Abstract
Background
Spinal muscular atrophy (SMA) is an autosomal recessive hereditary disorder caused by mutations of the survival motor neuron 1 (SMN1) gene. Recently, high-resolution DNA melting analysis (HRMA) with saturation LC Green dyes has become a powerful post-PCR technique for genotyping or mutation scanning. So far, no studies have applied HRMA to the molecular analysis of SMA.
Methods
The exon 7 and the flanking area of the SMN1 and SMN2 genes of 55 SMA patients and 46 unrelated normal individuals were amplified with asymmetric PCR with unlabeled probe and symmetric PCR without probe, respectively. The saturation LC Green dyes were added to the PCR system. The PCR products were loaded onto the LightScanner system and were melted from 60°C to 95°C slowly. The melting curves were acquired and analyzed by the LightScanner software.
Results
Three types of melting curves that correlated with the presumed genotype of SMA patients and controls were clearly separated on the HRMA chromatogram with the unlabeled probe. The 55 SMA patients and 46 non-SMA controls were identified with HRMA with a 100% clinical sensitivity.
Conclusion
The HRMA with saturation LC Green dyes and unlabeled probe appears to be a suitable, alternative method for the diagnosis of SMA, with high sensitivity and specificity.
Similar content being viewed by others
Background
Spinal muscular atrophy (SMA) is a common autosomal recessive hereditary disease characterized by degeneration of the anterior horn α-motor neurons in the spinal cord, leading to paralysis and atrophy of proximal muscles. The survival motor neuron (SMN) gene is the disease-causing gene of SMA [1], and it exists as two nearly identical copies, SMN1 and SMN2. SMN1 is the critical gene involved in SMA, as more than 90% of SMA patients have SMN1 exon 7 homozygous deletions [1–3]. Homozygous absence of SMN2 genes does not cause SMA and is found in about 5% of normal individuals. In SMA patients, the number of SMN2 copies is inversely correlated with disease severity [4–6]. Only five nucleotides differ between SMN1 and SMN2. SMN1 can be distinguished from SMN2 by two nucleotide changes in exon 7 and 8, which can be used to detect the deletion of SMN1 and establish the diagnosis of SMA. To date, four assays have been described for detecting the absence of SMN1: single-stranded conformation polymorphism (SSCP), restriction enzyme digestion analysis, denaturing high-performance liquid chromatography (DHPLC) analysis and liquid microbead arrays. SSCP is a very time-consuming procedure [1, 3]. Restriction enzyme digestion analysis is the traditional method for SMA, but its accuracy can be hindered by incomplete digestion [2]. DHPLC is a more recent technology for SMN1 mutation screening [7–9], but it is confined to the WAVE Nucleic Acid Fragment Analysis System. While the liquid microbead arrays is a sensitive, high-throughput approach that can be used to detect SMN1 exon 7 deletions from blood spots, the cost may prohibit its application in many laboratories [10].
High-resolution DNA melting analysis (HRMA) with saturation LC Green dyes is becoming a powerful post-PCR technique for genotyping and mutation scanning. Apparent advantages of HRMA include speed of analysis and convenience, as there is no need for processing or separation of PCR products. Recent reports indicate that the sensitivity and specificity is 100% for amplicons less than 400 bp, and those for amplicons between 400 to 1000 bp are 96.1% and 99.4%, respectively [11]. In this report, we present the results of a study using the HRMA method to analyze a cohort of SMA patients and controls. To date, there have been no reports using the HRMA method for the molecular analysis of SMA.
Methods
Subjects
A total of 55 unrelated patients with SMA and 46 unrelated normal individuals were recruited from April 2005 to April 2008. All of the patients fulfilled the diagnostic criteria [12] and were confirmed to have SMN1 exon 7 homozygous deletions by RFLP and DHPLC analysis using the detailed procedure previously described [9, 13, 14]. Of the 46 normal controls analyzed by RFLP, 3 were homozygous for SMN2 exon7 deletions. The remaining 43 normal controls possessed both SMN1 and SMN2. Furthermore, 12 of the normal individuals were analyzed by real-time fluorescence quantitative PCR, as previously reported [6, 15]. Informed consent was obtained from each individual or parents of individuals younger than 18 years. The study was approved by the local ethics commission. Genomic DNA was isolated from peripheral blood lymphocytes using the QIAamp DNA Blood Minikit (QIAGEN, Hilden, Germany).
Asymmetric and symmetric PCR
The exon 7 and flanking area of the SMN1 and SMN2 genes were amplified by PCR, the primers were SMNF(5'-AGACTATCAACTTAATTTCTGATCA-3') and SMNR(5'-GATTCACTTTCATAATGCTGG-3'). The length of the PCR product was 241 base pair and contained 2 nucleotide differences between SMN1 and SMN2. For asymmetric PCR, the 20 μL reaction contained 40 ng of genomic DNA, 200 μM dNTPs, 1.25 U Taq DNA polymerase (Dichuan Biosystem, China), 2.0 ul LCGreen Plus dye (Idaho Technology), 0.05 μM SMN F primer and 0.5 μM excess SMN R primer, and 0.5 μM unlabeled probe to SMN1. The sequence of the SMN1 probe is 5'-TATAGCTATCTATGTCTATATAGCTAT-P-3', in which the underlined "G" is specific to SMN1 and "-P" indicates a 3' phosphate. For symmetric PCR, the reaction was similar to that of asymmetric PCR except that both the SMNF and SMNR primers were 0.5 μM, and the SMN1 probe was absent. The PCR reactions were performed in a 9700 Thermal Cycler (Applied Biosystems, Foster City, CA); the conditions included an initial denaturation at 94°C for 2 min, followed by 45 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 minutes.
Melting curve acquisition and analysis
The PCR products were transferred to a 96-well plate and put on the LightScanner (Idaho Technology). The samples were first denatured at 95°C and rapidly cooled to 40°C at a rate of 20°C/s, then melted from 60°C to 95°C with a slope of 0.1°C/s. The data was acquired every 1°C for a total of 25 readings. Melting curves were analyzed with LightScanner software (Idaho Technology Inc).
Results
HRMA with unlabeled probe
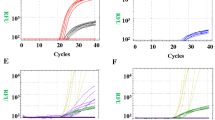
As illustrated in Figure 1A, HRMA displayed three types of melting curve shapes that correlated with the genotype of all the patients and controls. Samples from 55 SMA patients with a confirmed diagnosis of SMN1 exon 7 homozygous deletion had a lower melting temperature due to deletion of SMN1 (only possessing SMN2), inducing the probe-target mismatch. The three individuals with SMN2 deletion (only possessing SMN1) had a higher melting temperature, because PCR products were perfectly paired to the unlabeled probe. Melting curves of the control samples with SMN1 and SMN2 genes showed both duplexes. On the other hand, the results showed no correlation between the height or area of the curves and the copy number of the SMN1 or SMN2 gene as determined by real-time fluorescence quantitative PCR.
HRMA without probe
Melting curve profiles of HRMA without probes were available for 6 SMA patients with SMN1 deletion, 3 individuals with SMN2 deletion, and 6 normal controls without SMN1 or SMN2 deletion. It was easy to identify the controls with SMN1 and SMN2 genes according to the melting curve shape, while it was difficult to distinguish patients with a SMN1 deletion from normal individuals with a SMN2 deletion (Figure 1B).
Discussion
As already reported, SSCP, restriction enzyme digestion analysis, DHPLC and liquid microbead arrays could be used in detecting the absence of SMN1; all of these methods have advantages and disadvantages. Previously, one study showed that HRMA was superior to DHPLC in mutation scanning [16]. In the present study, we performed HRMA with and without an unlabeled probe for the SMA patients and controls. The exon 7 and flanking area of SMN gene were amplified. There are 2 nucleotide differences between SMN1 and SMN2 within this 241 bp product, which correspond to position intron6(-45) (G = SMN1, A = SMN2) and another to exon 7(+6) (C = SMN1, T = SMN2) [17]. According to a previous report [18], the unlabeled probe was designed to target SMN1, contained the intron6 (-45) difference in the center and was phosphorylated at the 3' end to prevent polymerase extension during amplification. Asymmetric PCR was applied to improve the rate of hybridization, and the primer ratio of 1:10 referred to in the previous report [18]. The symmetric PCR was performed routinely. Melting transitions were monitored in the presence of LCGreen I, a saturated dye originally designed to detect heteroduplexes [19]. The results showed three types of melting curve that correlate to the presumed genotype (SMN1, SMN2, SMN1/SMN2) were clearly separable on the chromatogram of HRMA with an unlabeled probe, and we succeeded in distinguishing the 55 SMA cases from the controls without any error. However, the results of the HRMA without the unlabeled probe were not as good as those with the probe, as it was difficult to distinguish patients with the SMN1 deletion from normal individuals with the SMN2 deletion.
HRMA with an unlabeled probe has many advantages in detecting the deletion of SMN1 compared with SSCP, restriction enzyme digestion analysis, DHPLC and liquid microbead arrays. First, HRMA is sensitive and accurate: the 55 patients with SMN1 exon 7 homozygous deletions were detected by HRMA with 100% sensitivity and specificity in accordance with a previous report using this technology [11]. Second, HRMA is easy and rapid to perform, with only four requirements: PCR, saturation LC Green dyes, unlabeled probe and melting instrumentation. Melting analysis can be finished within several minutes.
Conclusion
HRMA with saturation LC Green dyes and an unlabeled probe seems to be a useful alternative strategy for the diagnosis of SMA. This study has provided "proof of principle" data indicating the utility and sensitivity of HRMA when applied to diagnostic testing for SMA. However, our findings should be replicated in a much larger sampling of SMA patients to assess the specificity and sensitivity.
References
Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al: Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995, 80: 155-165. 10.1016/0092-8674(95)90460-3.
Steege van der G, Grootscholten PM, Vlies van der P, Draaijers TG, Osinga J, Cobben JM, Scheffer H, Buys CH: PCR-based DNA test to confirm the clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995, 345: 985-986. 10.1016/S0140-6736(95)90732-7.
Wu ZY, Wang N, Mu-Rong SX, Lin MT: Using polymerase chain reaction single strand conformation polymorphism to detect SMN Tgene deletions and to confirm clinical diagnosis of spinal muscular atrophy in Chinese. Chin J Neurol. 1998, 31: 289-291.
McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH: Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997, 60: 1411-1422. 10.1086/515465.
Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B: Quantitative analysis of SMN1 and SMN2 based on real-time lightcycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002, 70: 358-368. 10.1086/338627.
Chen WJ, Wu ZY, Wang N, Lin MT, Murong S-X: Study on the correlation between the SMN2 copies and the phenotype of spinal muscular atrophy. Chin J Neurol. 2005, 38: 673-676.
Sutomo R, Akutsu T, Takeshima Y, Nishio H, Sadewa AH, Harada Y, Mastsuo M: Rapid SMN1 deletion test using DHPLC to screen patients with spinal muscular atrophy. Am J Med Genet. 2002, 113 (2): 225-226. 10.1002/ajmg.10744.
Mazzei R, Conforti FL, Muglia M, Sprovieri T, Patitucci A, Magariello A, Gabriele AL, Quattrone A: A simple method for diagnosis of autosomal recessive spinal muscular atrophy by denaturing high-performance liquid chromatography. J Child Neurol. 2003, 18 (4): 269-271. 10.1177/08830738030180041301.
Chen WJ, Wu ZY, Wang N, Lin MT, Mu-Rong SX: Rapid diagnosis of spinal muscular atrophy using denaturing high-performance liquid chromatography. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005, 22: 291-293.
Pyatt RE, Mihal DC, Prior TW: Assessment of liquid microbead arrays for the screening of newborns for spinal muscular atrophy. Clin Chem. 2007, 53 (11): 1879-1885. 10.1373/clinchem.2007.092312.
Reed GH, Wittwer CT: Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 2004, 50 (10): 1748-1754. 10.1373/clinchem.2003.029751.
Munsat TL, Davies KE: International SMA consortium meeting. Neuromuscul Disord. 1992, 2: 423-428. 10.1016/S0960-8966(06)80015-5.
Wang N, Wu ZY, Mu-Rong SX, Lin MT: Rapid gene diagnosis of spinal muscular atrophy. Chin J Neuroimmunol & Neurol. 1999, 6: 124-126.
Chen WJ, Wu ZY, Lin MT, Su JF, Lin Y, Mu-Rong SX, Wang N: Molecular analysis and prenatal prediction of spinal muscular atrophy in Chinese patients by the combination of restriction fragment length polymorphism analysis, denaturing high-performance liquid chromatography, and linkage analysis. Archives of Neurology. 2007, 64 (2): 225-231. 10.1001/archneur.64.2.225.
Chen WJ, Wu ZY, Wang N, Lin MT, Murong S-X: Quantitative studies on SMN1 and carrier testing of spinal muscular atrophy. Zhong hua yi xue yi chuan xue za zhi. 2005, 22: 599-602.
Chou LS, Lyon E, Wittwer CT: A comparison of high-resolution melting analysis with denaturing high-performance liquid chromatography for mutation scanning: cystic fibrosis transmembrane conductance regulator gene as a model. Am J Clin Pathol. 2005, 124 (3): 330-338. 10.1309/BF3MLJN8J527MWQY.
Lorson CL, Hahnen E, Androphy EJ, Wirth B: A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999, 96 (11): 6307-6311. 10.1073/pnas.96.11.6307.
Zhou L, Myers AN, Vandersteen JG, Wang L, Wittwer CT: Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin Chem. 2004, 50 (8): 1328-1335. 10.1373/clinchem.2004.034322.
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ: High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003, 49 (6 Pt 1): 853-860. 10.1373/49.6.853.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/10/45/prepub
Acknowledgements
The authors thank Dr. Li Hong Biao from the Beijing Spectron Technology Company for his help and advice on HRMA. They sincerely thank the SMA families for their help and willingness to participate in this study, and they also thank the anonymous reviewers for improving this manuscript.
This work was supported by grant FMU-RT002 of the program for the Innovative Research Team in Science and Technology in the Fujian Province University, grant 30670730 from the National Natural Science Foundation of China, grant C0610008 from the Natural Science Foundation of Fujian Province of China, and grant 2005-1-4 from the Fujian Health Department. The roles of all of the funding organizations mentioned above are as follows: design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, and approval of the manuscript. Drs. Chen, Wu and Wang have received grants from all of the same funding organizations mentioned above for other research not reported in this article.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
WC and WD contributed equally to this work; the contributors are listed in the parentheses: study concept and design (WC, ZW and NW); acquisition of data (WC, WD, X-ZL, ZW, M-TL, SM and NW); analysis and interpretation of data (WC, WD, ZW, X-ZL and NW); drafting of the manuscript (WC, WD and NW); critical revision of the manuscript for important intellectual content (ZW and NW); obtaining of funding (WC, ZW and NW); administrative, technical, or material support (WC, WD, X-ZL, ZW and NW); study supervision (ZW and NW).
Wan Jin Chen, Wan Juan Dong contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chen, W.J., Dong, W.J., Lin, X.Z. et al. Rapid diagnosis of spinal muscular atrophy using High-Resolution Melting Analysis. BMC Med Genet 10, 45 (2009). https://doi.org/10.1186/1471-2350-10-45
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-10-45