Abstract
Background
Healthcare-associated infections caused by Klebsiella pneumoniae isolates are increasing and few effective antibiotics are currently available to treat patients. We observed decreased carbapenem susceptibility among K. pneumoniae isolated from patients at a tertiary private hospital that showed a phenotype compatible with carbapenemase production although this group of enzymes was not detected in any sample. The aim of this study was to describe the epidemiology and clinical outcomes associated with carbapenem-resistant K. pneumoniae and to determine the antimicrobial resistance mechanisms.
Methods
Risk factors associated with carbapenem-resistant K. pneumoniae infections were investigated by a matched case–control study from January 2006 through August 2008. A cohort study was also performed to evaluate the association between carbapenem resistance and in-hospital mortality. Bacterial identification and antimicrobial susceptibility were determined by Vitek 2 and Etest. Carbapenemase activity was detected using spectrophotometric assays. Production of beta-lactamases and alterations in genes encoding K. pneumoniae outer membrane proteins, OmpK35 and OmpK36, were analyzed by PCR and DNA sequencing, as well as SDS-Page. Genetic relatedness of carbapenem resistant isolates was evaluated by Pulsed Field Gel Electrophoresis.
Results
Sixty patients were included (20 cases and 40 controls) in the study. Mortality was higher for patients with carbapenem-resistant K. pneumoniae infections compared with those with carbapenem-susceptible K. pneumoniae (50.0% vs 25.7%). The length of central venous catheter use was independently associated with carbapenem resistance in the multivariable analysis. All strains, except one, carried bla CTX-M-2, an extended-spectrum betalactamase gene. In addition, a single isolate also possessed bla GES-1. Genes encoding plasmid-mediated AmpC beta-lactamases or carbapenemases (KPC, metallo-betalactamases or OXA-carbapenemases) were not detected.
Conclusions
The K. pneumoniae multidrug-resistant organisms were associated with significant mortality. The mechanisms associated with decreased K. pneumoniae carbapenem susceptibility were likely due to the presence of cephalosporinases coupled with porin alterations, which resulted from the presence of the insertion sequences in the outer membrane encoding genes.
Similar content being viewed by others
Background
Healthcare-associated infections, such as meningitis, pneumonia, and wound, surgical site and bloodstream infections caused by resistant gram-negative organisms such as Klebsiella pneumoniae are increasing [1]. This is a major public health threat and although antibiotics such as carbapenem can be used to treat ESBL Klebsiella, some strains of Klebsiella have developed resistance to carbapenem and polymyxin B is the option for this XDR Klebsiella[2, 3]. In 2003, the National Nosocomial Infection Surveillance System reported a 47% increase over 5 years in third-generation cephalosporin resistance in K. pneumoniae isolates recovered from patients in intensive care units (ICUs) [4]. Extended-spectrum β-lactamase (ESBL) enzymes are produced by bacteria that render them resistant to antibiotic treatment. Currently, there is a worldwide spread of ESBL-producing K. pneumoniae isolates in hospitals [5–8]. In response to this, carbapenem use has increased, resulting in increased bacterial carbapenem resistance [9]. During 2006/2007 in the US, high percentages of carbapenem-resistant K. pneumoniae isolates (4 to 11% of pathogenic isolates, with higher resistance found in isolates of primary bloodstream infections) were identified in ICUs by the National Healthcare Safety Network [10].
Carbapenem-resistant Enterobacteriaceae (CRE) pose a particular challenge as they cause numerous diseases, are hard to treat and have the potential to spread within healthcare facilities. Infections with these organisms are associated with high rates of morbidity and mortality [11–17]. Bacterial resistance to carbapenems may involve several combined mechanisms, such as the hyperproduction of AmpC β lactamases (cephalosporinases) and/or production of ESBLs and/or specific carbapenem hydrolyzing enzymes (carbapenemases) associated with alterations in the bacterial outer membrane proteins and hyperexpression of efflux systems [9]. The production of carbapenemase enzymes (KPC enzymes) is the most important mechanism of resistance to carbapenems in K. pneumoniae. The bla KPC gene is mostly plasmid-encoded and can be transferred to different K. pneumoniae clones and even to different bacterial genera [17].
Currently, K. pneumoniae is the most frequent species of CRE found in the United States and has recently been detected in Brazilian hospitals [18–20]. The outbreak and endemic dissemination of KPC-producing K. pneumoniae in hospitals is related to cross transmission with the predominance of few clones [12, 21]. Contact precautions and active surveillance are common measures employed for controlling the spread of these microorganisms in hospitals [22].
We observed a decrease in carbapenem susceptibility among K. pneumoniae in the Hospital Israelita Albert Einstein, São Paulo, Brazil. Interestingly, carbapenemase production was not detected in these strains and thus we investigated the epidemiology and clinical outcomes associated with these pathogens and determined their mechanism of resistance to beta-lactam antibiotics.
Methods
Patients and data collection
This study was conducted at the Hospital Israelita Albert Einstein (HIAE), a 620-bed private tertiary hospital in São Paulo, Brazil. Approximately 36,000 patients are admitted each year and most ward patients are hospitalized in private rooms.
All patients presenting K. pneumoniae healthcare-associated infections (HAIs) during the period from January 2006 to August 2008 were identified through the HIAE infection control database. Healthcare-associated infections were defined as those acquired after 48 hours of hospital admission. Active surveillance of all HAIs was performed by trained infection control nurses using Centers for Disease Control and Prevention (CDC) definitions [23] in the following units: bone marrow transplantation unit, step-down units, adult intensive care, pediatric and neonatal intensive care units. All episodes of catheter-associated bloodstream infections and surgical site infections were actively surveyed in all wards. When a patient had multiple episodes of K. pneumoniae infection, only the first occurrence was included in the study. A matched case–control study was performed to identify risk factors associated with carbapenem-resistant K. pneumoniae. Patients presenting carbapenem-resistant K. pneumoniae infections were considered as belonging to the case group or classified under the case group and compared to the control group (patients infected by carbapenem-susceptible K. pneumoniae). Matching was performed at a ratio of 1:2 according to infection date, anatomic site of infection, and the unit where infection was acquired. The association between carbapenem resistance and in-hospital mortality was evaluated by a cohort study and included all patients (case and control subjects). Both study strategies were approved by the Research Ethics Committee of the Hospital Israelita Albert Einstein.
K. pneumoniae isolates with imipenem and/or meropenem minimum inhibitory concentrations (MICs) ≥ 2 μg/ml as assessed by the Vitek 2 automated system (BioMérieux, St Louis, MO) were screened as possible carbapenemase producers. The reduced susceptibility or resistance to carbapenems was confirmed by Etest (BioMérieux, Marcy-l'Etoile, France). The interpretative criteria for antimicrobial susceptibility testing were according to the Clinical Laboratory Standards Institute (CLSI) M100-S19 document [24].
Clinical data were obtained from medical records. Variables analyzed as risk factors included sex, age, level of comorbidity [25], underlying disease severity [26], and treatments and procedures performed prior to the infection (such as steroid administration, neutropenia, dialysis, solid-organ or hematopoietic stem cell transplantation, surgery, use of invasive devices, and exposure to antibiotics). Length of hospital stay and previous ICU hospitalization before infection acquisition were also considered. Illness severity was assessed according to APACHE II and SOFA scores for patients hospitalized in ICUs [27, 28].
The Charlson score was divided into ≥ 3 (higher degree of underlying comorbidity), and ≤ 2 (lower degree). Antimicrobial usage was considered only when it occurred within 15 days prior to the infection diagnosis and its administration lasted for at least 48 hours [29]. Data regarding the antibiotics administered after isolation of K. pneumoniae, the time of administration, and whether the treatment was appropriate were collected. Adequate empiric antimicrobial treatment was defined as therapy administered within 24 hours after cultures samples were obtained that included the administration of an antimicrobial agent to which the K. pneumoniae isolate was resistant. Antimicrobial agents were considered adequate if the organism was susceptible, except when cephalosporins were used for the treatment of ESBL infections [30, 31]. Septic shock was defined as sepsis associated with hypotension unresponsive to intravenous fluid challenge or requiring a vasopressor agent [32].
Microbiological studies
Among the 20 K. pneumoniae isolates responsible for causing infection, 17 strains from 16 patients were viable after being stored at −80°C. For comparison purposes, a K. pneumoniae isolate susceptible to carbapenems was also included in the microbiological analysis because it was previously isolated from a patient who was subsequently infected by K. pneumoniae with reduced susceptibility to carbapenems .
Imipenem and meropenem MICs were confirmed by CLSI broth microdilution (TREK Diagnostic Inc. Westlake, OH). Quality control was performed by testing Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853.
Hydrolysis of imipenem was assessed by UV spectrophotometric assays as previously described [33]. Briefly, a full 10 μl loop of the test organism was inoculated into 500 μl of phosphate buffer 100 mM (pH 7.0) and then disrupted by sonication. Whole protein extracts were obtained after centrifugation. Hydrolytic activity of 20 μl of the crude extract was determined against 100 μM imipenem in 100 mM phosphate buffer (pH 7.0), and measurements were carried out at a 297 nm wavelength.
Specific primers under standard PCR conditions were used to detect ESBL- and carbapenemase encoding genes, namely, bla TEM , bla SHV, bla CTX-M , bla GES, bla KPC, bla IMP , bla VIM, bla SPM , bla GIM, bla SIM , bla OXA-2, bla OXA-10 , bla OXA-23 , bla OXA-24, bla OXA-48, bla OXA-51, bla OXA-58[34–37]. Amplified DNA fragments were purified with the Qiaquick PCR purification kit (Quiagen, Courtaboeuf, France) and sequenced on both strands with an automated ABI 337 sequencer (Applied Biosystems, Foster City, USA). The nucleotide and deduced protein sequences were analyzed with software available over the internet at the National Center for Biotechnology Information website. Alteration in the ompK35 and ompK36 genes that encode the K. pneumoniae major outer membrane porins were investigated by PCR and DNA sequencing [38]. Sixteen carbapenem-resistant K. pneumoniae isolates were sequenced for the ompk35 and ompk36 genes, as well as evaluated the OMP profiles by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [39]. Pulsed field gel electrophoresis (PFGE) was performed using SpeI. The results were interpreted following the criteria of Tenover et al. [40].
Statistical analysis
For continuous variables, mean values were compared using two sample t-tests for independent samples. The Mann–Whitney U-test was performed for non-normally-distributed continuous variables. Differences in proportions were compared using a Chi-square test or Fisher’s exact test when appropriate. Mean values are reported ± 1 SD. All tests of significance are two-tailed. When collinearity was identified between two variables, the one with the greatest clinical relevance associated with the event was included in the multivariate analysis. Odds ratios (OR) were calculated for independent variables associated with in-hospital mortality among patients with K. pneumoniae infections and for independent variables associated with carbapenem-resistant K. pneumoniae. The association of independent variables was expressed as OR with 95% confidence intervals. Alpha was set at 0.05. All statistical analyses were performed using the Statistical Package for the Social Sciences software (SPSS 17.0, Chicago, IL, USA).
Results
A total of 236 episodes of K. pneumoniae healthcare-associated infections were diagnosed in 175 patients during the study period. Twenty patients had carbapenem-resistant K. pneumoniae infections (8.5%), an overall rate of 0.7 episodes of carbapenem-resistant K. pneumoniae nosocomial infections per 10,000 patient-days. The carbapenem-resistant K. pneumoniae infections were: urinary tract infections (9), central venous catheter-associated bloodstream infections (5), surgical site infections (4) and skin and soft tissue infections (2).
There were no significant differences (p > 0.05) among cases and controls, with regard to baseline demographic and clinical characteristics. Among the 20 patients with imipenem and/or meropenem-resistant K. pneumoniae infections, 10 (50.0%) died during hospitalization, while 11 (27.5%) of the 40 patients with carbapenem-susceptible K. pneumoniae infections (p = 0.085) died.
By univariable analysis (Table 1) carbapenem-resistant K. pneumoniae infection was associated with previous ICU stay, central venous catheter catheterization, a longer central venous catheter use, and exposure to antimicrobials. By multivariable analysis only the length of central venous catheter use was independently associated with carbapenem resistance (OR 1.08 [95% CI, 1.01-1.16]).
Risk factors associated with in-hospital mortality among patients with K. pneumoniae infection measured by univariable analysis included receiving dialysis, elevated APACHE scores, and vasopressor drug administration and were predictors of death (Table 2). The prescription of an adequate initial antimicrobial regimen according to susceptibility testing results was not associated with patient survival. The appropriate definitive antimicrobial therapy was delayed in 3 days (median) in both groups, case and control patients.
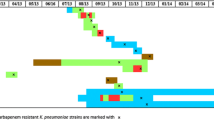
The results of the GES, CTX-M enzyme production, and the amplicon resulted of PCR amplification of the ompk35 and ompk36 are shown in Table 3. Among the 17 K. pneumoniae isolates evaluated, no carbapenem hydrolysis was detected by spectrophotometric assay or the presence of genes encoding plasmid-mediated AmpC beta-lactamase or carbapenemases. Only one strain revealed the presence of the bla GES-1 gene and all strains except one showed bla CTX-M-2, genes responsible for codifying ESBL. PCR analysis of OMP-encoding genes showed altered amplicons of at least one OMP-encoding gene, including a lack of amplification (8 isolates, 47.1%) or enhanced amplicon size (9 isolates, 53.0%). Through the DNA sequencing of some of these amplicons (data not shown), we observed that acquisition of insertion sequences (IS) were responsible for the unexpected, higher molecular size of the ompK35 or ompk36 amplification. Mutations on OmpK35- and/or OmpK36-encoding genes was observed in all isolates presenting only the constitutively OmpA in the SDS gels, such as a premature stop codon or a insertion sequence disrupting the porin gene. Seven different PFGE patterns were observed with a predominance of one subtype. This predominant subtype was observed in seven patients. Among these patients only two have been in the same intensive care unit for six days, although this period of companionship was about two months before the onset of the infection.
Analysis of the clinical isolate from a patient who initially harbored a K. pneumoniae-susceptible strain and then subsequently presented with a K. pneumoniae-resistant strain, demonstrated that although this strain belonged to the same PFGE subtype, it had no amplification of ompk36 gene. It suggests that the patient remained infected by the same K. pneumoniae clone that had become resistant to carbapenems probably due to the acquisition of an ESBL encoding gene and loss of OmpK36.
Discussion
Carbapenem resistance among the Enterobacteriaceae is an emerging phenomenon of vast clinical and public health importance. Controlling the spread of KPC enzymes is difficult once the gene encoding this enzyme reside on transmissible plasmids [23]. Current automated susceptibility testing methods have failed to reliably detect carbapenem resistance among K. pneumoniae isolates [38]. In this study, we obtained MICs for K. pneumoniae isolates by broth microdilution, thus avoiding the misclassification of some case patients as potential control subjects. During the study period, all K. pneumoniae isolates with MIC ≥ 2 μg/ml detected by the Vitek system were submitted for other susceptibility tests and screened for KPC production. K. pneumoniae isolates harboring KPC enzymes had MICs for carbapenem in a range that allowed K. pneumoniae to remain susceptible to carbapenem, and could therefore go unrecognized. Since 2010 CLSI have changed breakpoints and Enterobacteriaceae isolates with MICs for imipenem and/or meropenem ≥ 2 μg/ml have been categorized as intermediate or resistant (CLSI, M100-S20-U). Therefore, some strains included in our study would be classified as imipenem and/or meropenem resistant if the most recent CLSI breakpoints are applied.
The presence of a premature stop codon in the porin gene could explain why some K. pneumoniae isolates included in the present study presenting an ompK35 or ompK36 amplicon size of 1000 bp are resistant to carbapenens.
During the 32 months of the study period, 20 patients were diagnosed with healthcare-associated infections caused by carbapenem-resistant K. pneumoniae strains in our hospital. This represented 8.5% of the total episodes of K. pneumoniae healthcare-associated infections. Analysis of data on infectious disease outcomes of patients revealed that carbapenem-resistant K. pneumoniae patients had a higher mortality compared with patients infected with carbapenem-susceptible K. pneumoniae (50.0% and 27.5%, respectively), although it was not statistically significant (p=0.085). Similar harmful effects on patient outcomes have been observed in previous studies where carbapenem-resistant K. pneumoniae associated mortality was between 30 and 50% [11, 12, 15, 16, 21, 41, 42]. These studies examined the epidemiology of KPC producers during K. pneumoniae related-infections. Of interest, although patients included in our study were infected by carbapenem-resistant K. pneumoniae strains that did not produce KPC carbapenemase, they had similar outcomes in terms of mortality.
Evaluation of the factors that predict carbapenem resistance by univariable analysis, demonstrated that prior ICU stay, central venous catheterization, longer use of a central venous catheter, and exposure to antimicrobials were associated with carbapenem-resistant K. pneumoniae infection. In the multivariable analysis only the length of central venous catheter use was independently associated with K. pneumoniae carbapenem resistance. Previous studies reported similar risk factors for carbapenem-resistant K. pneumoniae infection and demonstrated associations with length of hospital stay, ICU admission, use of central venous catheter, recent solid-organ or stem-cell transplantation, receipt of mechanical ventilation, and exposure to broad-spectrum antibiotics [11, 12, 15]. Overestimation of the importance of antibiotic exposure as a risk factor is a common selection bias in case–control studies in which control subjects have susceptible isolates. Surprisingly, in this study carbapenem use was not an independent predictor for carbapenem resistance. This unexpected finding may be related to the small sample size of this study.
To further explore the risk of mortality in K. pneumoniae-infected patients (both case studies and controls), we evaluated the impact of patient characteristics and treatment interventions. Unexpectedly, the initial treatment of patients with antibiotics for clinical isolates that were in vitro susceptible to treatment was not associated with patient survival.
Therefore, poor patient outcomes cannot be fully explained by a delay in providing the appropriate therapy. Previous studies have suggested that removal of the focus of infection, such as a catheter, debridement, or drainage, is an effective way of improving survival among patients with carbapenem-resistant K. pneumoniae infections [11]. However, this adjunctive therapy was not evaluated in our study.
Besides observing clinical characteristics, we also performed molecular analysis of isolated K. pneumoniae strains to analyze the mechanisms of antimicrobial resistance, and to rule out the possibility of an outbreak during the study period. Although carbapenemase encoding genes including bla KPC were not identified in any of the K. pneumoniae isolates studied, bla CTX-M-2 and bla GES-1, ESBL encoding genes were detected in our collection. In addition, we observed changes in OMP-encoding genes amplicon size by PCR in 9 isolates suggesting the likelihood of altered porin functions. The amplicon size expected for ompK35 or ompk36 amplification is around 1000 bp. Through the DNA sequencing of some of these amplicons, we observed that acquisition of insertion sequences were responsible for the unexpected, higher molecular size of the ompK35 or ompk36 amplification. Therefore, we conclude that impermeability of outer membrane proteins contributed considerably to carbapenem decrease susceptibility in those K. pneumoniae isolates, especially when these isolates were ESBL producers (especially CTX-M-2- producing K. pneumoniae).
Most cases did not cluster in time and space. Molecular epidemiology revealed that the isolates belonged to seven distinct clones, although one subtype was predominant. However, most cases could not be linked to a specific patient-to-patient transmission event or to a common source.
Our results show that cephalosporinase production associated with porin modifications likely contributed to carbapenem resistance. This study focused on bacterial infection not colonization, and this allowed for a more accurate analysis of prognosis and mortality, as we only included patients with ongoing infections.
There were a number of limitations in this study. First, we had a low number of episodes of carbapenem-resistant K. pneumoniae infections in our hospital, suggesting the number of samples analyzed was small. Moreover this sample size may be underpowered to detect small significant differences. Second, we were not able to include prior colonization with carbapenem-resistant K. pneumoniae in our risk factor analysis for invasive infection, because the colonization status of each patient was unknown. At that time point we did not perform active surveillance for carbapenem-resistant Enterobacteriaceae (by rectal or peri-rectal swabs). Third, the case–control design for analyzing the risk factors for antimicrobial resistance has some limitations. The use of patients infected with carbapenem-susceptible K. pneumoniae as control subjects may be falsely inflated prior antimicrobial exposure (which was not observed in our study). The ability to match control-patients on important variables, such as time at risk and location, is problematic in case–control studies. Finally, because this study was performed at a single medical center, these results may not extrapolate to other hospitals.
Conclusions
In conclusion, the analysis of antimicrobial resistance and the molecular characterization of K. pneumoniae were useful for understanding preventive measures that should be implemented in the hospital.
References
Peleg AY, Hooper DC: Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010, 362: 1804-1813. 10.1056/NEJMra0904124.
Talbot GH, Bradley J, Edwards JE, Gilbert D, Scheld M, Bartlett JG: Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin Infect Dis. 2006, 42: 657-658. 10.1086/499819.
Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards JE, Infectious Diseases Society of America: The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008, 46: 155-164. 10.1086/524891.
Lockhart SR, Abramson MA, Beekmann SE, Gallagher G, Riedel S, Diekema DJ, Quinn JP, Doern GV: Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol. 2007, 45: 3352-3359. 10.1128/JCM.01284-07.
Hawkey PM: Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect. 2008, 14 (Suppl 1): 159-165.
Villegas MV, Kattan JN, Quinteros MG, Casellas JM: Prevalence of extended-spectrum beta-lactamases in South America. Clin Microbiol Infect. 2008, 14 (Suppl 1): 154-158.
Bush K: Extended-spectrum beta-lactamases in North America, 1987–2006. Clin Microbiol Infect. 2008, 14 (Suppl 1): 134-143.
Falagas ME, Karageorgopoulos DE: Extended-spectrum beta-lactamases-producing organisms. J Hosp Infect. 2009, 73: 345-354. 10.1016/j.jhin.2009.02.021.
Nordmann P, Cuzon G, Naas T: The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009, 9: 228-236. 10.1016/S1473-3099(09)70054-4.
Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK, National Healthcare Safety Network Team Participating National Healthcare Safety Network Facilities: NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemio. 2008, 29: 996-1011. 10.1086/591861.
Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP: Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008, 29: 1099-1106. 10.1086/592412.
Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H: An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis. 2010, 50: 364-373. 10.1086/649865.
Marchain D, Navon-Venezia S, Schwaber MJ, Carmeli Y: Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother. 2008, 52: 1413-1418. 10.1128/AAC.01103-07.
Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J: Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City. A new treat to our antibiotic armamentarium. Arch Intern Med. 2005, 165: 1430-1435. 10.1001/archinte.165.12.1430.
Falagas ME, Rafailidis PI, Kofteridis D, Virtzili S, Chelvatzoglou FC, Papaioannou V, Maraki S, Samonis G, Michalopoulos A: Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: a matched case control study. J Antimicrob Chemother. 2007, 60: 1124-1130. 10.1093/jac/dkm356.
Nguyen M, Eschenauer GA, Bryan M, O’Neil K, Furuya EY, Della-Latta P, Kubin CJ: Carbapenem-resistant Klebsiella pneumoniae bacteremia: factors correlated with clinical and microbiologic outcomes. Diagn Microbiol Infect Dis. 2010, 67: 180-184. 10.1016/j.diagmicrobio.2010.02.001.
Munoz-Price LS, Quinn JP: The spread of Klebsiella pneumoniae carbapenemases: a tale of strains, plasmids, and transposons. Clin Infect Dis. 2009, 49: 1739-1741. 10.1086/648078.
Monteiro J, Santos AF, Asensi MD, Peirano G, Gales AC: First report of KPC-2 producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009, 53: 333-334. 10.1128/AAC.00736-08.
Peirano G, Seki LM, Val Passos VL, Pinto MCFG, Guerra LR, Asensi MD: Carbapenem-hydrolysing β-lactamase KPC-2 in Klebsiella pneumoniae isolated in Rio de Janeiro, Brazi. J Antimicrob Chemother. 2009, 63: 265-268.
Pavez M, Mamizuka EM, Lincopan N: Early dissemination of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009, 53: 2702-10.1128/AAC.00089-09.
Hussein K, Sprecher H, Mashiach T, Oren I, Kassis I, Filkenstein R: Carbapenem resistance among Klebsiella pneumoniae isolates: risk factors, molecular characteristics, and susceptibility patterns. Infect Control Hosp Epidemiol. 2009, 30: 666-671. 10.1086/598244.
Centers for Diseases Control and Prevention: Guidance for control of infections with carbapenem-resistant or carbapenemase-produzing Enterobacteriaceae in acute care facilities. MMWR. 2009, 58: 256-260.
Horan TC, Andrus M, Dudeck MA: CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008, 36: 309-332. 10.1016/j.ajic.2008.03.002.
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S19. 2009, Wayne, PA: Clinical and Laboratory StandardsInstitute,
Charlson ME, Sax FL, MacKenzie CR, Fields SD, Braham RL, Douglas RGJ: Assessing illness severity: does clinical judgment work?. J Chronic Dis. 1986, 39: 439-452. 10.1016/0021-9681(86)90111-6.
McCabe WR, Jackson GG: Gram-negative bacteremia II: clinical, laboratory, and therapeutic observations. Arch Intern Med. 1962, 110: 856-864. 10.1001/archinte.1962.03620240038007.
Knaus WA, Draper EA, Wagner DP, Zimmerman JE: APACHE II: a severity of disease classification system. Crit Care Med. 1985, 13: 818-829. 10.1097/00003246-198510000-00009.
Vincent JL, De Mendonça A, Cantraine F, Moreno R, Takala J, Suter PM, Sprung CL, Colardyn F, Blecher S: Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: results of a multicenter, prospective study. Working group on "sepsis-related problems" of the European Society of Intensive Care Medicine. Crit Care Med. 1998, 26: 1793-1800. 10.1097/00003246-199811000-00016.
Marra AR, Wey SB, Castelo A, Gales AC, Cal RG, Filho JR, Edmond MB, Pereira CA: Nosocomial bloodstream infections caused by Klebsiella pneumoniae: impact of extended-spectrum beta-lactamase (ESBL) production on clinical outcome in a hospital with high ESBL prevalence. BMC Infect Dis. 2006, 6: 24-10.1186/1471-2334-6-24.
Byl B, Clevenbergh P, Jacobs F, Struelens MJ, Zech F, Kentos A, Thys JP: Impact of infectious diseases specialists and microbiological data on the appropriateness of antimicrobial therapy for bacteremia. Clin Infect Dis. 1999, 29: 60-66. 10.1086/520182.
Paterson DL, Ko W-C, Gotteberg A, Casellas JM, Mulazimoglu L, Klugman KP, Bonomo R, Rice LB, McCormack JG, Yu VL: Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum β-lactamases: implications for the clinical microbiology laboratory. J Clin Microbiol. 2001, 39: 2206-2212. 10.1128/JCM.39.6.2206-2212.2001.
American College of Chest Physicians/Society of Critical Care Medicine: Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992, 20: 864-874. 10.1097/00003246-199206000-00025.
Picão RC, Poirel L, Gales AC, Nordmann P: Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob Agents Chemother. 2009, 53: 3908-3913. 10.1128/AAC.00453-09.
Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S: Rapid detection and identification of metallo-beta-lactamase-encoding genes by multiplex real time PCR assay and melt curve analysis. J Clin Microbiol. 2007, 45: 544-547. 10.1128/JCM.01728-06.
Poirel L, Heritier C, Tolün V, Nordmann P: Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004, 48: 15-22. 10.1128/AAC.48.1.15-22.2004.
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM: Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006, 27: 351-353. 10.1016/j.ijantimicag.2006.01.004.
Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD: High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla ACT-1 beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob Agents Chemother. 2006, 50 (10): 3396-3406. 10.1128/AAC.00285-06.
Tenover FC, Kalsi RK, Williams PP, Carey RB, Stocker S, Lonsway D, Rasheed JK, Biddle JW, McGowan JE, Hannat B: Carbapenem resistant in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg Infect Dis. 2006, 12: 1209-1213. 10.3201/eid1708.060291.
Carvalhaes CG, Picão RC, Nicoletti AG, Xavier DE, Gales AC: Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010, 65: 249-251. 10.1093/jac/dkp431.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B: Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995, 33: 2233-2239.
Gasink LB, Edelstein PH, Lautenbach E, Synnestvedt M, Fishman NO: Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect Control Hosp Epidemio. 2009, 30: 1180-1185. 10.1086/648451.
Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M: Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol. 2009, 30: 972-976. 10.1086/605922.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2334/13/80/prepub
Acknowledgments
This work was supported by the Instituto Israelita de Ensino e Pesquisa Israelita Albert Einstein (project 449.08). The authors wish to thank ML Triunfol and JL Croxford for outstanding editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LC was the study’s main investigator, participated in its design and drafted the manuscript. MDVM and IS performed the phenotype analysis. ACG carried out the molecular genetic studies, performed the PFGE analysis and helped to review the manuscript. CVS and TZSC were responsible for reviewing the infection control database. PFS reviewed the medical records, collected the epidemiological data and participated in the statistical analysis. JP participated in the design and provided expert oversight. ARM conceived of the study, performed the statistical analysis and helped to draft the manuscript. All authors read and approved the final manuscript.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Correa, L., Martino, M.D.V., Siqueira, I. et al. A hospital-based matched case–control study to identify clinical outcome and risk factors associated with carbapenem-resistant Klebsiella pneumoniae infection. BMC Infect Dis 13, 80 (2013). https://doi.org/10.1186/1471-2334-13-80
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2334-13-80