits, we found a significant association between NPC1 rs1805081 and fasting serum insulin levels (A risk allele; β = −0.10[0.03] μU/mL; P = 1.21 × 10-3; Table 3). We also identified an effect of the obesity risk allele of MC4R rs17782313 on increased fasting plasma glucose levels (C risk allele; β = 0.36[0.11] mmol/L; P = 1.47 × 10-3; Table 3). A trend of association was found between FTO rs1421085 and both total cholesterol levels (C risk allele; β = 3.70[1.72] mg/dL; P = 0.032; Table 3) and fasting serum insulin levels (C risk allele; β = −0.06[0.04] μU/mL; P = 0.064; Table 3).
Discussion
From our study based on obese and nonobese Mexican children, we found a significant contribution of the minor allele of GNPDA2 rs10938397 to increased risk of obesity. The association between GNPDA2 and BMI had firstly been identified in a meta-analysis of several European GWASs performed by the GIANT consortium [7]. Subsequently, the association signal has been confirmed in other populations: in adults from East-Asia [18, 19] and in Chinese children [20]. To our knowledge, the present study is the first confirmation of the GNPDA2 association signal with risk of obesity in the Mexican population.
Furthermore, we found nominal associations between risk of obesity or BMI and the following SNPs: ENPP1 rs7754561, MC4R rs17782313 and NEGR1 rs2815752. Of note, the risk alleles for obesity or increased BMI were the same between Europeans and Mexicans, except for the risk allele of ENPP1 rs7754561 (in Europeans) that showed a protective effect in Mexicans. Although the association between both MC4R and NEGR1 and risk of obesity has been confirmed in a plethora of studies and populations, the ENPP1 association signal with obesity is more controversial [21]. To our knowledge, no other studies demonstrate a protective role of the risk allele of ENPP1 rs7754561. Recently, it has been shown that ENPP1 overexpression in human adipocyte cell lines resulted in defective adipocyte maturation [22]. If confirmed in other Mexican populations, the protective effect of the ENPP1 variant may be due to a loss-of-function of the protein.
We did not find any significant contribution of either NPC1 rs2815752 or FTO rs1421085 to obesity or BMI variation. Therefore, the FTO association signal with obesity which was found in Mexican adults by Villalobos-Comparán and colleagues [23], was not confirmed in children. Thus, the obesogenic effect of FTO would occur later in the Mexican population than in the Europeans [9], but as we lack some statistical power, additional genetic studies on Mexican children would be needed.
Importantly, we identified a significant effect of the risk allele of NPC1 rs1805081 on decreased fasting serum insulin levels. NPC1 encodes Niemann Pick disease type C1 protein that mediates intracellular cholesterol trafficking via binding of cholesterol to its N-terminal domain. Very recently, Jelinek and colleagues have reported that Npc1 haploinsufficiency developed abnormal metabolic features (including hyperinsulinemia) and increased susceptibility to weight gain in mice [24]. We could suspect that the variant may have a gain-of-function effect in Mexican children. Of note, we did not report any association between the NPC1 variant and obesity or BMI variation, and this association signal remains quite controversial according to population studies [20, 25, 26].
Furthermore, we identified an effect of the risk allele of MC4R variant and increased fasting plasma glucose levels. To our knowledge, no previous study has shown this association. However, two recent large meta-analyses of GWAS identified a significant association between the MC4R locus and type 2 diabetes risk, in European and Asian populations [27, 28]. Of note, our present association with fasting plasma glucose levels remained significant after adjustment for BMI (data not shown). Altogether, these findings would suggest a potential effect of MC4R polymorphisms on decreased pancreatic beta-cell function.
Conclusion
In summary, in our sample of Mexican children, we replicated four European obesity-related genes (GNPDA2, and nominally, NEGR1, MC4R and ENPP1) in the same direction of effect as previous findings, except for ENPP1. Interestingly, we found two novel association signals: between NPC1 variant and fasting serum insulin levels, and between MC4R variant and fasting plasma glucose levels. These findings should deserve some confirmatory studies in other Mexican populations.
It is noteworthy that these well established genetic associations with obesity explain very little of the genetic risk for pediatric phenotype, suggesting the existence of additional loci whose number and effect size remain unknown, which guarantee intense additional investigations in the near future.
Abbreviations
- β:
-
Effect-size
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- GWAS:
-
Genome-wide association study
- HOMA-IR:
-
Homeostasis model assessment of insulin resistance
- OR:
-
Odds ratio
- P:
-
P-value
- pc:
-
Percentile
- RA:
-
Risk-allele
- RAF:
-
Risk-allele frequency
- SE:
-
Standard error
- SNP:
-
Single nucleotide polymorphism.
References
Olaiz G, Rivera J, Shamah T, Rojas R, Villalpando S, Hernández M, Sepúlveda J: Encuesta Nacional de Salud y Nutrición. 2006, Instituto Nacional de Salud Pública, Mexico City, Mexico
Must A: Does overweight in childhood have an impact on adult health?. Nutr Rev. 2003, 61 (4): 139-142. 10.1301/nr.2003.apr.139-142.
Allison DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, Hayakawa K: The heritability of body mass index among an international sample of monozygotic twins reared apart. Int J Obes Relat Metab Disord. 1996, 20 (6): 501-506.
Bonora E, Kiechl S, Willeit J, Oberhollenzer F, Egger G, Targher G, Alberiche M, Bonadonna RC, Muggeo M: Prevalence of insulin resistance in metabolic disorders: the Bruneck Study. Diabetes. 1998, 47 (10): 1643-1649. 10.2337/diabetes.47.10.1643.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Magi R, et al: Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2011, 42 (11): 937-948.
Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, et al: Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008, 40 (6): 768-775. 10.1038/ng.140.
Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, et al: Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009, 41 (1): 25-34. 10.1038/ng.287.
Meyre D, Bouatia-Naji N, Tounian A, Samson C, Lecoeur C, Vatin V, Ghoussaini M, Wachter C, Hercberg S, Charpentier G, et al: Variants of ENPP1 are associated with childhood and adult obesity and increase the risk of glucose intolerance and type 2 diabetes. Nat Genet. 2005, 37 (8): 863-867. 10.1038/ng1604.
Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proenca C, et al: Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009, 41 (2): 157-159. 10.1038/ng.301.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al: A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007, 316 (5826): 889-894. 10.1126/science.1141634.
Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, Charles MA, Balkau B, Froguel P: Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007, 315 (5809): 187-author reply 187
Rosskopf D, Bornhorst A, Rimmbach C, Schwahn C, Kayser A, Kruger A, Tessmann G, Geissler I, Kroemer HK, Volzke H: Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007, 315 (5809): 187-author reply 187
Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, Zhu X, Thorleifsson G, Gunnarsdottir S, Walters GB, Thorsteinsdottir U, et al: The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007, 3 (4): e61-10.1371/journal.pgen.0030061.
Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z, Wei R, Curtin LR, Roche AF, Johnson CL: CDC Growth Charts for the United States: methods and development. Vital Health Stat 11. 2000, 2002 (246): 1-190.
Barlow SE: Expert committee recommendations regarding the prevention, assessment, and treatment of child and adolescent overweight and obesity: summary report. Pediatrics. 2007, 120 (Suppl 4): S164-S192.
Garcia Cuartero B, Garcia Lacalle C, Jimenez Lobo C, Gonzalez Vergaz A, Calvo Rey C, Alcazar Villar MJ, Diaz Martinez E: [The HOMA and QUICKI indexes, and insulin and C-peptide levels in healthy children. Cut off points to identify metabolic syndrome in healthy children]. An Pediatr (Barc). 2007, 66 (5): 481-490. 10.1157/13102513.
Juarez-Lopez C, Klunder-Klunder M, Medina-Bravo P, Madrigal-Azcarate A, Mass-Diaz E, Flores-Huerta S: Insulin resistance and its association with the components of the metabolic syndrome among obese children and adolescents. BMC Public Health. 2010, 10: 318-10.1186/1471-2458-10-318.
Cheung CY, Tso AW, Cheung BM, Xu A, Ong KL, Fong CH, Wat NM, Janus ED, Sham PC, Lam KS: Obesity susceptibility genetic variants identified from recent genome-wide association studies: implications in a chinese population. J Clin Endocrinol Metab. 2010, 95 (3): 1395-1403. 10.1210/jc.2009-1465.
Takeuchi F, Yamamoto K, Katsuya T, Nabika T, Sugiyama T, Fujioka A, Isono M, Ohnaka K, Fujisawa T, Nakashima E, et al: Association of genetic variants for susceptibility to obesity with type 2 diabetes in Japanese individuals. Diabetologia. 2011, 54 (6): 1350-1359. 10.1007/s00125-011-2086-8.
Wu L, Xi B, Zhang M, Shen Y, Zhao X, Cheng H, Hou D, Sun D, Ott J, Wang X, et al: Associations of six single nucleotide polymorphisms in obesity-related genes with BMI and risk of obesity in Chinese children. Diabetes. 2010, 59 (12): 3085-3089. 10.2337/db10-0273.
Loos RJ: Recent progress in the genetics of common obesity. Br J Clin Pharmacol. 2009, 68 (6): 811-829. 10.1111/j.1365-2125.2009.03523.x.
Liang J, Fu M, Ciociola E, Chandalia M, Abate N: Role of ENPP1 on adipocyte maturation. PLoS One. 2007, 2 (9): e882-10.1371/journal.pone.0000882.
Villalobos-Comparan M, Teresa Flores-Dorantes M, Teresa Villarreal-Molina M, Rodriguez-Cruz M, Garcia-Ulloa AC, Robles L, Huertas-Vazquez A, Saucedo-Villarreal N, Lopez-Alarcon M, Sanchez-Munoz F, et al: The FTO gene is associated with adulthood obesity in the Mexican population. Obesity (Silver Spring). 2008, 16 (10): 2296-2301. 10.1038/oby.2008.367.
Jelinek D, Millward V, Birdi A, Trouard TP, Heidenreich RA, Garver WS: Npc1 haploinsufficiency promotes weight gain and metabolic features associated with insulin resistance. Hum Mol Genet. 2011, 20 (2): 312-321. 10.1093/hmg/ddq466.
den Hoed M, Luan J, Langenberg C, Cooper C, Sayer AA, Jameson K, Kumari M, Kivimaki M, Hingorani AD, Grontved A, et al: Evaluation of common genetic variants identified by GWAS for early onset and morbid obesity in population-based samples. Int J Obes (Lond). 2012, 10.1038/ijo.2012.34.
Hotta K, Nakamura M, Nakamura T, Matsuo T, Nakata Y, Kamohara S, Miyatake N, Kotani K, Komatsu R, Itoh N, et al: Association between obesity and polymorphisms in SEC16B, TMEM18, GNPDA2, BDNF, FAIM2 and MC4R in a Japanese population. J Hum Genet. 2009, 54 (12): 727-731. 10.1038/jhg.2009.106.
Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al: Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012, 44 (9): 981-990. 10.1038/ng.2383.
Xi B, Takeuchi F, Chandak GR, Kato N, Pan HW, Zhou DH, Pan HY, Mi J: Common polymorphism near the MC4R gene is associated with type 2 diabetes: data from a meta-analysis of 123,373 individuals. Diabetologia. 2012, 55 (10): 2660-2666. 10.1007/s00125-012-2655-5.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/14/21/prepub
Acknowledgements
We are sincerely indebted to all participants of the present study. The study was supported by the EU funded EUROCHIP FP7 consortium (European Obesity Consortium studying the Hypothalamus and its Interaction with the Periphery). We also thank the IMSS Foundation A.C., the Gonzalo Rio Arronte Foundation I.A.P. and Cinvestav. The project was supported by CONACYT SALUD-2005-C02-14412, and Proyectos Prioritarios FIS/IMSS/PRIO/10/011. M.C. is a recipient of an IMSS Foundation Scholarship, Mexico. A. M.-B. was supported in part by Fellowship 233834 by CONACyT and by Cinvestav.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare no competing financial interests.
Authors’ contributions
AMB performed the genotyping, and contributed to statistical analyses and the writing of the paper; AB wrote the paper; DM, JGM, PF and MC revised the paper and contributed to discussion; LY and AB performed the statistical analyses; AMB, MKK, CA, EC, EPL, JMM, MEG, JPR, SFH, JGM and MC contributed to recruitment and clinical data of the Mexican children. All the authors approved the final version of the submitted draft.
Electronic supplementary material
12881_2012_1044_MOESM1_ESM.docx
Additional file 1: Table S1: Expected statistical power for reaching an odds ratio of 1.25 in the present obesity case-control study (N cases = 514; N controls = 949). (DOCX 15 KB)
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Mejía-Benítez, A., Klünder-Klünder, M., Yengo, L. et al. Analysis of the contribution of FTO, NPC1, ENPP1, NEGR1, GNPDA2 and MC4Rgenes to obesity in Mexican children. BMC Med Genet 14, 21 (2013). https://doi.org/10.1186/1471-2350-14-21
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-14-21