Abstract
Background
Acinetobacter baumannii is responsible for large epidemics in hospitals, where it can persist for long time on abiotic surfaces. This study investigated some virulence-related traits of epidemic A. baumannii strains assigned to distinct MLST genotypes, including those corresponding to the international clones I-III as well as emerging genotypes responsible for recent epidemics.
Methods
Genotyping of bacteria was performed by PFGE analysis and MLST according to the Pasteur’s scheme. Biofilm formation on polystyrene plates was assessed by crystal violet staining; resistance to desiccation was evaluated on glass cover-slips when kept at room-temperature and 31% relative humidity; adherence to and invasion of A549 human alveolar epithelial cells were determined by the analysis of viable bacteria associated with or internalized by A549 human alveolar epithelial cells; Galleria mellonella killing assays were used to analyze the virulence of A. baumannii in vivo.
Results
The ability to form biofilm was significantly higher for A. baumannnii strains assigned to ST2 (international clone II), ST25 and ST78 compared to other STs. All A. baumannii strains survived on dry surfaces for over 16 days, and strains assigned to ST1 (international clone I) and ST78 survived for up to 89 and 96 days, respectively. Adherence to A549 pneumocytes was higher for strains assigned to ST2, ST25 and ST78 than other genotypes; a positive correlation exists between adherence and biofilm formation. Strains assigned to ST78 also showed significantly higher ability to invade A549 cells. No significant differences in the killing of G. mellonella worms were found among strains.
Conclusions
Elevated resistance to desiccation, high biofilm-forming capacity on abiotic surfaces and adherence to A549 cells might have favoured the spread and persistence in the hospital environment of A. baumannii strains assigned to the international clones I and II and to the emerging genotypes ST25 and ST78.
Similar content being viewed by others
Background
Acinetobacter baumannii is an emerging bacterial pathogen responsible for widespread and persistent outbreaks among hospitalized patients. A. baumannii infections are difficult to control since many epidemic strains can resist several classes of antibiotics and disinfectants and are able to contaminate abiotic surfaces of the hospital environment, including the medical equipment [1, 2]. The vast majority of A. baumannii epidemics are caused by a limited number of strains worldwide, belonging to the initially named European clones, but now regarded to as International clonal lineages (IC) I, II and III [3–5]. Additional A. baumannii genotypes have recently emerged as epidemic clones in some regions, such as the closely related ST15 and ST84 genotypes from Europe [3, 4], the ST25 genotype from Eurasia [4, 6] and genotype ST78 isolated in several Italian hospitals [4, 7]. Since the majority of epidemic strains are multi-drug-resistant (MDR) or extensively-drug-resistant (XDR) [8], many studies have investigated the genetic and functional basis of antimicrobial resistance [1, 2, 4–6]. In contrast, few studies are available on the virulence traits and pathogenic potential of A. baumannii. It has been reported that A. baumannii can form biofilms on several abiotic surfaces, including polystyrene, polypropylene, polytetrafluoroethylene and glass [9], and ca. 70% of strains investigated in multicentre prospective cohort study performed in 28 Spanish hospitals were biofilm-formers [10]. While no difference in biofilm formation was observed between outbreak and sporadic strains, IC-II strains seem to form larger biofilms than IC-I strains [11]. Biofilm formation appeared to be positively correlated with multidrug resistance [10], as well as with the expression of several virulence factors, including the outer membrane protein OmpA, the extracellular polysaccharide poly-β-(1,6)-N-acetyl glucosamine (PNAG), type I pili, a homologue of the staphylococcal biofilm-associated protein (Bap), the outer membrane protein CarO, a quorum sensing system and proteins involved in histidine metabolism, such as urocanase [9, 12–17]. The ability of A. baumannii strains to survive for a long time on dry surfaces is likely to contribute to their persistence in hospitals [18]. In this respect, it has been recently reported that RecA protein is involved in general stress response and resistance to heat shock and desiccation in A. baumannii[19]. In addition, the ability of A. baumannii to adhere and invade epithelial cells has been investigated [20]. Among the virulence factors that may contribute to these processes, the Bap protein has been demonstrated to play a role during adherence to epithelial cells [21], and OmpA and phospholipase D have been shown to contribute to invasion of epithelial cells [15, 22].
The majority of the above studies, however, have been performed using a limited number of strains, often ATCC 19606T and/or ATCC 17978 reference strains [12, 15, 17, 23]. Only a few studies have compared the virulence traits of ICs I-III [11, 23, 24], while no studies have analyzed the virulence features of other emerging epidemic lineages. Although comparative genome analysis of twelve A. baumannii strains assigned to distinct genotypes [25, 26] and of 136 A. baumannii strains belonging to A. calcoaceticus-baumannii complex [27] has demonstrated that the genes encoding putative virulence factors are mostly conserved, phenotypic data on the expression of putative virulence factors disclosed variability among strains, even those belonging to the same IC, likely reflecting differences in expression levels [11, 23, 24].
The objective of the present study was to identify virulence-related traits of epidemic strains assigned to distinct genotypes of A. baumannii that could contribute to their ability to colonize and infect the human host and persist in the hospital environment. Biofilm formation, resistance to desiccation, adherence to and invasion of A549 human bronchial cells, and killing of Galleria mellonella caterpillars were evaluated for a diverse collection of MDR or XDR A. baumannii strains isolated during outbreaks in European hospitals, and in the reference strains ATCC 19606T and ATCC 17978.
Methods
Bacterial strains, cell line and culture conditions
Twenty-three A. baumannii strains isolated during outbreaks that occurred in several Mediterranean hospitals [4, 28–31], plus A. baumannii ATCC19606T and ATCC17978 reference strains [32, 33] were included in the study (Table 1). Epidemiological features of strains and number of patients involved in the outbreaks were in accordance to previous publications [4, 28–31]. No ethical approval was required for the study because there was no access to patient data. Bacteria were routinely cultured in Luria-Bertani broth (LB). Genotyping was performed by pulsed-field gel electrophoresis (PFGE) and multilocus sequencing typing (MLST) analyses as previously described [3, 31]. Antimicrobial susceptibilities were determined by a reference microdilution method [34]. MDR and XDR phenotypes were designed according to Magiorakos et al. [8]. The M9 minimal broth supplemented with magnesium sulphate and glucose as carbon source (M9) and Mueller Hinton broth (Oxoid, Milan, Italy) were used for biofilm growth [35] and pellicle formation [36], respectively. A549 human alveolar epithelial cells were cultured as previously described [15].
Measurement of biofilm formation
The ability of A. baumannii strains to form biofilm was measured using a microtiter plate assay [37]. One hundred microliter aliquots of overnight cultures were diluted to an optical density at 600 nm (OD600) of ~ 1.0. and dispensed in 96-well polystyrene microtiter plates. The number of bacterial cells in the supernatant was monitored by OD600 measurements during 24 h growth at 37°C. After removal of the culture, plates were washed with phosphate-buffer saline solution (PBS) and air-dried. Biofilm was stained with 0.1% crystal violet solution for 15 min. After washing with distilled water, the biofilm-associated dye was solubilised with 200 μl of 95% ethanol and the OD540 was measured. The OD540/OD600 ratio was used to normalize the amount of biofilm formed to the total cell content. In order to assess the induction of biofilm formation in the presence of antibiotics, the bacteria were grown for 24 h in microtiter plates in the presence of imipenem (Merck Research Laboratories, Rahway, N.J., USA) at sub-inhibitory concentrations as previously described [35]. Sub-inhibitory concentrations of imipenem were 0.5 mg/L for strains AYE, 700, 2979, LUH 5875, ATCC 19606T, and ATCC 17978, and of 4 mg/L for all other strains, which corresponded to 1/4 and 1/4 – 1/16 the MIC, respectively (Table 1). Three independent experiments, each one performed in triplicate, were conducted for each strain. For the pellicle formation at air-liquid interface assay, bacteria were grown for 72 h at 37°C without shaking, as previously described [36]. Positive air-liquid biofilm samples were identify visually; the isolates were considered positive when a pellicle was covering the whole liquid surface.
Desiccation survival assay
The desiccation assay was performed as previously described [18]. One ml aliquots of overnight LB cultures were centrifuged at 11,600 × g for 5 min in a microcentrifuge. The cell pellet was washed twice with PBS and suspended in distilled water to an OD600 of 1.0. Twenty microliters of each suspension were deposited onto a glass cover slip to produce an inoculum of ~2 × 107 colony-forming units (CFU). The coverslip was kept at 31% relative humidity by the presence of a saturated CaCl2 × 6H2O in an uncovered Petri dish, and stored at room temperature in an air tight transparent plastic box (17 × 11 × 5.5 cm) for up to 16 weeks. Strains were distributed into four separate boxes; each box stored up to six strains. Viable cell counts were determined once a week as previously described [23].
Adherence to and invasion of A549 cells
Adherence of A. baumannii strains to A549 cells (human type 2 pneumocytes) was determined as described previously [15], with minor modifications. In brief, ~ 105 A549 cells were infected with ~ 107 bacterial CFU and incubated for 60 min at 37°C in 5% CO2 (v/v) atmosphere. Non-adherent bacterial cells were removed by washing with PBS. Infected cells were lysed by the addition of 1 ml distilled water and serial 10-fold dilutions were plated on LB agar to determine the number of CFU of adherent bacteria. To determine adherent and invading bacteria, A549 cells were infected with A. baumannii strains as described above. The monolayers were then treated with 1 ml of fresh culture medium containing 5 mg/L of colistin sulfate (Sigma-Aldrich, Milan, Italy) for further 30 min, the shortest time point that resulted in the killing of all extracellular bacteria added to the monolayers. All strains included in the study were killed after a 30 min treatment with 5 mg/L of colistin. Afterwards, the cells were washed with PBS, harvested with trypsin, and lysed with sterile distilled water. Dilutions from harvested samples were inoculated on LB agar plates and bacterial colony counts were estimated after overnight incubation at 37°C. Each experiment was performed in triplicate.
G. mellonellakilling assay
The virulence of A. baumannii in vivo was assessed using the G. mellonella insect model of infection [23], with minor modifications. Ten G. mellonella caterpillars (average weight 500 ± 60 mg) were injected with 10 μl of serial ten-fold dilutions in PBS of A. baumannii cells grown overnight at 37°C in LB. Bacterial colony counts (CFU/ml) on LB agar plates were used to estimate the number of viable cells in each inoculum. Ten larvae that received no injection and ten larvae injected with 10 μl PBS were used as negative controls of each experiment. Two independent experiments, each one performed in triplicate (thirty larvae for each strain), were conducted for each strain. Larvae were incubated at 37°C in Petri dishes (ten larvae per dish) and monitored every 12 h for a total of 72 h. Survival curves were plotted using the Kaplan-Meier estimator and expressed in percentage. Lethal dose 50% (LD50) values were calculated using GraphPad Prism v.5.04 (GraphPad Software, La Jolla California USA) according to the following equation: Y= A + (1-A)/(1 + e(B–G×lnX)), where X is the number of viable bacterial cells injected, Y the fraction of larvae killed by a given bacterial inoculum, A is the fraction of larvae killed by the control solution, and B and G are curve-fitting constants automatically calculated by GraphPad Prism [23]. LD50 values were calculated as the value of X that corresponds to Y = 0.5.
Statistical analysis
Data were analysed using Statistical Package for the Social Sciences Version 13.0 (SPSS Inc., Chicago, IL, USA). The significance of the induction of biofilm formation in the presence of imipenem was analyzed with the Student’s t test and induction was considered significant when the P value was <0.05. The variability of strains assigned to the same ST was investigated using the One-Way Analysis of Variance (ANOVA), in which strains were grouped according to ST and differences were considered significant when the P value was <0.05. Correlations were evaluated by regression analysis using the Pearson’s correlation coefficient (r).
Results
Biofilm formation
Twenty-three A. baumannii strains isolated during outbreaks that occurred in several hospitals in France, Greece, Italy, Lebanon, Netherlands and Turkey [4, 28–31], and A. baumannii ATCC19606T and ATCC17978 reference strains [32, 33] were investigated. The strains had previously been assigned to 10 different MLST sequence types (STs) and to 17 different major PFGE types. The collection includes strains assigned to major international clones IC-I, IC-II and IC-III and additional emerging genotypes ST15, ST25 and ST78. The majority of the strains were representative of cross-transmission episodes and were isolated with identical PFGE type from more than two patients of the same or different institutions. Strains 3909 and 3957 assigned to ST78 were isolated with identical PFGE type Q from different patients during the same inter-hospital outbreak [31]. Seventeen strains exhibited an imipenem minimum inhibitory concentration (MIC) ≥16 mg/L and were considered to be carbapenem-resistant and XDR; four strains were classified as MDR (Table 1).
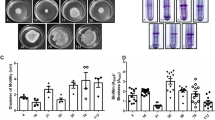
The ability to form biofilm on polystyrene microtiter plates was evaluated for outbreak A. baumannii strains assigned to different PFGE genotypes and STs (Table 1). Strains assigned to the four major PFGE types E-H (ST2, corresponding to IC-II), PFGE types L-N (ST25), and PFGE type Q (ST78) formed significantly higher biofilm levels than strains assigned to other PFGE types (including ST1, ST20, ST3, ST15, ST84, ST52, and ST77) (p <0.05) (Figure 1A). No relevant differences in growth yelds were observed among the strains, thus excluding the possibility that difference in biofilm formation were due to difference in bacterial growth. In addition, A. baumannii strains assigned to ST2, ST25 and ST78 formed a robust biofilm pellicle at the air-liquid interface when grown at 37°C for 72 h without shaking (data not shown).
Biofilm formation. Biofilm formation on 96-well polystyrene microtiter plates by A. baumannii grown statically in M9 medium for 24 hours in the absence (panel A) or presence (panel B) of imipenem concentrations 0.5 mg/L (strains AYE, 700, LUH 5875, 2979, ATCC 19606T and ATCC 17978), or 4 mg/L (all other strains). Strain and ST types are indicated on the bottom of the figure. Error bars represent standard deviations based on three independent experiments. Asterisks indicate statistically significant differences in biofilm formation among different STs (panel A) (p <0.05) and statistically significant induction in biofilm formation upon exposure to sub-MIC imipenem concentrations (panel B) (p <0.05).
Since exposure of A. baumannii to sub-inhibitory concentrations of imipenem has been demonstrated to stimulate biofilm growth [35], a biofilm formation assay was performed in the presence of sub-inhibitory concentrations of the antibiotic (Figure 1B). Growth yields were not affected by imipenem at the used concentrations for all strains tested. Compared with the untreated control, significantly higher biofilm levels were observed after exposure to imipenem for strains assigned to ST2 (except strain 3889), and ST78, which already produced high biofilm levels before treatment, and for strains assigned to ST1 and its single locus variant ST20 (IC-I), to ST3 (IC-III) and to ST52 (ATCC 19606T) (p <0.05) which, conversely, were weak producers when grown in the absence of antibiotic. On the other hand, no significant differences in biofilm formation were observed for the strains assigned to ST15, ST84 ST25 and ST77 (Figure 1A and B).
Resistance to desiccation
Strains showed an overall ability to resist desiccation, though survival times ranged from 16 to 96 days depending on the strain (Figure 2 and Additional file 1: Table S1). In particular, strains assigned to ST1 and ST78 showed extremely high resistance to desiccation, with survival times of 75 to 89 days and 68 to 96 days, respectively. Strains assigned to ST15, strain 4025 (ST3) and strain 3890 (ST25) showed a comparable trend of high resistance to desiccation, with survival times of 75 to 89 days. Strains assigned to ST2 and ST20, strains 4190 and 3865 (assigned to ST25) and strain ATCC 17978 were moderately resistant to desiccation with survival times of 33 to 54 days. Finally, strains 3871 (assigned to ST84), LUH 5875 (assigned to ST3) and ATCC 19606T were less resistant to dry conditions, and survived for < 29 days (Figure 2 and Additional file 1: Table S1).
Resistance to desiccation. Resistance to desiccation of A. baumannii strains. Strains were inoculated onto rounded glass cover slips and incubated at 31% relative humidity and room temperature. A starting bacterial inoculum of ~ 2 × 107 per cover slip was used. Values represent the mean of three independent experiments.
Adherence to and invasion of A549 human bronchial cells
Different steps of the interaction between A. baumannii and A549 human alveolar epithelial cells were investigated, including adherence to epithelial cells and subsequent invasion [15]. A. baumannii strains displayed different ability to adhere to epithelial cells (Figure 3A). In particular, all strains assigned to ST2, ST25, ST78 and strain ATCC 17978 adhered to epithelial cells at significantly higher levels than other strains (p <0.05). Moreover, a highly significant correlation was found between adhesiveness of A. baumannii to A549 human alveolar epithelial cells and biofilm formation for all strains, except ATCC 19606T and ATCC 17978 (r = 0.832, p <0.01).
Bacterial adherence to and invasion of A549 bronchial epithelial cells. (A) Cell surface-associated bacteria after 60 min incubation at 37°C. Statistically significant differences (p <0.05) among different STs are indicated with asterisks. (B) Intracellular bacterial counts (CFU/well) obtained after 60 min incubation of A549 cells with bacterial strains and subsequent treatment for 30 min at 37°C with colistin (5 mg/L). Statistically significant differences among different STs (p <0.05) are indicated with asterisks. Each value represents the mean ± standard deviation (error bar) of three independent experiments performed in duplicate.
Strains assigned to ST1, ST20, ST2, ST25 and ST78 and reference strain ATCC 19606T were all invasive, although only strains assigned to ST78 showed significantly high levels of invasiveness when compared with other genotypes (p <0.05) (Figure 3B). A similar number of bacteria adhered to A549 cells when the monolayers were incubated with A. baumannii strains for 60 min at 4°C, i.e. under conditions that do not allow for tissue invasion (data not shown).
G. mellonellakilling assays
The LD50 values were calculated for each strain at 24, 48 and 72 h post-infection (Table 2). For all strains, the LD50 decreased with post-infection time, attaining the final value at 72 h (Additional file 2: Figure S1). No relevant difference in larval death was observed at the subsequent time points (data not shown). Differences in LD50 between 24 h and 72 h provide a trend of the killing rapidity for individual strains (Table 2 and Additional file 2: Figure S1). The LD50 at 72 h were comprised between 8.1 × 104 and 4.7 × 106 CFU/larva, with the average LD50 being 5.8(±9.5) × 105 CFU/larva. These values are in the range of those determined in a previous study on clinical A. baumannii isolates [23]. Strain-specific differences between LD50 calculated at 72 h were significant (p <0.001). In particular, strains ATCC 19606T (ST52) and 3868 (ST15) had significantly (p <0.001) higher LD50 than all the other strains tested. In contrast, ST-specific differences between LD50 calculated at 72 h were not significant (p ≥0.05).
Discussion
Antibiotic resistance and long-term persistence in the hospital environment are two factors that most likely contribute to the success of A. baumannii as an opportunistic pathogen [1, 2, 5]. However, it is possible that the emergence of some epidemic strains or clones over others is the result of an improved ability to colonize patients and cause disease. In attempt to address this issue, biofilm formation, resistance to desiccation, adherence to and invasion of human epithelial cells and the ability of A. baumannii to kill G. mellonella catterpilars were investigated as possible virulence-related phenotypes among A. baumannii isolates belonging to the main ICs and to recently emerging clones [10, 15, 18, 23]. To this purpose, a collection of 21 A. baumannii strains assigned to the three main lineages circulating worldwide, i.e. IC-I, II and III, as well as to distinct genotypes recently identified as responsible for outbreaks, and two reference A. baumannii strains ATCC 19606T and ATCC 17978 were selected (Table 1).
Results demonstrate that epidemic A. baumannii genotypes differ in the ability to form biofilm on abiotic surfaces. In particular, strains assigned to genotypes ST2, ST25 and ST78 produce biofilm more efficiently and are able to form a robust biofilm pellicle at the air-liquid interface of the culture medium. In keeping with this result, it has been shown that strains belonging to the A. baumannii-calcoaceticus complex form higher biofilm pellicle than other Acinetobacter species [36] and that A. baumannii strains assigned to the IC-II form more biofilm than A. baumannii strains assigned to the IC-I [11]. It is also worth mentioning that the A. baumannii strain SMAL, responsible for two outbreaks in Northern Italy and shown to be an efficient biofilm producer [35], has recently been assigned to ST78 [7]. Biofilm could therefore be regarded as an important virulence feature for A. baumannii strains belonging to ST2, ST78 and also ST25. In consistence with previous findings [35], our study also demonstrates that exposure to sub-inhibitory concentrations of imipenem has an overall stimulating effect on biofilm formation as shown for epidemic strains assigned to ST1, ST2, ST3, ST20 and ST78. It is plausible that transition from planktonic to biofilm lifestyle upon imipenem exposure represents a defensive response of A. baumannii to the threat posed by the antibiotic challenge.
Most of the A. baumannii outbreak strains analyzed herein showed an impressive ability to survive on dry surfaces for long times, consistent with the overall capacity of this species to resist to drought [18, 23, 38]. Strain-dependent variation was found also for this character, since strains assigned to ST1 and ST78, as well as one strain assigned to ST25, showed extremely high resistance to desiccation; strains assigned to ST20, which belong to the same clonal complex as ST1, all ST2 strains, and two of three strains assigned to ST25 were moderately resistant to desiccation; in contrast, six other strains, including ATCC 19606T, were less resistant to drought. It has recently been reported that the survival times on dry surfaces of A. baumannii biofilm-forming strains are longer than those of non-biofilm-forming ones [38]. However, the collection analysed in this study did not reveal a strong correlation between biofilm formation and survival to desiccation, except for strains assigned to ST78 and two out of three strains assigned to ST25. A possible explanation for this discrepancy is that this correlation depends on the strains and/or culture conditions used.
The data obtained in this study also indicate that A. baumannii clonal lineages differ in the ability to adhere to alveolar epithelial cells. In particular, adherence to A549 cells was higher for strains assigned to ST2, ST25 and ST78, and a positive correlation exists between adherence to epithelial cells and biofilm formation in these strains. This is consistent with a previous report showing a positive association between biofilm production and adhesiveness to epithelial cells in MDR clinical isolates of A. baumannii[20]. Therefore, it can be speculated that common or partly overlapping cellular mechanisms might control biofilm growth on abiotic surfaces and adherence to epithelial cells. In partial support of this hypothesis, it was recently shown that the biofilm-associated protein Bap plays a role in both biofilm formation and adherence to human epithelial cells [21]. Our study also demonstrated that A. baumannii strains assigned to ST78 have a higher ability to invade A549 human bronchial epithelial cells than other genotypes. However, since the three strains assigned to ST78 belong to the same major PFGE type, it cannot be excluded that differences in the capacity to invade epithelial cells are strain-specific rather than lineage-specific.
No significant differences in the killing of G. mellonella larvae, measured as the LD50 at the 72 h post-infection time point, were found among strains assigned to the distinct genotypes, with the exception of strains ATCC 19606T (ST52) and 3868 (ST15), which showed significantly lower virulence. However, strain-specific differences were observed in the rate of killing, expressed as the ratio between LD50 values determined at 24 and 72 h. Killing was delayed for strains AYE (ST1) and 3990 (ST2) and, to a lesser extent, 3865, 4190 (both ST25) and 3911 (ST78) (LD50 ratio 5.3-12.2; Table 2 and Additional file 2: Figure S1). Overall, the LD50 values determined for our collection of isolates is in the same order of magnitude as that previously reported for prototypic A. baumannii strains of clinical origin (AYE, ACICU and ATCC 17978) [23].
The data just discussed indicate that, in spite of some interclonal variation, biofilm formation, adherence to epithelial cells and resistance to desiccation are virulence traits shared by A. baumannii clinical strains, irrespective of their belonging to the major or minor lineages. This is consistent with previous work [10, 18, 23, 38], including some recent publications showing that several genes associated with A. baumannii pathogenicity are more conserved in the A. calcoaceticus-baumannii (Acb) complex, and especially in A. baumannii, than in other Acinetobacter genomes, and that the most successful A. baumannii strains, although epidemiologically and genotypically different, share a common set of virulence-related traits in their core genome [25–27].
Conclusions
In conclusion, although epidemic A. baumannii clonal lineages show common virulence traits, they differ in the expression of virulence-associated phenotypes. In particular, A. baumannii strains belonging to IC-I showed higher resistance to desiccation, strains belonging to IC-II showed higher adherence to both biotic and abiotic surfaces, strains belonging to emerging genotypes ST25 and ST78 showed elevated resistance to desiccation, adherence to biotic and abiotic surfaces. Also, to the best of our knowledge, this is the first study to identify common A. baumannii virulence-associated phenotypes (resistance to desiccation, adherence to biotic and abiotic surfaces and alveolar epithelial cell invasion) in epidemic genotypes other than the three main ICs. Non-IC genotypes have increasingly been reported as responsible of hospital epidemics [5], and our findings are a first step towards the elucidation of virulence strategies that might have favoured the spread and persistence in the hospital environment of these emerging sequence types.
Abbreviations
- MLST:
-
Multilocus sequence typing
- ST:
-
Sequence type
- IC:
-
International clonal lineages
- MDR:
-
Multi-drug-resistant
- XDR:
-
Extensively-drug-resistant
- PFGE:
-
Pulsed-field gel electrophoresis
- LB:
-
Luria-Bertani broth
- M9:
-
Mueller Hinton broth
- OD:
-
Optical density
- PBS:
-
Phosphate-buffer saline solution
- CFU:
-
Colony-forming units
- LD50:
-
Lethal dose 50%
- r:
-
Pearson’s correlation coefficient.
References
Peleg AY, Seifert H, Paterson DL: Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008, 21: 538-582. 10.1128/CMR.00058-07.
Durante-Mangoni E, Zarrilli R: Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 2011, 6: 407-422. 10.2217/fmb.11.23.
Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S: The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One. 2010, 5: e10034-10.1371/journal.pone.0010034.
Di Popolo A, Giannouli M, Triassi M, Brisse S, Zarrilli R: Molecular epidemiology of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries using a multilocus sequence typing scheme. Clin Microbiol Infect. 2011, 17: 197-201. 10.1111/j.1469-0691.2010.03254.x.
Zarrilli R, Pournaras S, Giannouli M, Tsakris A: Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. 2013, 41: 11-19. 10.1016/j.ijantimicag.2012.09.008.
Karah N, Giske CG, Sundsfjord A, Samuelsen O: A diversity of OXA-carbapenemases and class 1 integrons among carbapenem-resistant Acinetobacter baumannii clinical isolates from Sweden belonging to different international clonal lineages. Microb Drug Resist. 2011, 17: 545-549. 10.1089/mdr.2011.0089.
Carretto E, Barbarini D, Dijkshoorn L, van der Reijden TJ, Brisse S, Passet V, Farina C, APSI Acinetobacter Study Group: Widespread carbapenem resistant Acinetobacter baumannii clones in Italian hospitals revealed by a multicenter study. Infect Genet Evol. 2011, 11: 1319-1326. 10.1016/j.meegid.2011.04.024.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL: Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012, 18: 268-281. 10.1111/j.1469-0691.2011.03570.x.
Tomaras AP, Dorsey CW, Edelmann RE, Actis L: Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003, 149: 3473-3484. 10.1099/mic.0.26541-0.
Rodríguez-Baño J, Martí S, Soto S, Fernández-Cuenca F, Cisneros JM, Pachón J, Pascual A, Martínez-Martínez L, McQueary C, Actis LA, Vila J, the Spanish Group for the Study of Nosocomial Infections (GEIH): Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin Microbiol Infect. 2008, 14: 276-278. 10.1111/j.1469-0691.2007.01916.x.
de Breij A, Dijkshoorn L, Lagendijk E, Van der Meer J, Koster A, Bloemberg G, Wolterbeek R, van den Broek P, Nibbering P: Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii?. PLoS One. 2010, 5: e10732-10.1371/journal.pone.0010732.
Choi C, Lee E, Lee Y, Park TI, Kim HJ, Hyun SH, Kim SA, Lee SK, Lee JC: Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol. 2005, 7: 1127-1138. 10.1111/j.1462-5822.2005.00538.x.
Loehfelm T, Luke N, Campagnari A: Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol. 2008, 190: 1036-1044. 10.1128/JB.01416-07.
Choi A, Slamti L, Avci F, Pier G, Maira-Litran T: The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetyl glucosamine PNAG that is critical for biofilm formation. J Bacteriol. 2009, 191: 5953-5963. 10.1128/JB.00647-09.
Gaddy JA, Tomaras AP, Actis LA: The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun. 2009, 77: 3150-3160. 10.1128/IAI.00096-09.
Kim S, Choi C, Moon D, Jin JS, Lee JH, Shin JH, Kim JM, Lee YC, Seol SY, Cho DT, Lee JC: Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol Lett. 2009, 301: 224-231. 10.1111/j.1574-6968.2009.01820.x.
Cabral MP, Soares NC, Aranda J, Parreira JR, Rumbo C, Poza M, Valle J, Calamia V, Lasa I, Bou G: Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J Proteome Res. 2011, 10: 3399-3417. 10.1021/pr101299j.
Jawad A, Seifert H, Snelling A, Heritage J, Hawkey P: Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol. 1998, 36: 1938-1941.
Aranda J, Bardina C, Beceiro A, Rumbo S, Cabral MP, Barbé J, Bou G: Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J Bacteriol. 2011, 193: 3740-3747. 10.1128/JB.00389-11.
Lee HW, Koh YM, Kim J, Lee JC, Lee YC, Seol SY, Cho DT, Kim J: Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect. 2008, 14: 49-54. 10.1111/j.1469-0691.2007.01842.x.
Brossard KA, Campagnari AA: The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun. 2012, 80: 228-233. 10.1128/IAI.05913-11.
Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM: Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun. 2010, 78: 1952-1962. 10.1128/IAI.00889-09.
Antunes LC, Imperi F, Carattoli A, Visca P: Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One. 2011, 6: e22674-10.1371/journal.pone.0022674.
de Breij A, Eveillard M, Dijkshoorn L, van den Broek PJ, Nibbering PH, Joly-Guillou ML: Differences in Acinetobacter baumannii strains and host innate immune response determine morbidity and mortality in experimental pneumonia. PLoS One. 2012, 7: e30673-10.1371/journal.pone.0030673.
Di Nocera PP, Rocco F, Giannouli M, Triassi M, Zarrilli R: Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol. 2011, 11: 224-10.1186/1471-2180-11-224.
Imperi F, Antunes LC, Blom J, Villa L, Iacono M, Visca P, Carattoli A: The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life. 2011, 63: 1068-1074. 10.1002/iub.531.
Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P: Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One. 2013, 8: e54287-10.1371/journal.pone.0054287.
Poirel L, Manuteau O, Agoli N, Cattoen C, Nordmann P: Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J Clin Microbiol. 2003, 41: 3542-3547. 10.1128/JCM.41.8.3542-3547.2003.
van Dessel H, Dijkshoorn L, van der Reijden T, Bakker N, Paauw A, van den Broek P, Verhoef J, Brisse S: Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res Microbiol. 2004, 155: 105-112. 10.1016/j.resmic.2003.10.003.
Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJ, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A: Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother. 2008, 52: 2616-2625. 10.1128/AAC.01643-07.
Giannouli M, Cuccurullo S, Crivaro V, Di Popolo A, Bernardo M, Tomasone F, Amato G, Brisse S, Triassi M, Utili R, Zarrilli R: Molecular epidemiology of multi-drug resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J Clin Microbiol. 2012, 48: 1223-1230.
Janssen P, Maquelin K, Coopman R, Tjernberg I, Bouvet P, Kersters K, Dijkshoorn L: Discrimination of Acinetobacter genomic species by AFLP fingerprinting. Int J Syst Bacteriol. 1997, 47: 1179-1187. 10.1099/00207713-47-4-1179.
Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M: New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21: 601-614. 10.1101/gad.1510307.
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: twentieth informational supplement. Document M100-S20. 2010, Wayne, PA: CLSI
Nucleo E, Steffanoni L, Fugazza G, Migliavacca R, Giacobone E, Navarra A, Pagani L, Landini P: Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a inmultidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol. 2009, 9: 270-10.1186/1471-2180-9-270.
Martì S, Rodríguez-Baño J, Catel-Ferreira M, Jouenne T, Vila J, Seifert H, Dé E: Biofilm formation at the solid–liquid and air-liquid interfaces by Acinetobacter species. BMC Res Notes. 2011, 4: 5-10.1186/1756-0500-4-5.
Merritt JH, Kadouri DE, O’Toole GA: Growing and analyzing static biofilms. Curr Protoc Microbiol. 2005, 00: 1B.1.1-1B.1.17.
Espinal P, Martì S, Vila J: Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect. 2012, 80: 56-60. 10.1016/j.jhin.2011.08.013.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2334/13/282/prepub
Acknowledgments
The Authors thank all colleagues who generously provided strains included in the study: Antonella Agodi, Matteo Bassetti, Susanna Cuccurullo, Ziad Daoud, Patrice Nordmann, Athanassios Tsakris, and Haluk Vahaboglu. The Authors also thank Elvira Crescenzi for kindly providing the A549 human alveolar epithelial cells.
LA was supported by a PhD fellowship from the Portuguese Fundação para a Ciência e a Tecnologia (FCT) (grant SFRH/BD/43420/2008).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: MG, LA, MT, PV, RZ. Performed the experiments: MG, LA, VM. Analyzed the data: MG, LA, PV, RZ. Wrote the paper: MG, LA, PV, RZ. All authors read and approved the final manuscript.
Electronic supplementary material
12879_2013_2487_MOESM1_ESM.xls
Additional file 1: Table S1: Resistance to desiccation of A. baumannii strains. Values are means ± standard deviations of three independent experiments done in duplicate. (XLS 36 KB)
12879_2013_2487_MOESM2_ESM.pdf
Additional file 2: Figure S1: Kaplan-Meier survival plots of G. mellonella larvae infected with the different A. baumannii strains. (PDF 112 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Giannouli, M., Antunes, L.C., Marchetti, V. et al. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis 13, 282 (2013). https://doi.org/10.1186/1471-2334-13-282
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2334-13-282