Abstract
Background
The interleukin-1 (IL-1) family of cytokines is involved in the inflammatory and repair reactions of skeletal muscle during and after exercise. Specifically, plasma levels of the IL-1 receptor antagonist (IL-1ra) increase dramatically after intense exercise, and accumulating evidence points to an effect of genetic polymorphisms on athletic phenotypes. Therefore, the IL-1 family cytokine genes are plausible candidate genes for athleticism. We explored whether IL-1 polymorphisms are associated with athlete status in European subjects.
Methods
Genomic DNA was obtained from 205 (53 professional and 152 competitive non-professional) Italian athletes and 458 non-athlete controls. Two diallelic polymorphisms in the IL-1β gene (IL-1B) at -511 and +3954 positions, and a variable number tandem repeats (VNTR) in intron 2 of the IL-1ra gene (IL-1RN) were assessed.
Results
We found a 2-fold higher frequency of the IL-1RN 1/2 genotype in athletes compared to non-athlete controls (OR = 1.93, 95% CI = 1.37-2.74, 41.0% vs. 26.4%), and a lower frequency of the 1/1 genotype (OR = 0.55, 95% CI = 0.40-0.77, 43.9% vs. 58.5%). Frequency of the IL-1RN 2/2 genotype did not differ between groups. No significant differences between athletes and controls were found for either -511 or +3954 IL-1B polymorphisms. However, the haplotype (-511)C-(+3954)T-(VNTR)2 was 3-fold more frequent in athletes than in non-athletes (OR = 3.02, 95% CI = 1.16-7.87). Interestingly, the IL-1RN 1/2 genotype was more frequent in professional than in non-professional athletes (OR = 1.92, 95% CI = 1.02-3.61, 52.8% vs. 36.8%).
Conclusions
Our study found that variants at the IL-1ra gene associate with athletic status. This confirms the crucial role that cytokine IL-1ra plays in human physical exercise. The VNTR IL-1RN polymorphism may have implications for muscle health, performance, and/or recovery capacities. Further studies are needed to assess these specific issues. As VNTR IL-1RN polymorphism is implicated in several disease conditions, athlete status may constitute a confounding variable that will need to be accounted for when examining associations of this polymorphism with disease risk.
Similar content being viewed by others
Background
Factors that affect athletic performance are only partially defined. However, there are accumulating data indicating that both pro- and anti-inflammatory responses can play a role. Prolonged strenuous exercise results in an activation of the acute phase response with some similarities to sepsis [1]. The induction of a systemic cytokine response, including elevated plasma levels of interleukin (IL)-1 receptor antagonist (IL-1ra), IL-6, IL-8, IL-10, and granulocyte colony-stimulating factor (G-CSF), following intense physical activity, is well documented [1–5]. IL-1ra is a member of the IL-1 family, which includes the classical IL-1 (α and β) cytokines, IL-18 and the newly described IL-1F 5-11 [6]. IL-1ra acts as an antagonist of the IL-1 receptor type I (IL-1RI) and prevents IL-1 (α and β)-dependent signaling. In the absence of IL-1ra, the activity of IL-1 is unopposed, promoting rampant inflammation. Recent research has shown that deficiency of IL-1ra in humans can cause IL-1-mediated systemic and local inflammation that affect infants soon after birth [7]. IL-1α is an IL-1RI agonist with proinflammatory action, that acts primarily as an intracellular transcriptional regulator; it is not found in high concentration in the circulation. In contrast, IL-1β can be secreted from cells in large quantities and thus it is commonly found in serum and secretions. In general, IL-1β acts synergistically with tumor necrosis factor (TNF)-α, activating proinflammatory responses in a wide range of cells and promoting the acute phase response [6]. For these reasons IL-1β levels have been studied in relation to physical activity. However, the circulating concentration of IL-1β is either unaffected by exercise, or it exhibits relatively small, delayed changes [1, 2]. In contrast to minimal changes in serum circulating IL-1β after exercise, there is evidence of increased local IL-1β levels after exercise within skeletal muscle; this is most likely associated with response to micro-injury of skeletal muscle caused by intense physical activity [8]. Interestingly, Mahoney and colleagues [9] demonstrated that elevated gene expression of IL-1RI is induced in skeletal muscle after eccentric exercise. Inflammatory and chemoattractant mediators likely promote phagocytosis of trauma-induced cellular debris by macrophages and these cells can continue to secrete IL-1β up to 5 days post-injury [8]. It is of note that accumulation and activation of muscle resident macrophages is a rich source of growth factors postulated to stimulate myogenesis [4]. Thus, inflammation may serve as a mechanism promoting hypertrophy. However, pre- and post-exercise levels of inflammatory factors display considerable variation among people, and this is likely influenced, at least partially, by genetic variation [10, 11].
IL-1β plays an important role in the regulation of acute inflammatory response [12]. After an insult to the body, IL-1β is one of the first pro-inflammatory cytokines to be activated. Intense physical activity can cause micro-injury to skeletal muscle, and cytokines such as IL-1β and TNF-α are thought to initiate and regulate the repair process. IL-1β is able to induce the secretion of several inflammatory factors, such as IL-6, IL-8, TNF-α, and GM-CSF, by many different cell types including myoblasts, smooth and skeletal muscle cells, fibroblasts, macrophages, peripheral blood mononuclear cells (PBMC) and endothelial cells [12–14]. IL-1β binds IL-1RI on the surface of a variety of cells and initiates a cascade of events leading to recruitment and activation of macrophages and neutrophils. However, inflammation must be strictly regulated to avoid secondary tissue damage and myopathy. The activity of IL-1β is modulated mainly by a receptor antagonist (IL-1ra) that inhibits IL-1 action by competing for its receptor [6, 15]. In healthy individuals, IL-1ra is easily detectable in plasma (concentrations are hundreds of pg/mL), but IL-1β levels are usually undetectable (very few or less than pg/mL) [15, 16]. In general, after an insult to the body both IL-1β and IL-1ra increase, and thus, plasma levels of these cytokines are positively correlated [17]. However, the final inflammatory activity of IL-1β is considered to be dependent on the actual ratio of IL-1β over IL-1ra [7, 18].
There is evidence that susceptibility to inflammation is influenced by genetic variation in cytokine genes [14, 19]. Several studies showed that polymorphisms in the IL-1β (IL-1B) and IL-1ra (IL-1RN) genes correlate with altered protein expression in vitro [14, 20] and in vivo [10, 16, 21, 22]. These two genes map close to each other on human chromosome 2 [12]. Polymorphisms in the IL-1 gene cluster associate with many clinical conditions, including inflammatory and infectious diseases (periodontal and arterial diseases, altered metabolic conditions, type 2 diabetes, type 2 diabetes complications, gastric cancer, rheumatoid arthritis, ankylosing spondylitis, systemic inflammation, myopathies, Alzheimer's disease, malaria, and bacterial vaginosis) [6, 12, 17, 19, 21, 23–31]. Mostly, two single nucleotide polymorphisms (SNPs) in IL-1B have been studied for disease predisposition, one at position -511 in the promoter region [32] and another at position +3954 in exon 5 (TaqI restriction site polymorphism) [18]. In addition, a pentaallelic polymorphic site in intron 2 of the IL-1RN gene consisting in a variable number tandem repeats (VNTR, 86 bp repeats) has been extensively investigated in relation to a variety of pathological conditions, including inflammatory myopathies [6, 18, 19, 29, 33, 34].
Recently, a study performed on 24 sedentary subjects (of unspecified ethnicity) selected on the basis of their haplotype pattern in the IL-1 gene cluster (based on specific combinations of +4845 IL-1A, +3954 IL-1B, -511 IL-1B, and -3737 IL-1B polymorphisms) showed that subjects C/C for IL-1B +3954 who carry allele 2 at IL-1RN +2018 were associated with the inflammation of skeletal muscle, following acute resistance exercise [10].
The aim of this study was to investigate whether polymorphisms in the IL-1β and IL-1ra genes affect athletic status, competitive athlete vs. non-athlete. We evaluated whether the genotype frequencies of -511 and +3954 SNPs of IL-1B, and intron 2 VNTR of IL-1RN vary between athletes and non-athletes. Athletes of National or Regional competitive standard were recruited. This analysis was performed in a relatively homogeneous ethnic group of Caucasian Italian subjects because associations between polymorphisms in immune system-related genes may be confounded by allele frequency differences between ethnic groups [35, 36]. We examined whether the polymorphisms of these genes are associated with being a professional (high-grade) or non-professional (medium-grade) athlete.
methods
Subjects and biological samples
The study population consisted of subjects attending the Galeazzi Hospital for routine blood testing. Exclusion criteria were age younger than 18 years and presence of a pathologic condition or chronic major diseases such as diabetes, autoimmune diseases, cardiovascular diseases and malignancies. Athletes were enrolled from teams of Italian professional or non-professional athletes during routine physical examinations. Controls were recruited from the general Italian population of unrelated subjects after the following questions: 1) are you or have you ever been a trained athlete?; 2) did you ever participate in an official athletic competition? All control subjects were without any competitive sport experience, they declared they had never been an athlete or participated in any athletic competition or athletic training. All subjects were Caucasian. The study was approved by the Institutional Review Board of Milan Azienda Sanitaria Locale (ASL). The methods used in this study were in accordance with the Helsinki Declaration of 1975 as revised in 1996. All eligible participants were enrolled consecutively after written informed consent.
Out of 688 subjects enrolled, 25 subjects were excluded from the study because 14 did not fulfill inclusion criteria and 11 had non-interpretable laboratory data. The final analyses were performed on 663 subjects, including 205 (30.9%) athletes and 458 (69.1%) non-athlete controls. The study population of 205 athletes included 53 high-grade professionals (50 current and 3 who had been professional athletes) who compete(d) at National level and 152 medium-grade non-professional athletes who participate in Regional athletic competitions. Specifically, high-grade athletes group included National Italian team grade (some of these athletes participated in Olympic Games, and some were medallists in International Games) or Third Division soccer players. The "caliber" of an Italian Third Division soccer team is similar to National teams of several European countries. The non-professional athlete group included athletes who are highly involved in sports activities (training and competitions >10 h/week).
Among the athletes there were 139 females and 66 males. Age range of study athletes was 18-53 years. A total of 458 healthy Italian non-athlete subjects of both sexes in the age range of 18-53 years (mean 33.6 ± 9.23 years) constituted the control group.
Blood samples were obtained from seated and fasting subjects in the morning from the antecubital vein with evacuated ethylenediamine tetra acetic acid (EDTA) tubes (Vacutainer Tubes, Becton-Dickinson, Franklin Lakes, NJ, USA). Samples were centrifuged at 2000 g for 10 min at 4°C and the cellular pellet was processed for DNA extraction. Personnel isolating and processing the DNA were blinded to the subjects' demographic characteristics.
IL-1B and IL-1RNpolymorphisms gene identification
Genomic DNA was extracted from the blood pellet fraction according to the standard proteinase-K digestion and ethanol extraction method. The extracted DNA was then stored at -20°C until further analysis.
To examine the -511 SNP (rs16944) [37], the promoter region of IL-1B was amplified by PCR, using the primers 5'-TGGCATTGATCTGGTTCATC-3' and 5'-GTTTAGGAATCTTCCCACTT-3' as described [32]. The protocol included 35 cycles at 94°C for 1 min, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C of 5 min. At the end of the procedure the amplicons were digested with AvaI at 37°C for 3 hours. Fragments were analyzed after electrophoresis on 10% acrylamide gels and visualized with ethidium bromide. This gave products of 190 bp and 114 bp (C allele) or 304 bp (T allele) [31]. Out of 663 study subjects, 4 data were not available for the -511 genotype.
To determine the +3954 (rs1143634) [37] SNP genotype, the polymorphic region containing the TaqI restriction site (designed as +3953 in older literature [30]) was amplified using the following primers: 5'-GTTGTCATCAGACTTTGACC-3' and 5'-TTCAGTTCATATGGACCAGA-3' [18]. The 249 bp products were digested with TaqI at 65°C for 1 hour, resulting in fragments that either remained intact (T allele) or were digested into 2 fragments of 135 and 114 bp (C allele). The restriction fragments were analyzed by electrophoresis on 10% acrylamide gels and visualized with ethidium bromide [31].
The IL-1RN intron 2 VNTR polymorphism (rs380092) [38] was analyzed using 5'-CTCAGCAACACTCCTAT-3' and 5'-TCCTGGTCTGCAGGTAA-3' as primers [18, 33]. The PCR products of 410 bp (allele 1 = 4 repeats of the 86 bp region), 240 bp (allele 2 = 2 repeats), 500 bp (allele 3 = 5 repeats), 325 bp (allele 4 = 3 repeats), 595 bp (allele 5 = 6 repeats) [18] were analyzed by electrophoresis on 8% acrylamide gel stained with ethidium bromide [31]. A blinded quality control was performed in a random subset of 20 samples, which were sequenced by use of ABI Prism 310 genetic analyzer (Applied Biosystems). The sample consisted of 11 subjects 1/1, 6 subjects 1/2, and 3 subjects 2/2 genotype; a 100% correspondence with gel data was obtained.
Statistical analysis
The genotype frequencies for each polymorphism were compared by 2-sided Pearson chi-square or Fisher's exact test as appropriate, using STATA statistical software version 10, College Station, TX, USA and Powermarker [39]. The odds ratio (OR) and the 95% confidence interval (CI) were calculated to evaluate the genotype effects of each genotype against all others. Tests for deviations from Hardy-Weinberg equilibrium (HWE) were performed in Powermarker, using a chi-square distribution in cases and controls separately for each SNP. Haplotype analysis was performed with the program Unphased [40]. Haplotypes estimated to be below 1% in frequency were not reported. Due to the low allele frequencies of the 3, 4 and 5 allele of IL-1RN, the intron 2 VNTR genotypes were grouped into the short allele (S, allele 2) and the long alleles (L, alleles 1, 3, 4, and 5) according to Machado et al. [41]. Linkage disequilibrium (LD) between the two SNPs in IL-1B and the VNTR short and long alleles in IL-1RN was determined using Haploview [42]. Differences in the haplotype distributions between cases and controls were determined by likelihood ratio tests. The reference distribution for which all the other haplotypes were compared against was the most common haplotype. The OR was considered significant if the CI did not cross unity. The P-value for each haplotype was determined by comparing the haplotype against all others. Therefore, a P-value may be slightly significant even if the OR is not.
Results
The study population consisted of 205 athletes; of these 53 were professional (high-grade, National level) and 152 were non-professional (medium-grade, Regional level, recreational athletes not paid for performing competitions) athletes regularly participating in athletic competitions. Controls were 458 healthy non-athletes. Demographic characteristics and sport disciplines practiced by the 205 athletes are shown in Table 1.
A total of 663 study subjects were genotyped for the three gene loci (IL-1B in position -511 and +3954; and IL-1RN intron 2 VNTR).
Neither case nor control samples deviated from Hardy Weinberg equilibrium (HWE) at site -511 or site +3954. The controls deviated from HWE at the VNTR in IL-1RN (P = 0.001); while athletes marginally deviated from HWE (P = 0.097). Athletes and controls did not differ in the distribution of -511 IL-1B genotypes; 38.8% athletes vs. 44.8% controls were homozygous CC, 48.8% athletes vs. 45.9% controls were heterozygous CT, and 12.4% athletes vs. 9.4% controls were homozygous TT (Table 2). The two groups also did not differ significantly in allele frequency (P = 0.11).
Genotype and allele distribution of IL-1B +3954 gene polymorphism are shown in Table 3. Athletes and controls did not differ; 60.0% athletes vs. 62.2% controls were homozygous CC, 36.1% athletes vs. 32.3% controls were heterozygous CT, and 3.9% athletes vs. 5.5% controls were homozygous TT. The two groups also did not differ significantly in allele frequency (P = 0.89).
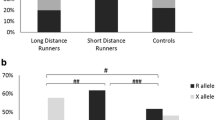
For the IL-1RN VNTR polymorphism three alleles (allele 1, 2 and 3) were common in our study; allele 4 was found in 3 subjects (one genotype 1/4 and two genotypes 2/4, all in the control group) and allele 5 was detected in only 1 subject (one genotype 1/5, in the athlete group). The most common allele, the IL-1RN*1 allele, was less frequent in athletes than in controls (65.4% vs. 74.0%, P = 0.001), whereas the second commonest allele, IL-1RN*2, had an allele frequency higher in athletes than controls (32.2% vs. 22.9%, P < 0.001), finally, IL-1RN*3 had similar frequency in athletes and controls (P = 0.57) (overall alleles, P = 0.001) (Table 4). Specifically, athletes were less likely to have the 1/1 genotype (OR = 0.55, 95% CI 0.40-0.77), and 2-fold more likely to be 1/2 (OR = 1.93, 95% CI 1.37-2.74). Athletes and controls did not differ in frequency of the 2/2 genotype. However, we observed increased frequencies of cumulative 1/2, 2/2, 2/3 and 2/4 genotypes in athletes (53.7%) compared with controls (36.7%), OR = 2.00 (95% CI = 1.43-2.79), suggesting a dominant effect of the 2 allele. These observations were significant in both male and female athletes when analyzed separately (data not shown, see additional File 1). Genotypes that included the 3 allele did not differ between athletes and controls.
In a secondary analysis (Table 5), we compared IL-1RN genotypes between the 53 professional athletes and the 152 non-professional athletes. Interestingly, we found that the 1/2 VNTR genotype was almost 2-fold more frequent in professional than in non-professional athletes (OR = 1.92, 95% CI = 1.02-3.61). No other significant differences were noted between professional and recreational athletes.
Comparison of professional athletes with non-athletes revealed that genotype 1/2 VNTR was 3-fold more frequent in high-grade athletes (OR = 3.12, 95% CI = 1.75-5.56) (Table 5). The same genotype was only 1.6-fold more frequent in medium grade/recreational athletes than in non-athletes (OR = 1.62, 95% CI = 1.10-2.40) (Table 5). The 1/1 genotype was 2.5-fold more frequent in non-athletes than in professional athletes (OR = 2.52, 95% CI = 1.40-4.56), but 1.6-fold more frequent in non-athletes than in recreational athletes (OR = 1.61, 95% CI = 1.11-2.33). Overall (frequencies already reported in Table 4), the 1/1 genotype was 1.8-fold more frequent in non-athletes than in all athletes (OR = 1.80, 95% CI = 1.29-2.51).
Haplotype analyses were performed, using all three polymorphisms (Table 6). There was virtually no LD between any of the three sites in athletes (Figure 1) or non-athletes (r2 ≤ 0.05) (Figure 2). The three site haplotype was differentially distributed between athletes and controls (P = 0.012). Individually only one haplotype associated with athlete status. Specifically, (-511)C-(+3954)T-(VNTR)2 was 3 times more common in athletes than non-athletes (OR = 3.02, 95%CI = 1.15-7.87). We also analyzed the three pair wise haplotypes and found that only those that included the VNTR were significant, indicating that the association was related to the VNTR or a marker in linkage disequilibrium with it.
Discussion
Polymorphisms and exercise
Our finding that athletes have significantly different genotypes than non-athletes reinforces the evidence that athletic performance is associated with genetic constitution in addition to environmental/behavioral factors. However, we want to emphasize that carriage of a specific genetic polymorphism is only a predisposing factor and does not by itself necessarily result in elite performance. Clearly, more than one biologic process is necessary for the development of high-grade athletic performance.
In general, genetic associations of athletic phenotype are only partially understood. Increased knowledge in this research may help to identify physiological and biochemical pathways related to exercise [43]. Genetic studies so far focused on candidate genes (over 200) mainly involved in cardiovascular performance, including angiotensin-converting enzyme (ACE) and proteins involved in skeletal-muscle activity such as α-actinins [44–46]. Immunity-related genes have been minimally explored [45]. A study that examined polymorphisms of the IL-15 receptor-α (IL-15RA) gene identified a SNP strongly associated with muscle hypertrophy in young subjects (76 men and 77 women) of mixed ethnicity who performed resistance exercise training [47]. Two studies performed in non-athletes reported that the -174G/C variant in the IL-6 gene affected the response to exercise by modulating changes in serum concentrations of IL-6 [48] and high-density lipoprotein cholesterol (HDL-C) [49]. However, Walston and colleagues [50] identified no significant relationship between IL-6 SNPs or IL-6 haplotypes and serum IL-6 concentration, grip, knee, or hip strength, or frailty in 463 older women.
IL-1β and IL-1ra roles in human disease and exercise
The inflammatory response after intensive exercise is a well documented phenomenon; the coordinated action of cytokines is considered crucial for regulation of inflammatory reactions responsible for immune defenses and muscle repair. Conceivably, a delay in the removal of dead cells increases the likelihood of a slow recovery.
IL-1β is a crucial proinflammatory cytokine involved in initiating and amplifying the inflammatory response; it stimulates the production of prostaglandins and nitric oxide, both of which are highly inflammatory, and it induces the synthesis of several chemokines [6, 12]. IL-1β is a highly active pleiotropic cytokine with several known functions. It has many systemic effects in the protection of the body, including response to stress and the metabolism of insulin, lipids and bone [12]. Injecting a few nanograms per kilogram of IL-1β results in an increase of acute phase proteins, plasma IL-6, neutrophilia and thrombocytosis [12]. Recent evidence suggests that IL-1β plays a key pathogenic role in many human conditions, including cardiovascular diseases [23, 51], and in the central nervous system effects of exercise [52, 53]. The bioactivity of IL-1β is finely modulated as it is a potentially harmful factor able to promote tissue damage and fever. The main cytokine that counteracts the effects of IL-1β is IL-1ra, a naturally occurring inhibitor of IL-1β that binds to IL-1 receptor type I, but does not elicit post-receptor signaling [7]. In addition, IL-1ra seems to modulate the IL-1 decoy receptor, IL-1 receptor type II [54]. IL-1ra protein is produced by many cells that can synthesize IL-1β, mainly by hepatocytes, adipocytes, neutrophils and macrophages. The IL-1ra plasma levels are co-coordinately regulated by both the IL-1RN and IL-1B genes [16, 22]. The biological functions of IL-1ra are not completely understood. Interestingly, at present a recombinant IL-1ra (anakinra) is approved for use in humans as an anti-inflammatory therapy in several conditions, such as rheumatic diseases, ankilosing spondylitis, chronic Achilles tendinopathy, type 2 diabetes mellitus and systemic inflammatory diseases [6, 7, 51, 55].
Several studies found that serum IL-1β concentration does not change after physical exercise, whereas IL-1ra concentration significantly increases [1, 5]. In this respect, the profile of systemic cytokines' changes in response to exercise differs from that observed in sepsis, where there is a relevant increase in both serum IL-1β and IL-1ra [1]. The rise of circulating anti-inflammatory cytokines induced by exercise has been suggested as one of the pathways involved in protection against several diseases, primarily against cardiovascular disease and type 2 diabetes mellitus exerted by regular exercise [1].
VNTR polymorphism in the intron 2 of the IL-1RNgene
We demonstrated that the genotype -511 in the promoter region and +3954 in exon 5 of IL-1B gene have no influence on the athletic phenotype. These findings seem to fit with the in vivo observation that neither circulating IL-1β, nor IL-1β mRNA levels are significantly changed by physical activity [1, 9, 56].
In contrast, we showed that a specific genotype (1/2) of the IL-1ra gene, a VNTR in intron 2 of IL-1RN, is almost 2-fold more frequent in athletes than in non-athletes. Additionally, we observed a dose-effect relationship with the 1/2 IL-1RN genotype being 2-fold more frequent in professional than in recreational athletes and 3-fold more frequent in professional athletes than in non-athletes. Thus, our study suggests the 1/2 IL-1RN frequency is increasing in parallel with the level of athletic grade. We did not observe gender effects.
The precise molecular effects of the IL-1RN polymophisms are not totally clear [22, 27]. Possibly, the 1/2 IL-1RN genotype could favor muscle repair and hypertrophy in athletes; however, further studies are needed to clarify this issue [4, 47]. This can occur through an influence on circulating or local levels of IL-1ra in humans [22, 27]. Consistent with this is that the IL-1RN*2 allele is associated with a higher production of inflammatory reactions, and therefore, an overall increase of IL-1 activity, meaning less effective IL-1ra inhibitory activity [19, 27]. However, contrasting findings were obtained for the association of IL-1RN*2 with IL-1ra levels; some authors found that the IL-1RN*2 allele is associated with reduced IL-1ra [14, 57, 58], but other authors observed no association [29] or even increased IL-1ra [16, 18, 21]. Some of these discrepancies can be due to different experimental designs and/or by the presence of heterogeneity with other polymorphisms in the IL-1RN gene [27].
In our study athletes were not more likely to be homozygous for the allele 2, a genotype that has been associated with several pathological conditions of inflammatory or autoimmune nature [18, 19, 26, 28, 34, 59, 60]. It is of note that the IL-1RN 2/2 genotype was more likely associated with disease than the IL-1RN 1/2, especially for gastric cancer development [19, 34, 41]. Based on the in vitro observation that VNTR allele 2 has been associated with a decreased production of IL-1ra [14, 58]; and increased production of IL-1β [20] in a dose-dependent fashion, these findings, taken together, suggest that a moderate increase of IL-1-dependent inflammation, as conferred by one IL-1RN allele 2 can favor athletic performance, but not an excessive increase as conferred by the IL-1RN 2/2 genotype.
It is known that there are different IL-1 gene cluster genetic profiles between ethnic groups, and particularly between black and white populations [35]. Therefore, we considered only white Italian subjects, a homogeneous Caucasian ethnic group. A limitation of this approach is that our results may not be applicable to non-Caucasian subjects.
Conclusions
Our study supports the notion that the IL-1 family of genes is significantly associated with physical performance. The finding of a higher frequency of allele 2, and specifically of the 1/2 genotype VNTR IL-1RN, in athletes with respect to non-athletes highlights that immune genetic variants are involved in athletic predisposition. Based on previous evidence of inflammatory effects of the VNTR IL-1RN allele 2, the association of IL-1 related genes with athlete status appears to associate a genetic predisposition of a moderately enhanced inflammatory response. However, we cannot exclude much more indirect effects modulated by IL-1ra. Immunity in physical exercise is likely regulated by subtle tuning of cytokine levels; for example, a partial reduction in IL-1ra could reinforce immune defenses against exercise-associated micro-injuries.
It was beyond the scope of this study to investigate the effects of IL-1 polymorphisms on exercise-induced systemic and local inflammation, recovery capacity and type of sport activity (aerobic vs. anaerobic; endurance vs. sprint), which should be clarified in future studies. Of major interest will be the comparison of aerobic vs. anaerobic athletes; however, in the present study this kind of evaluation was not possible due to small number of athletes participating in purely anaerobic sports. Future studies relating IL-1RN genotypes and cytokine levels should focus preferably on immune genetic profiles and inflammatory markers evaluated before and after exercise. Although our results indicate an association between the IL-1 family of cytokines and athlete status, it is not clear how, specifically, the VNTR IL-1RN polymorphism affects the phenotype promoting a relationship between this allele and grade of athletic performance.
In this study we focused on 2 SNPs of the IL-1B gene and 1 VNTR in the IL-1RN gene because these are the most frequently studied polymorphisms of the IL-1β and IL-1ra cytokines, which have been associated with a variety of human conditions. Interestingly, athlete status may constitute a confounding factor contributing to variability of results concerning the association of the IL-1RN polymorphisms with pathologic conditions [61].
Our present results should prompt larger studies on additional polymorphisms in the IL-1 family in relation to physical performance and elite athletic grade, particularly, those located in the IL-1ra gene, as the SNP at position +2018.
In conclusion, we demonstrated in a cross-sectional study that a polymorphism potentially able to modulate the levels of the IL-1ra cytokine, and indirectly IL-1 (α and β) activity, is associated with athlete status. Further research into immune factors that increase athletic performance will help to inform the design of how immune modulators may improve recovery or implement cure interventions after injuries. Our finding suggests that clinical trials performed to assess efficacy of various treatments for athlete's recovery should probably differentiate subjects on a genetic basis in addition to etiology to maximize the chances of observing statistically significant and reproducible results. Our study suggests cytokines are important molecules to further explore in relation to sports activity. Overall, this study may contribute to an increased understanding of the genetic basis of athletic performance. It may also increase our understanding of inflammatory mechanisms associated with intensive exercise.
References
Petersen AM, Pedersen BK: The anti-inflammatory effect of exercise. J Appl Physiol. 2005, 98: 1154-1162. 10.1152/japplphysiol.00164.2004.
Nieman DC, Henson DA, Smith LL, Utter AC, Vinci DM, Davis JM, Kaminsky DE, Shute M: Cytokine changes after a marathon race. J Appl Physiol. 2001, 91: 109-114.
Gleeson M: Immune function in sport and exercise. J Appl Physiol. 2007, 103: 693-699. 10.1152/japplphysiol.00008.2007.
Tidball JG: Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol. 2005, 288: R345-353.
Peake JM, Suzuki K, Hordern M, Wilson G, Nosaka K, Coombes JS: Plasma cytokine changes in relation to exercise intensity and muscle damage. Eur J Appl Physiol. 2005, 95: 514-521. 10.1007/s00421-005-0035-2.
Dinarello CA: Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009, 27: 519-550. 10.1146/annurev.immunol.021908.132612.
Dinarello CA: Interleukin-1beta and the autoinflammatory diseases. N Engl J Med. 2009, 360: 2467-2470. 10.1056/NEJMe0811014.
Fielding RA, Manfredi TJ, Ding W, Fiatarone MA, Evans WJ, Cannon JG: Acute phase response in exercise. III. Neutrophil and IL-1 beta accumulation in skeletal muscle. Am J Physiol. 1993, 265: R166-172.
Mahoney DJ, Safdar A, Parise G, Melov S, Fu M, MacNeil L, Kaczor J, Payne ET, Tarnopolsky MA: Gene expression profiling in human skeletal muscle during recovery from eccentric exercise. Am J Physiol Regul Integr Comp Physiol. 2008, 294: R1901-1910.
Dennis RA, Trappe TA, Simpson P, Carroll C, Huang BE, Nagarajan R, Bearden E, Gurley C, Duff GW, Evans WJ, et al: Interleukin-1 polymorphisms are associated with the inflammatory response in human muscle to acute resistance exercise. J Physiol. 2004, 560: 617-626. 10.1113/jphysiol.2004.067876.
Duff GW: Evidence for genetic variation as a factor in maintaining health. Am J Clin Nutr. 2006, 83: 431S-435S.
Dinarello CA: Biologic basis for interleukin-1 in disease. Blood. 1996, 87: 2095-2147.
Gallucci S, Provenzano C, Mazzarelli P, Scuderi F, Bartoccioni E: Myoblasts produce IL-6 in response to inflammatory stimuli. Int Immunol. 1998, 10: 267-273. 10.1093/intimm/10.3.267.
Dewberry R, Holden H, Crossman D, Francis S: Interleukin-1 receptor antagonist expression in human endothelial cells and atherosclerosis. Arterioscler Thromb Vasc Biol. 2000, 20: 2394-2400.
Dinarello CA: Blocking IL-1 in systemic inflammation. J Exp Med. 2005, 201: 1355-1359. 10.1084/jem.20050640.
Hurme M, Santtila S: IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1beta genes. Eur J Immunol. 1998, 28: 2598-2602. 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K.
Buchs N, di Giovine FS, Silvestri T, Vannier E, Duff GW, Miossec P: IL-1B and IL-1Ra gene polymorphisms and disease severity in rheumatoid arthritis: interaction with their plasma levels. Genes Immun. 2001, 2: 222-228. 10.1038/sj.gene.6363766.
Bioque G, Crusius JB, Koutroubakis I, Bouma G, Kostense PJ, Meuwissen SG, Pena AS: Allelic polymorphism in IL-1 beta and IL-1 receptor antagonist (IL-1Ra) genes in inflammatory bowel disease. Clin Exp Immunol. 1995, 102: 379-383.
El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, et al: Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000, 404: 398-402. 10.1038/35006081.
Santtila S, Savinainen K, Hurme M: Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol. 1998, 47: 195-198. 10.1046/j.1365-3083.1998.00300.x.
Strandberg L, Lorentzon M, Hellqvist A, Nilsson S, Wallenius V, Ohlsson C, Jansson JO: Interleukin-1 system gene polymorphisms are associated with fat mass in young men. J Clin Endocrinol Metab. 2006, 91: 2749-2754. 10.1210/jc.2005-2786.
Rafiq S, Stevens K, Hurst AJ, Murray A, Henley W, Weedon MN, Bandinelli S, Corsi AM, Guralnik JM, Ferrucci L, et al: Common genetic variation in the gene encoding interleukin-1-receptor antagonist (IL-1RA) is associated with altered circulating IL-1RA levels. Genes Immun. 2007, 8: 344-351. 10.1038/sj.gene.6364393.
Isoda K, Ohsuzu F: The effect of interleukin-1 receptor antagonist on arteries and cholesterol metabolism. J Atheroscler Thromb. 2006, 13: 21-30.
Achyut BR, Srivastava A, Bhattacharya S, Mittal B: Genetic association of interleukin-1beta (-511C/T) and interleukin-1 receptor antagonist (86 bp repeat) polymorphisms with Type 2 diabetes mellitus in North Indians. Clin Chim Acta. 2007, 377: 163-169. 10.1016/j.cca.2006.09.012.
Marculescu R, Endler G, Schillinger M, Iordanova N, Exner M, Hayden E, Huber K, Wagner O, Mannhalter C: Interleukin-1 receptor antagonist genotype is associated with coronary atherosclerosis in patients with type 2 diabetes. Diabetes. 2002, 51: 3582-3585. 10.2337/diabetes.51.12.3582.
Blakemore AI, Cox A, Gonzalez AM, Maskil JK, Hughes ME, Wilson RM, Ward JD, Duff GW: Interleukin-1 receptor antagonist allele (IL1RN*2) associated with nephropathy in diabetes mellitus. Hum Genet. 1996, 97: 369-374. 10.1007/BF02185776.
Reiner AP, Wurfel MM, Lange LA, Carlson CS, Nord AS, Carty CL, Rieder MJ, Desmarais C, Jenny NS, Iribarren C, et al: Polymorphisms of the IL1-receptor antagonist gene (IL1RN) are associated with multiple markers of systemic inflammation. Arterioscler Thromb Vasc Biol. 2008, 28: 1407-1412. 10.1161/ATVBAHA.108.167437.
Paardt van der M, Crusius JB, Garcia-Gonzalez MA, Baudoin P, Kostense PJ, Alizadeh BZ, Dijkmans BA, Pena AS, Horst-Bruinsma van der IE: Interleukin-1beta and interleukin-1 receptor antagonist gene polymorphisms in ankylosing spondylitis. Rheumatology (Oxford). 2002, 41: 1419-1423. 10.1093/rheumatology/41.12.1419.
Rider LG, Artlett CM, Foster CB, Ahmed A, Neeman T, Chanock SJ, Jimenez SA, Miller FW: Polymorphisms in the IL-1 receptor antagonist gene VNTR are possible risk factors for juvenile idiopathic inflammatory myopathies. Clin Exp Immunol. 2000, 121: 47-52. 10.1046/j.1365-2249.2000.01266.x.
Walley AJ, Aucan C, Kwiatkowski D, Hill AV: Interleukin-1 gene cluster polymorphisms and susceptibility to clinical malaria in a Gambian case-control study. Eur J Hum Genet. 2004, 12: 132-138. 10.1038/sj.ejhg.5201084.
Cauci S, Di Santolo M, Casabellata G, Ryckman K, Williams SM, Guaschino S: Association of interleukin-1beta and interleukin-1 receptor antagonist polymorphisms with bacterial vaginosis in non-pregnant Italian women. Mol Hum Reprod. 2007, 13: 243-250. 10.1093/molehr/gam002.
di Giovine FS, Takhsh E, Blakemore AI, Duff GW: Single base polymorphism at -511 in the human interleukin-1 beta gene (IL1 beta). Hum Mol Genet. 1992, 1: 450-10.1093/hmg/1.6.450.
Tarlow JK, Blakemore AI, Lennard A, Solari R, Hughes HN, Steinkasserer A, Duff GW: Polymorphism in human IL-1 receptor antagonist gene intron 2 is caused by variable numbers of an 86-bp tandem repeat. Hum Genet. 1993, 91: 403-404. 10.1007/BF00217368.
Witkin SS, Gerber S, Ledger WJ: Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin Infect Dis. 2002, 34: 204-209. 10.1086/338261.
Pillay V, Gaillard MC, Halkas A, Song E, Dewar JB: Differences in the genotypes and plasma concentrations of the Interleukin-1 receptor antagonist in black and white South African asthmatics and control subjects. Cytokine. 2000, 12: 819-821. 10.1006/cyto.1999.0637.
Tishkoff SA, Williams SM: Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002, 3: 611-621. 10.1038/ni0702-611.
Kim SH, Mok JW, Kim HS, Joo CK: Association of -31T>C and -511 C>T polymorphisms in the interleukin 1 beta (IL1B) promoter in Korean keratoconus patients. Mol Vis. 2008, 14: 2109-2116.
Sainz J, Perez E, Gomez-Lopera S, Jurado M: IL1 gene cluster polymorphisms and its haplotypes may predict the risk to develop invasive pulmonary aspergillosis and modulate C-reactive protein level. J Clin Immunol. 2008, 28: 473-485. 10.1007/s10875-008-9197-0.
Liu K, Muse SV: PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005, 21: 2128-2129. 10.1093/bioinformatics/bti282.
Dudbridge F: Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008, 66: 87-98. 10.1159/000119108.
Machado JC, Pharoah P, Sousa S, Carvalho R, Oliveira C, Figueiredo C, Amorim A, Seruca R, Caldas C, Carneiro F, et al: Interleukin 1B and interleukin 1RN polymorphisms are associated with increased risk of gastric carcinoma. Gastroenterology. 2001, 121: 823-829. 10.1053/gast.2001.28000.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265. 10.1093/bioinformatics/bth457.
Roth SM: Perspective on the future use of genomics in exercise prescription. J Appl Physiol. 2008, 104: 1243-1245. 10.1152/japplphysiol.01000.2007.
Bray MS, Hagberg JM, Perusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C: The human gene map for performance and health-related fitness phenotypes: the 2006-2007 update. Med Sci Sports Exerc. 2009, 41: 35-73.
Rankinen T, Bray MS, Hagberg JM, Perusse L, Roth SM, Wolfarth B, Bouchard C: The human gene map for performance and health-related fitness phenotypes: the 2005 update. Med Sci Sports Exerc. 2006, 38: 1863-1888. 10.1249/01.mss.0000233789.01164.4f.
Macarthur DG, North KN: Genes and human elite athletic performance. Hum Genet. 2005, 116: 331-339. 10.1007/s00439-005-1261-8.
Riechman SE, Balasekaran G, Roth SM, Ferrell RE: Association of interleukin-15 protein and interleukin-15 receptor genetic variation with resistance exercise training responses. J Appl Physiol. 2004, 97: 2214-2219. 10.1152/japplphysiol.00491.2004.
Oberbach A, Lehmann S, Kirsch K, Krist J, Sonnabend M, Linke A, Toenjes A, Stumvoll M, Bluher M, Kovacs P: Long term exercise training decreases IL-6 serum levels in subjects with impaired glucose tolerance: effect of the -174G/C variant in interleukin-6 gene. Eur J Endocrinol. 2008, 159: 129-136. 10.1530/EJE-08-0220.
Halverstadt A, Phares DA, Roth S, Ferrell RE, Goldberg AP, Hagberg JM: Interleukin-6 genotype is associated with high-density lipoprotein cholesterol responses to exercise training. Biochim Biophys Acta. 2005, 1734: 143-151.
Walston J, Arking DE, Fallin D, Li T, Beamer B, Xue Q, Ferrucci L, Fried LP, Chakravarti A: IL-6 gene variation is not associated with increased serum levels of IL-6, muscle, weakness, or frailty in older women. Exp Gerontol. 2005, 40: 344-352. 10.1016/j.exger.2005.01.012.
Abbate A, Salloum FN, Vecile E, Das A, Hoke NN, Straino S, Biondi-Zoccai GG, Houser JE, Qureshi IZ, Ownby ED, et al: Anakinra, a recombinant human interleukin-1 receptor antagonist, inhibits apoptosis in experimental acute myocardial infarction. Circulation. 2008, 117: 2670-2683. 10.1161/CIRCULATIONAHA.107.740233.
Chennaoui M, Drogou C, Gomez-Merino D: Effects of physical training on IL-1beta, IL-6 and IL-1ra concentrations in various brain areas of the rat. Eur Cytokine Netw. 2008, 19: 8-14.
Carmichael MD, Davis JM, Murphy EA, Brown AS, Carson JA, Mayer EP, Ghaffar A: Role of brain IL-1beta on fatigue after exercise-induced muscle damage. Am J Physiol Regul Integr Comp Physiol. 2006, 291: R1344-1348.
Colotta F, Saccani S, Giri JG, Dower SK, Sims JE, Introna M, Mantovani A: Regulated expression and release of the IL-1 decoy receptor in human mononuclear phagocytes. J Immunol. 1996, 156: 2534-2541.
Larsen CM, Faulenbach M, Vaag A, Volund A, Ehses JA, Seifert B, Mandrup-Poulsen T, Donath MY: Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007, 356: 1517-1526. 10.1056/NEJMoa065213.
Suzuki K, Peake J, Nosaka K, Okutsu M, Abbiss CR, Surriano R, Bishop D, Quod MJ, Lee H, Martin DT, et al: Changes in markers of muscle damage, inflammation and HSP70 after an ironman triathlon race. Eur J Appl Physiol. 2006, 98: 525-534. 10.1007/s00421-006-0296-4.
Andus T, Daig R, Vogl D, Aschenbrenner E, Lock G, Hollerbach S, Kollinger M, Scholmerich J, Gross V: Imbalance of the interleukin 1 system in colonic mucosa-association with intestinal inflammation and interleukin 1 receptor antagonist [corrected] genotype 2. Gut. 1997, 41: 651-657.
Tountas NA, Casini-Raggi V, Yang H, Di Giovine FS, Vecchi M, Kam L, Melani L, Pizarro TT, Rotter JI, Cominelli F: Functional and ethnic association of allele 2 of the interleukin-1 receptor antagonist gene in ulcerative colitis. Gastroenterology. 1999, 117: 806-813. 10.1016/S0016-5085(99)70338-0.
Crusius JB, Pena AS, Van Oosten BW, Bioque G, Garcia A, Dijkstra CD, Polman CH: Interleukin-1 receptor antagonist gene polymorphism and multiple sclerosis. Lancet. 1995, 346: 979-10.1016/S0140-6736(95)91607-5.
Langdahl BL, Lokke E, Carstens M, Stenkjaer LL, Eriksen EF: Osteoporotic fractures are associated with an 86-base pair repeat polymorphism in the interleukin-1-receptor antagonist gene but not with polymorphisms in the interleukin-1beta gene. J Bone Miner Res. 2000, 15: 402-414. 10.1359/jbmr.2000.15.3.402.
Kamangar F, Cheng C, Abnet CC, Rabkin CS: Interleukin-1B polymorphisms and gastric cancer risk-a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006, 15: 1920-1928. 10.1158/1055-9965.EPI-06-0267.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/11/29/prepub
Acknowledgements
Many thanks to Prof. Giuseppe Damante, Institute of Genetics, University Hospital of Udine, Udine, Italy for blinded quality control of genotyping.
This study was supported in part by year 2006, 2007 grants from University of Udine.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Conceived and designed the experiments: SC. Performed the experiments: MDS, SC. Analyzed the data and performed statistical analysis: KKR, SMW. Wrote the paper: SC, SMW, GB. All authors read and approved the final manuscript.
Electronic supplementary material
12881_2009_590_MOESM1_ESM.DOC
Additional file 1: Table 7 Genotype frequencies of IL-1B promoter at position -511 in female and male athletes. Table 8 Genotype frequencies of IL-1B exon 5 at position +3954 in female and male athletes. Table 9 Genotype frequencies of IL-1RN VNTR in female and male athletes. Frequencies of IL-1B and IL-1RN genotype frequencies stratified by athlete gender. (DOC 172 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Cauci, S., Di Santolo, M., Ryckman, K.K. et al. Variable number of tandem repeat polymorphisms of the interleukin-1 receptor antagonist gene IL-1RN: a novel association with the athlete status. BMC Med Genet 11, 29 (2010). https://doi.org/10.1186/1471-2350-11-29
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-11-29