Abstract
Background
Biofilm production has been established as a virulence factor which allows Staphylococcus to adhere and persist in medical devices. The objective was to determine whether therapeutic failure in patients infected with Staphylococcus spp. is linked to biofilm production, the presence of the ica operon, and the bacterial insertion sequence element IS256.
Methods
Staphylococcus spp. isolates from patients with device-related infections were collected. Therapeutic failure with proper antimicrobial treatment was registered. Biofilm phenotype was determined by Congo red test agar and Christensen assay. Presence of the ica operon genes A-D and IS256 was detected by PCR. Differences were compared through x 2.
Results
100 isolates from staphylococcal infections episodes were included: 40 sepsis/bacteremia, 32 ependymitis, and 28 peritonitis. 73.77% of CoNS and 79.5% of S. aureus isolates harbored the icaD gene, 29% of all isolates IS256-A+ IS256-D genes, icaA and icaB genes were only found in CoNS (27.8% and 21.3% respectively). Therapeutic failure occurred in 95.4.% of patients with a positive IS256-A+ IS256-D S. epidermidis isolate, RR 5.49 (CI 95% 2.24-13.44 p ≤ 0.0001), and 85.76% in CoNS isolates, RR 2.57 (CI 95% 0.97-6.80, p = 0.05). Although none S. aureus was positive for IS256-A + IS256-D, therapeutic failure was observed in 35.8%.
Conclusions
The presence of icaA/D genes along with the sequence element IS256 was associated with therapeutic failure in most CoNS infections, even though its absence in S. aureus isolates does not ensure therapeutic success.
Similar content being viewed by others
Background
Staphylococci inhabit the skin and mucous membranes of animals and humans. Due to the ability to produce biofilm, they are also a common cause of device-associated infections [1–4]. Biofilm formation capacity is associated with antimicrobial resistance, and considered widely as a virulence factor. Invasive isolates are more prone to produce biofilm than carriage isolates of healthy individuals [5, 6]. Staphylococcus epidermidis was the first species to be described as a biofilm producer; however, the same ability is encountered in S. aureus and other coagulase-negative Staphylococcus species [7, 8].
The principal component of biofilm is a polysaccharide intercellular adhesin {PIA} [9, 10]. PIA is composed of a beta-1,6-N-acetylglucosamine polymer synthesized by an enzyme codified by the ica operon found on the bacterial chromosome, that includes a regulating element of four genes (A, B, C, and D), and a transposable element, IS256 [11]. It is known that the A gene codifies the N-acetylglucosamyl transferase enzyme responsible for synthesizing PIA. This enzyme is not very active in vitro, but co-expression of the D gene increases the activity. IcaB is the deacetylase responsible for the de-acetylation of mature PIA and the transmembrane protein IcaC seems to be involved in externalization and elongation of the growing polysaccharide [9, 12].
The expression of the icaADBC genes is controlled by a complex variety of conditions and factors; one of them is the excision or insertion of the bacterial sequence element IS256 at various locations on the operon. The molecular basis for the regulation process is still unclear; however, the insertion element is definitely responsible for up to 33% of the activated portion of the operon that allows its expression [1, 10, 13, 14]. Other regulator genes (RsbU, σB, Tca-R, agr, sarA) for PIA production have also been reported [15].
Biofilm production usually occurs in two steps, 1) the initial step of adhesion to the surface, facilitated by adhesive polysaccharides and/or a polysaccharide with multiple proteins (including autolysins), and 2) the accumulation of cells due to PIA production, synthesized after activation of the ica operon [9]. Initial findings assumed that the presence of icaADBC genes will contribute to the persistence of infection and therapeutic failure in presence of a medical device. Some authors have tried to establish the presence of the AD genes as a prognosis biomarker in device associated infections [8, 16]. Many other environmental conditions and the phenotype of the Staphylococcus isolate also participate in the regulation of biofilm production [17]. By understanding the different mechanisms of biofilm production it will be possible to support the development of therapeutic strategies [5].
The objective of this study was to describe the association between biofilm production, the presence of icaADBC genes, the bacterial insertion sequence IS256 and therapeutic failure, in isolates from patients with medical device-related infections.
Methods
Clinical staphylococci isolates were collected prospectively from hospitalized patients with a staphylococcal medical device-related infection from September 2002 to July 2003. Isolates were obtained from blood, cerebrospinal fluid and peritoneal fluid. The protocol was approved by the Institutional Review Board. Patient's data were obtained from clinical records. Infection was defined in accordance with internationally proposed criteria [18].
Empirical antimicrobial treatment was prescribed for all patients, and was modified according to susceptibility patterns.
Therapeutic failure was considered:
-
1)
For central venous catheter (CVC) related bacteremia/sepsis, persistence of positive blood cultures collected through the CVC after 72 hours of administering proper antimicrobial treatment.
-
2)
For peritonitis, persistence of positive peritoneal fluid culture collected through peritoneal dialysis catheter or a cellularity ≥ 100 cells per mm3 after 72 hours of administering proper antimicrobial treatment.
-
3)
For ependymitis, after removal of the infected shunt, persistence of positive cerebrospinal fluid (CSF) culture collected through external ventriculostomy, after 72 hours of systemic and/or intraventricular antimicrobial treatment.
Microbiology
Staphylococcus species were identified by API-Staph system (bioMérieux). Antimicrobial susceptibility was determined in accordance with CLSI recommendations, oxacillin resistant isolates were tested for mecA gene by PCR [19, 20].
The isolates were characterized phenotipically by culture on Congo red agar plates (CRA) as described by Freeman et al. [21], with modifications as described by Arciola et al [22]: agar plates were prepared with 0.8 g of CRA (Sigma) and 36 g of saccharose (Sigma) to 1 liter of brain heart infusion agar (Dibico, Mexico) and incubated 24 h at 37°C and subsequently overnight at room temperature. For S. aureus CRA were kept up to 72 h.
The quantitative assay by Christensen was used to test the ability to produce biofilm [23]. Briefly, 1:100 dilutions of overnight cultures in trypticase soy broth were used to inoculate wells in a microtiter polystyrene plate (Falcon, Becton Dickinson, Labware, NJ, USA). After incubation for 24 h at 37°C, the plates were gently washed two times with phosphate-buffered saline (PBS, 10 mM potassium phosphate, 0.15 M NaCl pH 7.0), and stained with 1% (w/v) crystal violet solution; the excess stain was washed off with demineralised water. The adherent cells were resuspended in acid-isopropanol (5% v/v 1 M HCl in isopropanol), and the absorbance (A) was measured at 492 nm in a microplate reader. A biofilm producer strain was defined as an optical density at 492 nm of ≥ 0.17. Clonal relatedness of strains was excluded using Pulsed Field Gel Electrophoresis (PFGE) [24].
The presence of the A, B, C, and D genes of the ica operon and the position of the bacterial sequence element IS256 were detected by PCR according to the protocol described by de Silva G. [6] with some modifications: DNA extraction was performed with the Promega® Wizard Genomic Kit (Madison, WI, USA). Primer sequences were taken from the GenBank sequence database of the National Center for Biotechnology Information [GenBank accession number U43366 for S. epidermidis, EF546621 for S. lugdunensis]. The sequence for the bacterial insertion element was taken from the publication by Ziebuhr [25].
The PCR cycling conditions used were 30 cycles of 1 min of denaturation at 94°C and 2.5 min of elongation at 72°C for all reactions, with annealing for 1 min at 60°C (icaA), 59°C (icaB), 45°C (icaC), 59°C (icaD), or 59°C (IS256). After amplification in a T-personal thermocycler (Whatman Biometra GmbH, Göttingen, Germany), 5 μl of the PCR mixture was used in analysis for horizontal electrophoresis in agarose gel of up to 2% tris-borate-EDTA. A 100 bp ladder DNA molecular weight marker was used. Samples with positive amplification for the A and D fragments were analyzed for the presence of the insertion element IS256, amplifying the sequence by PCR (Table 1). S. aureus ATCC 29247 and S. epidermidis ATCC 35984 (RP62A) were used as negative and positive controls.
Only one strain of each infection episode was included for analysis.
Statistical analysis
To compare differences between groups Mantel-Haenszel x 2 or Fisher exact test were employed, a value of p < 0.05 was considered to be significant. Statistical analysis was performed with SPSS for Windows software version 11.0.
Results
During the study period, one hundred isolates from patients with diagnosis of device-related infections were included. The episodes were divided in 40 CVC-related bacteremias, 32 ependymitis, and 28 peritonitis. S. epidermidis was isolated in 45 episodes, S. aureus in 39 and diverse coagulase negative Staphylococcus (CoNS)non-epidermidis in 16 (S. hominis, S. haemolyticus, S. lugdunensis, S. auricularis, S. warnerii and S. sciurii). 33% of the isolates were oxacillin-resistant, 31% were resistant to amikacin, 26% to norfloxacin and 0% to vancomycin. According to the PFGE results, all the strains included corresponded to a different genotype.
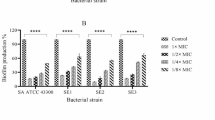
Biofilm production was confirmed in 22 strains by the Christensen assay, of these, 19 had a positive phenotype on the CRA plates. IcaADBC genes and the bacterial insertion sequence IS256 were detected in most of the strains, except for two strains of S. auricularis, one S. warnerii and one S. sciurii. 73.77% of CoNS and 79.5% of S. aureus isolates harbored the icaD gene; icaA, icaB and icaC genes were present in 27.8%, 21.3% and 9.8% of CoNS isolates (Table 2, figure 1). Only 4/39 S. aureus isolates were positive for icaA+IcaD genes but none for the bacterial insertion sequence. IS256-A+ IS256-D genes were detected in 21/45 S. epidermidis and 6/16 non-epidermidis CoNS. (figure 2, figure 3).
Electrophoresis of PCR products with primers for IS 256 and icaA. Lane 1, 100 bp molecular weight marker; lane 2, negative control; lane 3, 814-bp band from icaA positive control; lane 4 and lane 5, negative samples; lane 6, 814-bp band from S. aureus strain; lane 7, 1102-bp band (IS256) and 814-bp band (icaA) from S. epidermidis strain; lane 8, 1102-bp band from IS256 positive control; lane 9, lane 10 and lane 11, 1102-bp bands (IS256) and 814-bp bands (icaA) from S. epidermidis strains.
Electrophoresis of PCR products with primers for IS 256 and i caD. Lane 1, 100 bp molecular weight marker; lane 2, 371-bp band from icaD positive control; lane 3 and lane 4, 371-bp bands (icaD) from S. haemolyticus strains; lane 5, 1102-bp band (IS256) and 371-band (icaD) from S. haemolyticus strain; lane 6, 371-bp band (icaD) from S. haemolyticus strain; lane 7, 1102-bp band (IS256) and 371-band (icaD) from S. haemolyticus strain; lane 8, 371-band (icaD) from S. haemolyticus strain.
Therapeutic failure in episodes caused by an IS256-A+ IS256-D positive strain was 95.4% for S. epidermidis, RR 5.49 (CI 95% 2.24-13.44, p ≤ 0.0001), and 85.76% in CoNS isolates, RR 2.57 (CI 95% 0.97-6.80, p = 0.05). Although none S. aureus was positive for IS256-A + IS256-D, therapeutic failure was observed in 35.8%. (Table 3).
Discussion
Biofilm production has been clearly linked to infections in the presence of foreign bodies for various decades [26], it has been demonstrated in vivo tests with laboratory animals and in vitro, that biofilm hampered thorough penetration of the antimicrobial and the concentrations require to eradicate biofilm-producing strains are higher than those required to eradicate strains that did not produce biofilm [27, 28].
In this study, as reported by other authors, it was found that the genes of the Ica operon frequently appeared in strains of Staphylococcus epidermidis [4, 6, 29]. The number of S. aureus isolates with a positive biofilm phenotype or producing biofilm was low (4/39), and in none of them the bacterial insertion element IS256 was present. Experimental studies have also shown that biofilm formation is possible in icaADBC operon S. aureus mutants [30], but we did not find S. aureus negative for the operon that was a biofilm producer. S. epidermidis strains with the genetic determinant for biofilm formation were clearly associated to a high risk of therapeutic failure, which was not corroborated with non-epidermidis CoNS and S. aureus, perhaps due to a smaller number of collected isolates.
As previously stated, adhesion to the plastic device could be a factor linked to weaker expression of symptoms of infection, due to the existence of a smaller number of circulating bacteria, most of them remaining inside the catheter. This mild presentation could support the non-standard recommendation to preserve the medical device in patients with bacteremia. This is necessary in some cases due to the critical condition of the patient. However, the risk of failure to proper antimicrobial treatment is so high, that this conduct should be avoided and the CVC removed as soon as possible, even if a negative icaA +IcaD isolate is detected.
The present work has several limitations. Infections included are difficult to compare to elicit general recommendations, in particular for the ependymitis and peritonitis episodes. The ica operon genes have been widely described in Staphylococcus epidermidis and Staphylococcus aureus, several authors have found similarity in other CoNs species [6, 7] but results cannot be extended to all pathogenic species. Recently biofilm formation has been described in detail in S. haemolyticus isolates [31]. Phenotypic variation as well as clonal lineage must be included in future studies to develop alternative therapeutic strategies.
Conclusions
The presence of icaA/D genes along with the sequence element IS256 was associated with therapeutic failure in most CoNS infections, even though its absence in S. aureus isolates does not ensure therapeutic success.
References
Dietrich M, Holger R, Dobinsky S, Riedewald J, Nedelmann M, Knobloch JM: Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect Immun. 2000, 68: 3799-3807. 10.1128/IAI.68.7.3799-3807.2000.
Otto M: Virulence factors of the coagulase-negative staphylococci. Front Biosci. 2004, 9: 841-863. 10.2741/1295.
Rohde H, Mack D, Christner M, Burdelski C, Franke G, Knobloch JM: Pathogenesis of staphylococcal device-related infections: from basic science to new diagnostic, therapeutic and prophylactic approaches. Rev Med Microbiol. 2006, 17: 45-54.
Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J: Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J Infect Dis. 2003, 188: 730-737. 10.1086/377452.
Peetermans WE, Merckx R, Rijnders A, Van Eldere J: Reliability of the ica, aap and AtlE genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infection. Clin Microbiol Infect. 2003, 9: 114-119. 10.1046/j.1469-0691.2003.00544.x.
de Silva G, Kantzanou M, Justice A, Massey R, Wilkinson A, Day N, Peacock S: The ica operon and biofilm production in coagulase-negative Staphylococci associated with carriage and disease in a neonatal intensive care unit. J Clin Microb. 2002, 40: 2382-2388. 10.1128/JCM.40.02.382-388.2002.
Cramton S, Gerke C, Schnell N, Nichols W, Götz F: The intercellular adhesin (Ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999, 67: 5427-5433.
Götz F: Staphylococcus and biofilms. Mol Microbiol. 2002, 43: 1367-1378. 10.1046/j.1365-2958.2002.02827.x.
Lappin-Scott H, Bass C: Biofilm formation: attachment, growth, and detachment of microbes from surfaces. Am J Infect Control. 2001, 29: 250-251. 10.1067/mic.2001.115674.
Ziebuhr W, Heilmann F, Götz F, Meyer K: Detection of the intercellular adhesin gene cluster (Ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997, 65: 890-896.
Kozitskaya S, Seung-Hak Ch, Dietrich K, Marre R, Naber K, Ziebuhr W: The bacterial insertion sequence element IS 256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates:association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004, 72: 1210-1215. 10.1128/IAI.72.2.1210-1215.2004.
Gerke C, Kraft A, Süßmuth R, Schweitzer O, Götz F: Characterization of the n-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998, 273: 18586-18593. 10.1074/jbc.273.29.18586.
Cafiso V, Bertuccio T, Santagati M, Campanile G, Amicosante G: Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect. 2004, 10: 1081-1088. 10.1111/j.1469-0691.2004.01024.x.
Cramton S, Ulrich M, Götz F, Döring G: Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001, 69: 4079-4085. 10.1128/IAI.69.6.4079-4085.2001.
Knobloch JK, Jäger S, Horstkotte M, Rohde H, Mack D: Rsb-U dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene ica R. Infect Immun. 2004, 72: 3838-3848. 10.1128/IAI.72.7.3838-3848.2004.
Arciola CR, Collimati S, Donati E, Montanaro L: A rapid PCR method for detection of slime-producing strains of Staphylococcus epidermidis and S. aureus in periprothesis infections. Diagn Mol Pathol. 2001, 10: 130-137. 10.1097/00019606-200106000-00010.
Dobinsky S, Kile K, Rohde H, Bartscht K, Knobloch JK, Horstkotte MA, Mack D: Glucose-related dissociation between Ica ADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J Bacteriol. 2003, 185: 2879-2886. 10.1128/JB.185.9.2879-2886.2003.
Horan TC, Andrus M, Dudeck MA: CDC/NHSH surveillance definition of health-care associated infection and criteria for specific types of infection in the acute care setting. Am J Infect Control. 2008, 36: 309-332. 10.1016/j.ajic.2008.03.002.
CLSI (Clinical and Laboratory Standards Institute): Performance Standards for Antimicrobial Susceptibility testing. Fifteenth Informational Supplement. CLSI document M100-S15. Pennsylvania. 19087, -1898. 2005. USA
Kohner F, Kolbert C, Persing D, Cockerill F: Comparison of susceptibility testing methods with mec A gene analysis for determining oxacillin (methicillin) resistance in clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus spp. J Clin Microbiol. 1999, 37: 2952-2961.
Freeman J, Falkiner F, Keane C: New method for detecting slime production by coagulase-negative staphylococci. J Clin Pathol. 1989, 42: 872-874. 10.1136/jcp.42.8.872.
Arciola CR, Baldassarri L, Montanaro L: Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microb. 2001, 39: 2151-2156. 10.1128/JCM.39.6.2151-2156.2001.
Christensen GD, Baddour LM, Simpson WA: Phenotypic variation of Staphylococcus epidermidis slime production in vitro and in vivo. Infect Immun. 1987, 55: 2870-2877.
Amato D, Ventura MJ, Miranda G, Leaños B, Alcántara G, Hurtado ML, Paniagua R: Staphylococcal peritonitis in continuous ambulatory peritoneal dialysis: colonization with identical strains at exit site, nose and hands. Am J Kidney Dis. 2001, 37: 43-48. 10.1053/ajkd.2001.20576.
Ziebuhr W, Krimmer V, Rachid S, Löâner I, Götz F, Hacker J: A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: Evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS 256. Mol Microbiol. 1999, 32: 345-356. 10.1046/j.1365-2958.1999.01353.x.
Rupp ME, Archer GL: Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994, 19: 231-245.
Curtin J, Cormican M, Fleming G, Keelehan J, Colleran E: Linezolid compared with eperezolid, vancomycin, and gentamicin in an in vitro model of antimicrobial lock therapy for Staphylococcus epidermidis central venous catheter-related biofilm infections. Antimicrob Agents Chemother. 2003, 47: 3145-3148. 10.1128/AAC.47.10.3145-3148.2003.
Gagnon RF, Richards GK, Kostiner GB: Time-kill efficacy of antibiotics in combination with rifampin against Staphylococcus epidermidis biofilms. Adv Perit Dial. 1994, 10: 189-192.
Vandecasteele SJ, Peetermans WE, Merckx R, Rijnders BJA, Van Eldere J: Reliability of the ica, aap, and atl E genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infections. Clin Microbiol Infect. 2003, 9: 114-119. 10.1046/j.1469-0691.2003.00544.x.
Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, Lasa I: Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the ar IRS two-component system. J Bacteriol. 2005, 187: 5318-29. 10.1128/JB.187.15.5318-5329.2005.
Fredheim EGA, Klingenberg C, Rohde H, Frankenberger S, Gaustad P, Trond F, Ericson Sollid J: Biofilm formation by Staphylococcus haemolyticus. J Clin Microbiol. 2009, 47: 1172-1180. 10.1128/JCM.01891-08.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2334/10/68/prepub
Acknowledgements
This paper was supported by FOFOI grant number 2003/012 from the Instituto Mexicano del Seguro Social.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BDH participated in data collection, analysis, interpretation, and literature research; FSS performed a critical review; BLM participated in data collection and laboratory assays; LPB helped to review the manuscript; and GMN conceived the study and participated in design, analysis, supervision of laboratory assays and corrected the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Diemond-Hernández, B., Solórzano-Santos, F., Leaños-Miranda, B. et al. Production of icaADBC-encoded polysaccharide intercellular adhesin and therapeutic failure in pediatric patients with staphylococcal device-related infections. BMC Infect Dis 10, 68 (2010). https://doi.org/10.1186/1471-2334-10-68
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2334-10-68