Abstract
Background
The role of mutations in the serine protease inhibitor Kazal type 1 (SPINK1) gene in chronic pancreatitis is still a matter of debate. Active SPINK1 is thought to antagonize activated trypsin. Cases of SPINK1 mutations, especially N34S, have been reported in a subset of patients with idiopathic chronic pancreatitis. However, the inheritance pattern is still unknown. Some cases with N34S heterozygosity have been reported with and without evidence for CP indicating neither an autosomal recessive nor dominant trait. Therefore SPINK1 mutations have been postulated to act as a disease modifier requiring additional mutations in a more complex genetic model. Familial hypocalciuric hypercalcemia (FHH) caused by heterozygous inactivating mutations in the calcium sensing receptor (CASR) gene is considered a benign disorder with elevated plasma calcium levels. Although hypercalcemia represents a risk factor for pancreatitis, increased rates of pancreatitis in patients with FHH have not been reported thus far.
Methods
We studied a family with a FHH-related hypercalcemia and chronic pancreatitis. DNA samples were analysed for mutations within the cationic trypsinogen (N29I, R122H) and SPINK1 (N34S) gene using melting curve analysis. Mutations within CASR gene were identified by DNA sequencing.
Results
A N34S SPINK1 mutation was found in all screened family members. However, only two family members developed chronic pancreatitis. These patients also had FHH caused by a novel, sporadic mutation in the CASR gene (518T>C) leading to an amino acid exchange (leucine->proline) in the extracellular domain of the CASR protein.
Conclusion
Mutations in the calcium sensing receptor gene might represent a novel as yet unidentified predisposing factor which may lead to an increased susceptibility for chronic pancreatitis. Moreover, this family analysis supports the hypothesis that SPINK1 mutations act as disease modifier and suggests an even more complex genetic model in SPINK1 related chronic pancreatitis.
Similar content being viewed by others
Background
Chronic pancreatitis (CP) remains one of the big challenges among the GI-tract disorders. Since 1996 the understanding of genetic risk factors for CP extended with the discovery of different gene defects causing hereditary pancreatitis (HP). A French and two American groups were the first to identify chromosome 7q35 as a putative locus for HP [1–3]. The disease gene was mapped and a heterozygous point mutation A>T in exon 3 (R122H) of the protease serine 1 (PRSS1) was detected in a majority (~70%) of cases with cationic trypsinogen mutations [4]. Subsequently the PRSS1 mutations N29I (25%), A16V (<3%), K23R, D22G, R122C and N29T (several kindreds) were discovered [4–9]. The second gene that was characterized to be clearly associated with idiopathic chronic pancreatitis (ICP) was the cystic fibrosis transmembrane conductance regulator (CFTR) gene [10, 11].
The third gene that was postulated to be associated with ICP is the serine protease inhibitor Kazal type 1 (SPINK1). Initially an autosomal recessive inheritance pattern was suspected for the N34S SPINK1 mutation resulting in CP in homozygous patients while another heterozygous subgroup seems to suffer from disease due to a combination of genetic defects of SPINK1 and yet unidentified genes [12]. Pfützer et al. showed that neither an autosomal dominant disorder with a low penetrance pattern (observed frequency of N34S mutation in the population [0.77%] versus ICP prevalence [0.0066%]) nor an autosomal recessive trait can be used as a model for disease inheritance [13]. The prevalence for the N34S mutation in tested healthy controls stemming from these countries ranges in between 1.6 % (USA) [13], 1.58% (France) [14] and 0.36% (Germany) [12]. Thus the role of SPINK1 mutations in ICP which have a much lower disease prevalence (<0.01%) remains to be further defined and the hypothesis of a modifying but not causative factor for SPINK1 mutations was proposed [13, 15].
One of the toxic metabolic risk factors for CP is hypercalcemia. The correlation was first observed in patients with hyperparathyroidism while trypsin activation, secretion stimulus and stabilization are suspected mechanisms [16].
Therefore, mutations in genes involved in calcium homeostasis might be associated with an increased risk for CP. We describe a family with members suffering from both CP and familial hypocalciuric hypercalcemia (FHH) to investigate the association of calcium sensing receptor gene (CASR) and SPINK1 mutations with CP.
Case Presentation
We report on a 35-year-old caucasian male (II:4, Fig. 3) who was admitted to the hospital with an acute exacerbation of chronic pancreatitis in 2002. In 1993, he had been hospitalized for a first episode of acute pancreatitis that was followed by 40 recurrent exacerbations necessitating 27 periods of prolonged hospitalization. While the aetiology of pancreatitis remained undefined between 1993 and 1996, prolonged hypercalcemia in combination with an elevated level of parathormone (PTH) were suspected as the dominating causes in 1996.
A familial hypocalciuric hypercalcemia (FHH) was diagnosed biochemically by measuring a persisting high serum calcium level and a low urinary calcium excretion. The ratio of the creatinine and calcium clearance was below 0.01. The fact that his older brother (II:2, Fig. 3) had the same complex of symptoms (recurrent pancreatitis, elevated serum calcium level and low urinary calcium excretion) supported the hypothesis of a genetic cause and the diagnosis of FHH. In the younger brother, the disease onset occurred at the age of 27 years. In the patient, treatment with bisphosphonates and octreotide (200 μg TID) was initiated in 1995 and continued through 2001 with little to none clinical improvement.
In 1996, the two lower parathyroid glands were removed surgically because of persistent elevated PTH serum levels and calcitonin substitution was started (100 μg BID). This therapy also failed to suppress the recurrent episodes of pancreatitis.
In December 1999, the patient was the first time admitted to our clinic with another exacerbation of his recurrent pancreatitis. After endoscopic investigation (ERCP and endoscopic ultrasound), indicating neither signs of chronic pancreatitis nor any evidence for the pathology of his disease, we suspected that additive genetic alterations might cause the disease. An analysis for PRSS1 mutations was performed which revealed neither an R122H or N29I mutation.
In 2001, during another episode of acute pancreatitis, clinical evaluation revealed marked signs of chronic pancreatitis according to the Marseilles-Rome criteria. The endoscopic ultrasound was consistent with chronic calcifying pancreatitis with intraductal stones within the head of the pancreas. Persistent pain and impaired exocrine function with an abnormal fecal elastase accompanied the disease progress. A heterozygous N34S mutation was detected by DNA sequence analysis of the SPINK1 gene.
Blood samples and DNA studies
We collected blood samples for clinical chemistry and 24 h urine of all family members who accepted testing. Blood samples for mutation analysis were taken after counselling according to the guidelines of the Consensus Committees of the European Registry of Hereditary Pancreatic Diseases [17].
Informed consent was given by the patient's mother (62), the elderly brother (43), while a half-sister of the index patient (24) denied genetic testing. The daughter (8) of the index patient was tested giving her and her parents informed consent. We extracted genomic DNA from whole blood according to established protocols.
Coded DNA samples of the patients were analyzed for PRSS1 mutations (N29I, R122H) and the SPINK1 variant N34S by melting point curve analysis. DNA sequencing of the CASR gene were performed using PCR products of the exons.
Polymerase Chain Reaction and melting point curve analysis
Primers flanking the designated coding regions of the CASR, PRSS1 and SPINK1 genes were designed according to the nucleotide sequences published (CASR: GenBank #U20759 [18], PRSS1: GenBank #U66061 [19] and SPINK1: GenBank #AF286028 [12]).
Melting curve analyses for the N29I, R122H PRSS1 mutations were performed using specific pairs of fluorescence resonance energy transfer (FRET) probes (N29I: LC-Red attached to 5'-CTGTCCCCTACCAGGTGTCC and FL attached to 5'-GGGCTACAACTGTGAG GAGAA-FL ; R122H: LC-Red 5'-TCTCTGCCCACCGCCCCTCCAGCC and 5'-CAACGCCCACGTGTCCACCA-FL) synthesized by TIB MOLBIOL (Berlin, Germany) and the LightCycler (Roche Diagnostics, Germany). Primers for the SPINK1 melting curve analysis have been described previously [12].
DNA sequence analysis
Genomic DNA was prepared from whole blood using standardized protocols. The patient was screened for published mutations in the 6 translated exons (exon 2–7) of the CASR gene. PCR products of all 6 exons were gel-purified and sequenced on an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster, CA) using the listed primers (Table 1). DNA from relatives of the patient was screened for mutations in exon 4 identified in the index patient's DNA c.518T>C.
Results
Family history
The family history revealed no other family members with pancreatitis, with the exception of the two previously mentioned brothers (36 and 39 years), who both experienced a first episode at the age of 27. The father had died of cardiac arrest in 1971 and never had any documented illness or symptoms indicating pancreatitis. The daughter of the index patient is asymptomatic but still at risk to develop pancreatitis. The brother who had a documented history of recurrent pancreatitis and FHH could not be located. Looking at the family tree and his medical history of recurrent episodes of pancreatitis, a N34S mutation can only be suspected.
Clinical chemistry
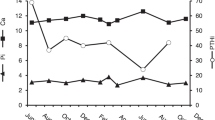
Serum plasma levels of Ca2+, creatinine and urinary calcium excretion were normal in the mother and asymptomatic brother. The ratio of creatinine clearance and calcium clearance was within normal limits and thus not indicating FHH. The index patient (12/2001) had a normal PTH serum concentration of 19.0 ng/l (normal 12–72 ng/l) and an elevated plasma calcium between 2.75 and 3.25 mmol/l. Urinary calcium excretion was 180 mg/24 h, creatinine clearance was 164 ml/min and the ratio of creatinine and calcium clearance was 0.014.
Polymerase Chain Reaction, melting point curve analysis and DNA sequencing
The data of the melting point curve analysis revealed a heterozygous N34S mutations for the index patient, the unaffected brother, mother and daughter (Figure 1).
Melting curve analysis ( SPINK1 N34S) of the index patient and family members To assess the SPINK1 genotype, representative gene fragments of the SPINK1 gene were amplified by PCR. The melting point (Tm) of each amplified double-stranded DNA fragment was determined using the LightCycler thermocycling device and designated probes. Curves with a melting point at 58,53°C represent the SPINK1 (N34S) control, index patient (II:4), daughter of the index patient (III:1), healthy brother (II:3) and healthy mother of the index patient (I:2). The melting point curve with the higher melting point at 63,17°C, due to a more stable complex (donor probe was complimentary to the wild type) represents the SPINK1 wild type.
The sequence similarity search of the CASR gene showed a point mutation in exon 4 at position c.518T>C leading to a heterozygous missense mutation L173P in the extracellular domain of the calcium sensing receptor (CaR) protein in the index patient and his daughter (Figure 1 and 2).
Calcium-sensing receptor gene mutation L173P The DNA sequence electropherogram of the index patient revealed a heterozygous mutation of nucleotide 518T>C. At the bottom, the amino acid sequence corresponding to this DNA region is shown. The identified mutation at base 518 results in a T to C transition (518T>C) which leads to a substitution of leucine 173 by proline in exon 4 (L173P). This corresponds to the extracellular domain of the CaR protein.
Family tree and clinical parameters of the family with SPINK1 (N34S) and CASR (518T>C) gene mutations Two family members were diagnosed with chronic pancreatitis and FHH (II:2 and II:4). Mutational analysis of the index patient (36 years) and three other individuals, respectively, mother (62), unaffected brother (43) and daughter (8) (I:2, II:3, II:4 and III:1) showed a heterozygous SPINK1 mutation (N34S). In addition, the index patient and his daughter (II:4, III:1) had a point mutation in exon 4 at position 518 (518T>C) of the CASR gene. As for the individual II:3 in generation I no family member complained about abdominal pain (I:1 deceased in 1971). DNA from the symptomatic brother (39 years) was unavailable for genetic testing, but CP and FHH was diagnosed clinically.
The other family members who had no clinical signs for FHH (normal plasma and urinary Ca2+ excretion; Tab.2) did not have a CASR gene mutation in exon 4. The results of the mutational analysis are summarized in the family tree (Fig. 3).
Discussion
Hypercalcemia is considered a risk factor of acute pancreatitis. Elevated serum calcium levels can lead to premature activation of trypsinogen inducing autodigestion of the pancreatic parenchyma [20]. Clinical examples for this association are patients with endocrine disorders such as primary hyperparathyroidism (pHPT) characterized by hypercalcemia with an elevated risk for pancreatitis. However, this interrelation is questioned by some investigators and for the specific case of pancreatitis in patients with primary hyperparathyroidism the prevalence rates range from 1 to 19 % [21]. In our case, FHH-associated hypercalcemia was thought to be required for recurrent episodes of acute pancreatitis but even parathyroidectomy and treatment with bisphosphonates did not bring the expected result. FHH is a benign disorder with a slight elevation of plasma Ca2+. It is inherited in an autosomal dominant order and shows almost 100% penetrance. In 1992, the disease gene (CASR) was mapped to the long arm of chromosome 3 [22] and today more than 70 mutations have been described [23]. The CaR plays a key role in calcium homeostasis and is widely expressed in tissues including kidney, parathyroid glands and bone cells which are involved in calcium metabolism [24]. Recently the CaR was characterized in human pancreatic acinar and duct cells [25]. However, the functional significance of CaR expression in pancreatic acinar and duct cells in the pathogenesis of pancreatitis remains to be defined.
While a heterozygous inactivating CASR mutation leads to FHH, homozygous mutations result in severe neonatal pHPT. In contrast to pHPT, pancreatitis is an unusual complication of FHH.
Interestingly, there are a few reports of as yet unexplained cases with FHH and recurrent episodes of pancreatitis [26, 27]. Pearce et al. (1996) suspected in their collective of three FHH kindreds [26] that patients with FHH and CP may represent a distinct subgroup. Subsequently they identified three different CASR mutations, suggesting that they may represent distinct FHH variants with pancreatitis. From our point of view, it cannot be ultimately ruled out, that other genetic factors may have contributed to the development of pancreatitis in the presented cases, as the researchers did not exclude additional mutations in other genes including the SPINK1, PRSS1 and CFTR genes. However, our observation of an individual with a combination of heterozygous SPINK1 (N34S) and CASR gene (L173P) mutations, who developed chronic pancreatitis, may indicate the specific pathogenetic background of CP within this family.
There is still an ongoing controversy about the relevance of SPINK1 mutations in CP although different groups postulated an association of N34S mutations in patients with ICP [12, 13]. Pfützer et al. reported that approximately 40 % of their collective of patients with ICP (n = 57) had a N34S mutation (12 % were homozygous) [13] while Witt and coworkers detected N34S mutations in 23 % of their cases (n = 96) in a similar study [12].
Even though these observations support the importance of N34S mutations in ICP, it still remains to be determined whether N34S mutations are causative. Threadgold et al. postulated that SPINK1 mutations "rather act as a disease modifying factor but not causing the disease" [15]. Today their role is further substantiated as independent groups showed a strong association of SPINK1 mutations (N34S) in patients with tropical pancreatitis [28–30].
Conclusion
Family and clinical history may indicate that the combination of the N34S SPINK1 and CASR gene mutation induces CP in this kindred. Our observation supports the hypothesis that SPINK1 mutations rather modify the disease phenotype than actually cause CP.
This is therefore the first report to show an association between CASR mutations and SPINK1 related CP. Larger association studies or animal models are required to elucidate whether the CASR gene plays a more important role in SPINK1 related CP. The current investigation may thus represent another part of the mosaique in the complex model of genetic causes for CP.
Note
Table 1: Sequences of oligonucleotide primers used for the PCR of the CASR gene The patient was screened for published mutations in exons 2–6 (exon 1 is untranslated) of the CASR gene. PCR products were gel-purified and sequenced on an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster, CA) using the listed primers for PCR and sequencing.
Abbreviations
- CaR:
-
calcium sensing receptor
- CASR :
-
calcium receptor gene
- CP:
-
chronic pancreatitis
- FHH:
-
familial hypocalciuric hypercalcemia
- HP:
-
hereditary pancreatitis
- ICP:
-
idiopathic chronic pancreatitis
- PRSS1 :
-
cationic trypsinogen gene
- SPINK1 :
-
serine protease inhibitor Kazal type 1 gene.
References
Whitcomb DC, Preston RA, Aston CE, Sossenheimer MJ, Barua PS, Zhang Y, Wong-Chong A, White GJ, Wood PG, Gates LK, Ulrich C, Martin SP, Post JC, Ehrlich GD: A gene for hereditary pancreatitis maps to chromosome 7q35. Gastroenterology. 1996, 110: 1975-80.
Le Bodic L, Bignon JD, Raguénès O, Mercier B, Georgelin T, Schnee M, Soulard F, Gagne K, Bonneville F, Muller JY, Bachner L, Férec C: The hereditary pancreatitis gene maps to long arm of chromosome 7. Hum Mol Genet. 1996, 5: 549-54. 10.1093/hmg/5.4.549.
Pandya A, Blanton SH, Landa B, Javaheri R, Melvin E, Nance WE, Markello T: Linkage studies in a large kindred with hereditary pancreatitis confirms mapping of the gene to a 16-cM region on 7q. Genomics. 1996, 38: 227-30. 10.1006/geno.1996.0620.
Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK, Amann ST, Toskes PP, Liddle R, McGrath K, Uomo G, Post JC, Ehrlich GD: Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996, 14: 141-5.
Gorry MC, Gabbaizedeh D, Furey W, Gates LK, Preston RA, Aston CE, Zhang Y, Ulrich C, Ehrlich GD, Whitcomb DC: Mutations in the cationic trypsinogen gene are associated with recurrent acute and chronic pancreatitis. Gastroenterology. 1997, 113: 1063-1068.
Férec C, Raguénès O, Salomon R, Roche C, Bernard JP, Guillot M, Quéré I, Faure C, Mercier B, Audrézet MP, Guillausseau PJ, Dupont C, Munnich A, Bignon JD, Le Bodic L: Mutations in the cationic trypsinogen gene and evidence for genetic heterogeneity in hereditary pancreatitis. J Med Genet. 1999, 36: 228-232.
Witt H, Luck W, Becker M: A signal peptide cleavage site mutation in the cationic trypsinogen gene is strongly associated with chronic pancreatitis. Gastroenterology. 1999, 117: 7-10.
Teich N, Ockenga J, Hoffmeister A, Manns M, Mössner J, Keim V: Chronic pancreatitis associated with an activation peptide mutation that facilitates trypsin activation. Gastroenterology. 2000, 119: 461-465.
Pfützer R, Myers E, Applebaum-Shapiro S, Finch R, Ellis I, Neoptolemos J, Kant JA, Whitcomb DC: Novel cationic trypsinogen (PRSS1) N29T and R122C mutations cause autosomal dominant hereditary pancreatitis. Gut. 2002, 50: 271-2. 10.1136/gut.50.2.271.
Sharer N, Schwarz M, Malone G: Mutations of the cystic fibrosis gene in patients with chronic pancreatitis. N Engl J Med. 1998, 339: 645-652. 10.1056/NEJM199809033391001.
Cystic Fibrosis Genetic Analysis Consortium. [http://www.genet.sickkids.on.ca/cftr]
Witt H, Luck W, Hennies HC, Classen M, Kage A, Lass U, Landt O, Becker M: Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000, 25: 213-216. 10.1038/76088.
Pfützer RH, Barmada MM, Brunskill AP, Finch R, Hart PS, Neoptolemos J, Furey WF, Whitcomb DC: SPINK1/PSTI polymorphisms act as disease modifiers in familial and idiopathic chronic pancreatitis. Gastroenterology. 2000, 119: 615-23.
Chen JM, Mercier B, Audrézet MP, Férec C: Mutational analysis of the human pancreatic secretory trypsin inhibitor (PSTI) gene in hereditary and poradic chronic pancreatitis. J Med Genet. 2000, 37: 67-9. 10.1136/jmg.37.1.67.
Threadgold J, Greenhalf W, Ellis I, Howes N, Lerch MM, Simon P, Jansen J, Charnley R, Laugier R, Frulloni L, Olah A, Delhaye M, Ihse I, Schaffalitzky de Muckadell OB, Andren-Sandberg A, Imrie CW, Martinek J, Gress TM, Mountford R, Whitcomb D, Neoptolemos JP: The N34S mutation of SPINK1 (PSTI) is associated with a familial pattern of idiopathic chronic pancreatitis but does not cause the disease. Gut. 2002, 50: 675-81. 10.1136/gut.50.5.675.
Etemad B, Whitcomb DC: Chronic pancreatitis: diagnosis, classification, and new genetic developments. Gastroenterology. 2001, 120: 682-707.
Ellis I, Lerch MM, Whitcomb DC: Consensus Committees of the European Registry of Hereditary Pancreatic Diseases, Midwest Multi-Center Pancreatic Study Group, International Association of Pancreatology. Genetic testing for hereditary pancreatitis: guidelines for indications, counselling, consent and privacy issues. Pancreatology. 2001, 1: 405-15. 10.1159/000055840.
Garrett JE, Capuano IV, Hammerland LG, Hung BC, Brown EM, Hebert SC, Nemeth EF, Fuller F: Molecular cloning and functional expression of human parathyroid calcium receptor cDNAs. J Biol Chem. 1995, 270: 12919-12925. 10.1074/jbc.270.21.12919.
Rowen L, Koop BF, Hood L: The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science. 1996, 272: 1755-62.
Krüger B, Albrecht E, Lerch M: The role of intracellular calcium signaling in premature protease activation and the onset of pancreatitis. Am J Pathol. 2000, 157: 43-50.
Koppelberg T, Bartsch D, Printz H, Hasse C, Rothmund M: Pancreatitis in primary hyperparathyroidism (pHPT) is a complication of advanced pHPT. Dtsch Med Wochenschr. 1994, 119: 719-24.
Chou YH, Brown EM, Levi T, Crowe G, Atkinson AB, Arnqvist HJ, Toss G, Fuleihan GE, Seidman JG, Seidman CE: The gene responsible for familial hypocalciuric hypercalcemia maps to chromosome 3q in four unrelated families. Nat Genet. 1992, 1: 295-300.
Calcium Sensing Receptor Database. [http://www.casrdb.mcgill.ca/]
Brown EM, MacLeod RJ: Extracellular calcium sensing and extracellular calcium signaling. Physiol Rev. 2001, 81: 239-297.
Racz GZ, Kittel A, Riccardi D, Case RM, Elliott AC, Varga G: Extracellular calcium sensing receptor in human pancreatic cells. Gut. 2002, 51: 705-11. 10.1136/gut.51.5.705.
Pearce SH, Wooding C, Davies M, Tollefsen SE, Whyte MP, Thakker RV: Calcium-sensing receptor mutations in familial hypocalciuric hypercalcemia with recurrent pancreatitis. Clin Endocrinol (Oxf). 1996, 45: 675-80. 10.1046/j.1365-2265.1996.750891.x.
Stuckey BG, Gutteridge DH, Kent GN, Reed WD: Familial hypocalciuric hypercalcemia and pancreatitis: no causal link proven. Aust N Z J Med. 1990, 20: 718-9.
Chandak GR, Idris MM, Reddy DN, Bhaskar S, Sriram PV, Singh L: Mutations in the pancreatic secretory trypsin inhibitor gene (PSTI/SPINK1) rather than the cationic trypsinogen gene (PRSS1) are significantly associated with tropical calcific pancreatitis. J Med Genet. 2002, 39: 347-51. 10.1136/jmg.39.5.347.
Bhatia E, Choudhuri G, Sikora SS, Landt O, Kage A, Becker M, Witt H: Tropical calcific pancreatitis: strong association with SPINK1 trypsin inhibitor mutations. Gastroenterology. 2002, 123: 1020-5. 10.1053/gast.2002.36028.
Schneider A, Suman A, Rossi L, Barmada MM, Beglinger C, Parvin S, Sattar S, Ali L, Khan AK, Gyr N, Whitcomb DC: SPINK1/PSTI mutations are associated with tropical pancreatitis and type II diabetes mellitus in Bangladesh. Gastroenterology. 2002, 123: 1026-30. 10.1053/gast.2002.36059.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-230X/3/34/prepub
Acknowledgements
We like to thank the family for their contribution to this study. Written consent was obtained from the index patient for publication of this report.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
None declared.
Authors' contributions
All authors contributed to treatment and characterization of this kindred. PF and HE carried out the CASR gene analysis, while FS gave his broad support concerning all aspects. All authors read and approved the final manuscript
An erratum to this article is available at http://dx.doi.org/10.1186/1471-230X-4-6.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Felderbauer, P., Hoffmann, P., Einwächter, H. et al. A novel mutation of the calcium sensing receptor gene is associated with chronic pancreatitis in a family with heterozygous SPINK1 mutations. BMC Gastroenterol 3, 34 (2003). https://doi.org/10.1186/1471-230X-3-34
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-230X-3-34