Abstract
Background
Addison's disease (AD) is caused by an autoimmune destruction of the adrenal cortex. The pathogenesis is multi-factorial, involving genetic components and hitherto unknown environmental factors. The aim of the present study was to investigate if gene dosage in the form of copy number variation (CNV) could add to the repertoire of genetic susceptibility to autoimmune AD.
Methods
A genome-wide study using the Affymetrix GeneChip® Genome-Wide Human SNP Array 6.0 was conducted in 26 patients with AD. CNVs in selected genes were further investigated in a larger material of patients with autoimmune AD (n = 352) and healthy controls (n = 353) by duplex Taqman real-time polymerase chain reaction assays.
Results
We found that low copy number of UGT2B28 was significantly more frequent in AD patients compared to controls; conversely high copy number of ADAM3A was associated with AD.
Conclusions
We have identified two novel CNV associations to ADAM3A and UGT2B28 in AD. The mechanism by which this susceptibility is conferred is at present unclear, but may involve steroid inactivation (UGT2B28) and T cell maturation (ADAM3A). Characterization of these proteins may unravel novel information on the pathogenesis of autoimmunity.
Similar content being viewed by others
Background
Primary adrenal insufficiency (Addison's disease (AD)) is most commonly caused by an autoimmune destruction of the adrenal cortex, resulting in failure to produce corticosteroids. AD can appear alone, but is frequently part of autoimmune polyendocrine syndromes (APS). APS type I is caused by mutations in the gene autoimmune regulator (AIRE), which has a function in developing immunological tolerance in the thymus (for review see [1]). Isolated AD, or AD in combination with other endocrine components where APS I has been ruled out, is thought to be caused by a combination of genetic and environmental factors, and stochastic events.
Using the candidate gene approach, several genes have been associated with autoimmune AD. The human leukocyte antigen (HLA) haplotypes DR3-DQ2 and DR4-DQ8 is the strongest predisposing genetic factors [2, 3]. Other susceptibility genes are MHC-class I related chain A and B (MICA and MICB) [4, 5], cytotoxic T lymphocyte antigen-4 (CTLA-4) [6], tyrosine-protein phosphatase non-receptor type 22 (PTPN22) [7], the gene encoding MHC class II transactivator (CIITA) [8], C-type lectin domain family 16, member A (CLEC16A) [9], Programmed death ligand 1 (PD-L1) [10], and the gene coding for NACHT leucine rich repeats protein 1 (NALP1) [11]. All these genes are also associated with other autoimmune diseases and can be viewed as autoimmunity genes. So far a disease-specific AD gene has not been identified, with the possible exception of the DRB1*0404 HLA-subtype [2].
The recent identification of common copy number variation (CNV) in the human genome as a source of genetic modification could possibly explain some of the susceptibility to autoimmunity. In support of this notion is the finding that low copy number of FCGR3B (Fc fragment of IgG, low affinity IIIb, receptor (CD16b)), has been coupled to the systemic autoimmune diseases systemic lupus erythematosus (SLE), vasculitis, microscopic polyangiitis and Wegener's granulomatosis [12]. Low FCGR3B copy number possibly contributes to impaired clearance of immune complexes, which is of pathogenic importance in these diseases [12]. Organ-specific diseases like Graves' disease and AD, on the other hand, fail to show this association [12]. A similar gene, FCGR2C, is associated with idiopathic thrombocytopenic purpura [13]. Furthermore, low copy number of the complement associated genes C4A and C4B have been described in SLE-patients (reviewed in [14]). Finally, aberrant copy number of the gene chemokine ligand 3-like 1 (CCL3L1), a potent binder of several pro-inflammatory cytokine receptors, has been reported in SLE [15], while high copy number has been reported in rheumatoid arthritis and type 1 diabetes [16].
These studies show that CNVs in immunoregulatory genes could add to the susceptibility to autoimmune diseases, and that identification of such genes can contribute to the understanding of pathogenesis. Only a few reports on genome-wide CNV have been reported in organ-specific autoimmune diseases so far [17]. Here, we have conducted the first genome-wide (GW) CNV study in autoimmune AD.
Methods
Subjects
A total of 352 patients with autoimmune AD (63% females, 37% males; mean age 57 years, range 13 - 93 years; 52% of whom had APS type II) recruited from the National Norwegian Registry of patients with AD [2], and 353 blood donors (35% females, 65% males, mean age 46 years, range 19 - 71 years) were available for CNV analyses. For comparison we had access to an in-house database with CNV-results from 361 non-Addison's patients analysed with the same GW array. These samples comprise individuals which were analysed for CNVs in the daily routine analyses at the Medical Genetics Department, Haukeland University Hospital, for a diversity of dysfunctions. An overview of the subjects used in the present study is given in Figure 1.
Ethical considerations
All included patients and blood donors signed a written consent form. The study was approved by the Regional Committee for Medical Ethics of Western Norway, and performed according to the Helsinki Declaration.
GW CNV gene chip analyses
Twenty eight unrelated patients with isolated autoimmune AD (39% females, 61% males, mean age 63 years, ranging from 26 to 96 years) were randomly selected for GW CNV analyses. The genome-wide copy number screening was performed using the Affymetrix GeneChip® Genome-Wide Human SNP Array 6.0, Part# 901182, (Affy 6.0) following the procedure described in Affymetrix® Cytogenetics Copy Number User Guide. The arrays were scanned by the Gene Chip Scanner 3000 7G, controlled by the AGCC software. The array data were analyzed by the Affymetrix Genotyping Console™ version 3.0.1. The Contrast quality control (QC) was set to higher than 0.4 and the Median Absolute Pair wise Difference (MAPD) threshold was set to 0.30 or less for each sample. CNV analysis was performed by comparing the files from the AD patients with a reference file of 44 healthy controls (anonymous material used in the daily routine analyses at the Medical Genetics Department, Haukeland University Hospital). CNV were further evaluated in the Affymetrix® Genotyping Console Browser version 1.0.11 and the Affymetrix Chromosome Analysis Suite version 1.0.1. The CNVs were flagged using a cut off filter of minimum 20 markers per 100 Kbp segment. Flagged CNVs were checked against the Database of Genomic Variants (DGV) (http://projects.tcag.ca/variation/)©[18]. Findings that varied from the reference file were compared with the in-house database from 361 non-AD individuals. In addition, a search for CNVs in candidate genes of Addison's disease was performed on the chip data without using cut off filters (see Additional file 1).
Additional CNV assays
Individual CNV assays were performed by duplex Taqman real-time PCR assays in order to confirm findings on new candidates from the GW analysis in a larger patient and blood donor material. In addition, an individual CN assay was conducted for FCGR3B since it is not included on the Affy 6.0 chip. FCGR3B, KIAA1267 and ADAM3A copy number assays were supplied from Applied Biosystems (Assay ID: Hs04211858_cn, Hs03983160_cn, and Hs03269461_cn, respectively). Copy number assay for UGT2B28 was designed by Applied Biosystems based on the gene's sequence (ugt2b28_CCGJPAM). RNaseP (TaqMan copy number reference assay, part # 4403328) and Desmoplakin (DSP) (adapted from Parajes et al, 2007) [19] were used as reference genes. All primers and probes were supplied from Applied Biosystems, and duplex real-time PCR assays were performed according to the TaqMan® copy number assay protocol (Applied Biosystems). All data was further analyzed using the 7300 System SDS software version 2.3. The genes were quantified by calibration curves for each gene. The reference genes are known to occur in two copies in the genome. Hence, the copy number was determined by the relative relationship between the quantity of the candidate gene and the reference gene.
Statistics
Fisher's Exact test was applied for comparing the frequencies of CNs of the different genes in AD patients and controls, using the statistical work package PASW Statistics 18. The data was considered to be significant when P < 0.05.
Results
Quality control of the Affy 6.0 gene chips
Twenty-six of twenty-eight arrays in the patient group and all controls (N = 44) were in accordance with Affymetrix' recommendations, with MAPD values ranging from 0.194-0.299 and QC call rate ranging from 88.2-99.0%. Two patient arrays failed these quality criteria and were excluded from further analyses.
Frequent CNVs associated with AD
The gene regions with most frequent CNVs in AD patients are given in Table 1 all of which are reported as normal CN polymorphisms in the DGV [18]. Except for the DEFB cluster region, a number of genes within this region were found to have CNV that differed significantly from 361 in-house non-AD controls (Table 1). Low CN of KIAA1267 (coding for an uncharacterised protein) and UGT2B28 (encoding uridine diphosphate glucuronosyltransferase (UGT) 2 family, polypeptide B28), and high CN of ADAM3A (encoding a disintegrin and metalloproteinase domain 3A), occurred more frequently (> 30%) in AD patients. These were selected for replication studies in the large Addison and control cohorts.
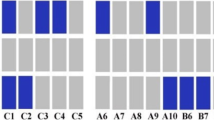
The Affy 6.0 gene chip experiment revealed that a heterozygous deletion (CN = 1) in the UGT2B28 gene occurred more often in the AD patients (30%) than in the in-house reference material (10%). This association was confirmed (25% vs 14% P = 0.0008) in the large AD patient and blood donor materials using the gene specific CN assay (Figure 2a). On the other hand, the heterozygous deletion (CN = 1) in the KIAA1267 gene found to be more frequently in AD patients (38% vs 9% respectively, Table 1), was no longer significant (55% vs 59%, P = 0.48) (Figure 2b) in the replication study.
Copy number variations in genes associated with AD by the Affy 6.0 chip. Copy number frequencies in Addison patients and controls for the three genes associated with AD by the Affy 6.0 chip (AD chip) and validated by duplex RT-qPCR. UGT2B28 (a), KIAA1267 (b), and ADAM3A (c). P-values for differences of copy number variation between Addison patients (AD Taqman) and healthy controls (HC Taqman) were calculated by Fishers's Exact test.
High copy numbers (CN > 2) of the ADAM3A gene were detected more frequently in AD patients compared to the in-house reference database (42% vs 10% respectively) by the Affy 6.0 gene chip analysis (Table 1). A significant association (P = 0.0002) of CNVs, with respect to CN = 3, was still evident in the replication experiment on the larger cohort with duplex Taqman real-time PCR assays (Figure 2c).
The differences in CNVs for UGT2B28 and ADAM3A were still evident when patients with isolated AD and APS II were analysed separately. With respect to sex, the association to UGT2B28 was only significant for males (P = 0.002), while the association to ADAM3A was significant for both females (P = 0.004) and males (P = 0.004).
Rare CNV in AD patients detected by Affy 6.0
We also performed a GW scan for rare CNVs (Table 2). These CNVs are not listed in the DGV [18], nor detected in our in-house database. Eight novel CNV gene regions were found in the AD patients, with the longest segment consisting of a 700 Kbp heterozygote deletion which was found in patient #23 on chromosome 7p14.3. This segment covers only one gene, BMP binding endothelial regulator (BMPER), an inhibitor of bone morphogenetic protein (BMP) function. Patient #3 had two large CNVs regions, one heterozygote deletion of 103 Kbp in 5q23.1 covering the last third part of the 17-beta-hydroxysteroid dehydrogenase IV gene (HSD17B4), and a single copy gain of 115 Kbp in 14q13.2, covering the NFKBIA gene (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha). Patient #20 had two regions of single copy gain, one in 3q21.3 (102 Kbp), and one in 8p21.1 (103 Kbp) covering the gene SCARA5 (Scavenger receptor class A, member 5).
CNV in candidate genes of AD
A number of candidate autoimmune genes of which some have been reported to have CNV associated with autoimmunity (e.g. FCGR3B), were tested for association to AD. Of these, only CNVs in the candidate genes CCL3L1 and HLA-DRB5 were detected by the gene chip (see Additional file 1). No significant differences in CNV were found in the selected candidates, including FCGR3B (see Additional file 2).
Discussion
The present study is the first effort to search for common CNVs as a source for genetic susceptibility in AD. We found two novel CNVs associated with AD, UGT2B28 and ADAM3A. Both have previously been reported as regions with copy number polymorphisms [18].
The enzyme UGT2B28 is expressed in liver and mammary glands, and it might play a role in steroid inactivation [20]. In the present study we found an increased frequency of CN = 1 of the gene UGT2B28 in our AD cohort. It has previously been reported that this heterozygote deletion occurs in 23% of healthy Caucasians, and deletions in this gene region, which also includes the UGT2B17 gene, are very common [21]. The connection to autoimmunity, if any, is at present not clear.
We also found an association with CNV and ADAM3A in AD patients. Three different transcript variants of ADAM3A (Entrez Gene; http://www.ncbi.nlm.nih.gov/gene; Gene ID 1587) have been reported, and the gene is embedded in a cluster of other genes in the ADAM family [18]. Several proteins of the ADAM family are believed to be involved in the inflammation process, and recent studies have demonstrated their role in T cell maturation and development [22, 23]. The function of ADAM3A has mainly been associated with sperm-egg binding and fusion [24], but it has also been linked to pro-inflammatory activity and survival of macrophages [25]. Thus a role in autoimmunity and immunological tolerance can be envisioned.
Recent evidence shows that common genetic variants do not explain all genetic susceptibility to autoimmune diseases. Rare variants in a number of genes, e.g. PTPN22, have been shown to be associated with autoimmune diseases such as AD [7] and diabetes [26–28]. One could hypothesize that rare CNV could play a role for AD susceptibility. Thus, as an alternative approach we looked for rare or private CNVs. We found that one patient had a single copy gain of the NFKBIA gene, previously shown to be related to Crohn's diseases [29] and multiple sclerosis [30]. This gene produces an inhibitor of NFKB1, which encodes the NFκB subunit 1. Gene polymorphisms in NFκB have been associated with type 1 diabetes [31]. The same patient also had a heterozygous deletion in the HSD17B4 gene. Mutations and deletions in this gene have been reported to cause D-bifunctional protein deficiency (http://www.ncbi.nlm.nih.gov/omim ID 601860). High copy number of the gene SCARA5 was also detected in one patient. This gene might be of interest because of its expression in murine thymus and adrenals, and its function in innate immune activities of epithelial cells [32].
When searching for CNVs in candidate gene regions, we only detected aberrant CN in the CCL3L1 and HLA-DRB5 genes in some of the AD patients, but not significantly different from controls. CNVs of these two genes have been also reported previously as normal variants [18]. Moreover, we did not find any association between CNVs in the FCGR3B gene and AD, which is in agreement to the results of Fanciulli et al (2007) [12].
Conclusions
We have identified two novel CNV associations to ADAM3A and UGT2B28 in AD. The mechanism by which this susceptibility is conferred is at present unclear, but may involve steroid inactivation (UGT2B28) and T cell maturation (ADAM3A). Characterization of these proteins may unravel novel information on the pathogenesis of autoimmunity.
References
Husebye ES, Perheentupa J, Rautemaa R, Kampe O: Clinical manifestations and management of patients with autoimmune polyendocrine syndrome type I. J Intern Med. 2009, 265 (5): 514-529. 10.1111/j.1365-2796.2009.02090.x.
Erichsen MM, Lovas K, Skinningsrud B, Wolff AB, Undlien DE, Svartberg J, Fougner KJ, Berg TJ, Bollerslev J, Mella B, et al: Clinical, immunological, and genetic features of autoimmune primary adrenal insufficiency: observations from a Norwegian registry. J Clin Endocrinol Metab. 2009, 94 (12): 4882-4890. 10.1210/jc.2009-1368.
Myhre AG, Undlien DE, Lovas K, Uhlving S, Nedrebo BG, Fougner KJ, Trovik T, Sorheim JI, Husebye ES: Autoimmune adrenocortical failure in Norway autoantibodies and human leukocyte antigen class II associations related to clinical features. J Clin Endocrinol Metab. 2002, 87 (2): 618-623. 10.1210/jc.87.2.618.
Park YS, Sanjeevi CB, Robles D, Yu L, Rewers M, Gottlieb PA, Fain P, Eisenbarth GS: Additional association of intra-MHC genes, MICA and D6S273, with Addison's disease. Tissue Antigens. 2002, 60 (2): 155-163. 10.1034/j.1399-0039.2002.600206.x.
Gambelunghe G, Falorni A, Ghaderi M, Laureti S, Tortoioli C, Santeusanio F, Brunetti P, Sanjeevi CB: Microsatellite polymorphism of the MHC class I chain-related (MIC-A and MIC-B) genes marks the risk for autoimmune Addison's disease. J Clin Endocrinol Metab. 1999, 84 (10): 3701-3707. 10.1210/jc.84.10.3701.
Blomhoff A, Lie BA, Kemp EH, Weetman AP, Akselsen HE, Myhre AG, Husebye ES, Undlien DE: Polymorphisms in the CTLA4 gene region confer susceptibility to Addison's disease. J Clin Endocrinol Metab. 2004.
Skinningsrud B, Husebye ES, Gervin K, Lovas K, Blomhoff A, Wolff AB, Kemp EH, Egeland T, Undlien DE: Mutation screening of PTPN22: association of the 1858T-allele with Addison's disease. Eur J Hum Genet. 2008, 16 (8): 977-982. 10.1038/ejhg.2008.33.
Ghaderi M, Gambelunghe G, Tortoioli C, Brozzetti A, Jatta K, Gharizadeh B, De Bellis A, Pecori Giraldi F, Terzolo M, Betterle C, et al: MHC2TA single nucleotide polymorphism and genetic risk for autoimmune adrenal insufficiency. J Clin Endocrinol Metab. 2006, 91 (10): 4107-4111. 10.1210/jc.2006-0855.
Skinningsrud B, Husebye ES, Pearce SH, McDonald DO, Brandal K, Wolff AB, Lovas K, Egeland T, Undlien DE: Polymorphisms in CLEC16A and CIITA at 16p13 are associated with primary adrenal insufficiency. J Clin Endocrinol Metab. 2008, 93 (9): 3310-3317.
Mitchell AL, Cordell HJ, Soemedi R, Owen K, Skinningsrud B, Wolff AB, Ericksen M, Undlien D, Husebye E, Pearce SH: Programmed death ligand 1 (PD-L1) gene variants contribute to autoimmune Addison's disease and Graves' disease susceptibility. J Clin Endocrinol Metab. 2009, 94 (12): 5139-5145. 10.1210/jc.2009-1404.
Magitta NF, Boe Wolff AS, Johansson S, Skinningsrud B, Lie BA, Myhr KM, Undlien DE, Joner G, Njolstad PR, Kvien TK, et al: A coding polymorphism in NALP1 confers risk for autoimmune Addison's disease and type 1 diabetes. Genes Immun. 2009, 10 (2): 120-124. 10.1038/gene.2008.85.
Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, Heward JM, Gough SC, de Smith A, Blakemore AI, et al: FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007, 39 (6): 721-723. 10.1038/ng2046.
Breunis WB, van Mirre E, Bruin M, Geissler J, de Boer M, Peters M, Roos D, de Haas M, Koene HR, Kuijpers TW: Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008, 111 (3): 1029-1038.
Ptacek T, Li X, Kelley JM, Edberg JC: Copy number variants in genetic susceptibility and severity of systemic lupus erythematosus. Cytogenet Genome Res. 2008, 123 (1-4): 142-147. 10.1159/000184701.
Mamtani M, Rovin B, Brey R, Camargo JF, Kulkarni H, Herrera M, Correa P, Holliday S, Anaya JM, Ahuja SK: CCL3L1 gene-containing segmental duplications and polymorphisms in CCR5 affect risk of systemic lupus erythaematosus. Ann Rheum Dis. 2008, 67 (8): 1076-1083. 10.1136/ard.2007.078048.
McKinney C, Merriman ME, Chapman PT, Gow PJ, Harrison AA, Highton J, Jones PB, McLean L, O'Donnell JL, Pokorny V, et al: Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008, 67 (3): 409-413.
Grayson BL, Smith ME, Thomas JW, Wang L, Dexheimer P, Jeffrey J, Fain PR, Nanduri P, Eisenbarth GS, Aune TM: Genome-wide analysis of copy number variation in type 1 diabetes. PLoS One. 2010, 5 (11): e15393-10.1371/journal.pone.0015393.
The Database of Genomic Variants (DGV). [http://projects.tcag.ca/variation/]
Parajes S, Quinterio C, Dominguez F, Loidi L: A simple and robust quantitative PCR assay to determine CYP21A2 gene dose in the diagnosis of 21-hydroxylase deficiency. Clin Chem. 2007, 53 (9): 1577-1584. 10.1373/clinchem.2007.087361.
Levesque E, Turgeon D, Carrier JS, Montminy V, Beaulieu M, Belanger A: Isolation and characterization of the UGT2B28 cDNA encoding a novel human steroid conjugating UDP-glucuronosyltransferase. Biochemistry. 2001, 40 (13): 3869-3881. 10.1021/bi002607y.
Menard V, Eap O, Harvey M, Guillemette C, Levesque E: Copy-number variations (CNVs) of the human sex steroid metabolizing genes UGT2B17 and UGT2B28 and their associations with a UGT2B15 functional polymorphism. Hum Mutat. 2009, 30 (9): 1310-1319. 10.1002/humu.21054.
Gossens K, Naus S, Hollander GA, Ziltener HJ: Deficiency of the metalloproteinase-disintegrin ADAM8 is associated with thymic hyper-cellularity. PLoS One. 2010, 5 (9): e12766-10.1371/journal.pone.0012766.
Tian L, Wu X, Chi C, Han M, Xu T, Zhuang Y: ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int Immunol. 2008, 20 (9): 1181-1187. 10.1093/intimm/dxn076.
Primakoff P, Myles DG: The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000, 16 (2): 83-87. 10.1016/S0168-9525(99)01926-5.
Yang ZF, Ho DW, Lau CK, Lam CT, Lum CT, Poon RT, Fan ST: Allograft inflammatory factor-1 (AIF-1) is crucial for the survival and pro-inflammatory activity of macrophages. Int Immunol. 2005, 17 (11): 1391-1397. 10.1093/intimm/dxh316.
Onengut-Gumuscu S, Ewens KG, Spielman RS, Concannon P: A functional polymorphism (1858C/T) in the PTPN22 gene is linked and associated with type I diabetes in multiplex families. Genes Immun. 2004, 5 (8): 678-680. 10.1038/sj.gene.6364138.
Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, et al: A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004, 36 (4): 337-338. 10.1038/ng1323.
Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, Vella A, Nutland S, Rance HE, Maier L, et al: Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004, 53 (11): 3020-3023. 10.2337/diabetes.53.11.3020.
Klein W, Tromm A, Folwaczny C, Hagedorn M, Duerig N, Epplen JT, Schmiegel WH, Griga T: A polymorphism of the NFKBIA gene is associated with Crohn's disease patients lacking a predisposing allele of the CARD15 gene. Int J Colorectal Dis. 2004, 19 (2): 153-156. 10.1007/s00384-003-0531-y.
Miterski B, Bohringer S, Klein W, Sindern E, Haupts M, Schimrigk S, Epplen JT: Inhibitors in the NFkappaB cascade comprise prime candidate genes predisposing to multiple sclerosis, especially in selected combinations. Genes Immun. 2002, 3 (4): 211-219. 10.1038/sj.gene.6363846.
Hegazy DM, O'Reilly DA, Yang BM, Hodgkinson AD, Millward BA, Demaine AG: NFkappaB polymorphisms and susceptibility to type 1 diabetes. Genes Immun. 2001, 2 (6): 304-308. 10.1038/sj.gene.6363776.
Jiang Y, Oliver P, Davies KE, Platt N: Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J Biol Chem. 2006, 281 (17): 11834-11845. 10.1074/jbc.M507599200.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/12/111/prepub
Acknowledgements
Dr. Atle Brendehaug is acknowledged for help with the analyses of the GWA study. The study was funded by the Regional Health Authorities of Western Norway, Bergen Medical Research Foundation and the EU FP7 project Euradrenal (grant number 201167).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
IB carried out the genome-wide CN study and the real-time PCR copy number assays, and performed the statistical analysis. ABSW and ESH were responsible for supplying the DNA material from AD patients and healthy controls. KL performed the statistical design and analysis. PMK was responsible for coordinating the gen chip analysis and supplying data from gene chip controls. All the authors have contributed to design the experiment, participated in writing the manuscript, critically read and approved the final manuscript.
Electronic supplementary material
12881_2011_842_MOESM1_ESM.XLS
Additional file 1: Candidate genes of Addison's disease. List of selected candidate genes of Addison's disease, and CNVs in candidate genes detected by the Affymetrix 6.0 gene chip. (XLS 16 KB)
12881_2011_842_MOESM2_ESM.PDF
Additional file 2: Copy number variations in FCGR3B determined by duplex RT-qPCR. Copy number frequencies of the FCGR3B gene in AD patients and healthy controls. P-values for differences of copy number variation between Addison patients (AD Taqman) and healthy controls (HC Taqman) were calculated by Fishers's exact test. (PDF 17 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Brønstad, I., Wolff, A.S., Løvås, K. et al. Genome-wide copy number variation (CNV) in patients with autoimmune Addison's disease. BMC Med Genet 12, 111 (2011). https://doi.org/10.1186/1471-2350-12-111
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-12-111