Abstract
Background
The wild herb Swertia mussotii is a source of the anti-hepatitis compounds swertiamarin, mangiferin and gentiopicroside. Its over-exploitation has raised the priority of producing these compounds heterologously. Somatic hybridization represents a novel approach for introgressing Swertia mussotii genes into a less endangered species.
Results
Protoplasts derived from calli of Bupleurum scorzonerifolium and S. mussotii were fused to produce 194 putative hybrid cell lines, of which three (all derived from fusions where the S. mussotii protoplasts were pre-treated for 30 s with UV light) later differentiated into green plants. The hybridity of the calli was confirmed by a combination of isozyme, RAPD and chromosomal analysis. The hybrid calli genomes were predominantly B. scorzonerifolium. GISH analysis of mitotic chromosomes confirmed that the irradiation of donor protoplasts increased the frequency of chromosome elimination and fragmentation. RFLP analysis of organellar DNA revealed that mitochondrial and chloroplast DNA of both parents coexisted and recombined in some hybrid cell lines. Some of the hybrid calli contained SmG10H from donor, and produced swertiamarin, mangiferin and certain volatile compounds characteristic of S. mussotii. The expression of SmG10H (geraniol 10-hydroxylase) was associated with the heterologous accumulation of swertiamarin.
Conclusions
Somatic hybrids between B. scorzonerifolium and S. mussotii were obtained, hybrids selected all contained introgressed nuclear and cytoplasmic DNA from S. mussotii; and some produced more mangiferin than the donor itself. The introgression of SmG10H was necessary for the accumulation of swertiamarin.
Similar content being viewed by others
Background
Somatic hybridization provides a means to bypass the problem of sexual incompatibility which prevents the production of many wide hybrids in the plant kingdom. The technique has been successfully demonstrated in a number of intra- and inter-specific, intergeneric, intertribal and even inter-familial combinations [1–4]. The possibility of introgression from exotic sources is of interest not just in the applied field, but also because it provides opportunities for the discovery of novel synthetic pathways for secondary metabolites and signalling compounds.
The medicinal herb Swertia mussotii Franch is native to Tibet, where it has enjoyed a long history of use as a curative for hepatitis [5, 6]. Its major active compounds have been shown to be swertiamarin, mangiferin and gentiopicroside [7]. The economic value of the species is such that there is now a real risk of species extinction as a result of over-exploitation. Swertiamarin and gentiopicroside are both iridoid monoterpenoids, but their synthetic pathway has not as yet been characterized in any detail [8, 9]. However, many of the reactions in this pathway are known to be catalyzed by P450 proteins [10, 11]. Members of this highly diverse protein family are involved in the synthesis of pigments, antioxidants and defense compounds [12], and one of particular importance for the synthesis of swertiamarin is the enzyme geraniol 10-hydroxylase (G10H) [13]. Recently, we isolated a full length cDNA clone of S. mussotii G10H (SmG10H), which has the catalytic activity of hydroxylating geraniol [14].
Bupleurum scorzonerifolium Willd (2n = 12), as a member of the Umbelliferae family, also is a very useful herb in Chinese traditional medicine, where it is used to treat acesodyne, diminish inflammation, ease fever and increase resistance to hepatic injury and promote immunity [15]. We previously reported plant regeneration from cultured B. scorzonerifolium protoplasts [16]. And these cultured cell lines with the fast-growing capacities have remained viable for at least 16 years, showing a chromosome numbers of 2n = 12 in over 90% of cells [3].
Our aim was to obtain somatic hybrids between S. mussotii and B. scorzonerifolium. The latter was chosen as the other biparent because it had a rapid growth and similar many secondary metabolic pathways [15, 16]. We have used a number of fingerprinting methods to characterize the introgression events achieved by applying this process, and in particular have focussed on the presence of SmG10H. Finally we sought to establish the relationship between the accumulation of swertiamarin and gentiopicroside and the level of expression of SmG10H.
Results
Growth and development of somatic fusion nuclei
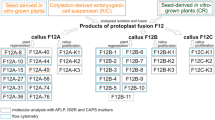
Granular calli were formed from combinations A-D after about two months of culture in liquid P5 medium in the dark, but no callus was observed in combination E. Once the clones had reached a diameter of 1.5-2 mm (Figure 1A), they were transferred to the proliferation medium MB2 (Figure 1B), where they were maintained through three rounds of sub-culturing before their final transfer to the differentiation medium MB3. A population of 194 clones was obtained by this process (Table 1); of these only three, all from combination B, were successfully regenerated into plants. These all developed narrow and long leaves, resembling those of B. scorzonerifolium (Figure 1C-E).
Genetic characterization of the somatic hybrid calli
Esterase isozyme analysis of 194 clones indicated that 104 had the partial characteristic band(s) of both parents and novel bands and were verifiable as hybrid (Additional file 1). A set of 88 RAPD primers was applied to generate DNA fingerprints of the presumptive hybrid calli (Additional file 2). Fragments from both biparents, as well as fragments not present in either of them, were observed in all of the hybrid clones tested (Figure 2). As 91% of the fragments in the hybrids were derived from the B. scorzonerifolium biparent, and only 0.2-2.4% from the S. mussotii biparent, the hybrid genomes were dominated by the recipient species (Table 2). The construction of donor nuclear genome DNAs in these hybrids were similar except hybrid C10 (Additional file 3). RAPD analysis showed in hybrids exposure of UV for 30 s, there were 1.7% donor characteristic bands and 1.2% new bands (2.9% in total), however the numbers were raised to 4.5% and 3.2% (7.7% in total) in hybrids exposure of UV for 1 min (Additional file 3).
RAPD analysis of hybrid clones. A, Primer H19; B, Primer Q15; C, Primer Q8; D, Primer N20. Sm, S. mussotii; Bs, B. scorzonerifolium. Lanes 2, 3, 10, 13 and 14 refer, respectively to hybrid clones B9, B24, B52, C59 and C10. →, Distinctive bands inherited from the donor or recipient. ►, Bands not present in either the donor or the recipient. *, The regenerated hybrid clones.
Karyotypes of somatic hybrid clones
The chromosome numbers of B. scorzonerifolium and S. mussotii calli were 11-12 and 17-20, respectively (Figure 3A, B and Table 3). In the clones derived from combination A, the number was no lower than 15, with most carrying 17-20 (Table 3). Combination B clones carried 11-16 chromosomes, and combination C ones carried 11-14 (Table 3). Combination D clones had 11-14 (Table 3). When analysed using GISH, the biparental genomes were readily distinguishable from one another (Figure 4A, B). The three regenerable hybrid clones had chimera cells with different chromosome numbers, carrying 11-13 intact B. scorzonerifolium, none intact S. mussotii, and 1-3 recombined chromosomes (Figure 4C). In contrast, the non-regenerable clones carried 11-13 intact B. scorzonerifolium, none intact S. mussotii and 5-9 recombined chromosomes (Figure 4D).
Analysis of the cytoplasmic genomes of somatic hybrids
The RFLP profiles of mitochondrial DNA obtained using restriction enzymes HindIII and hybridized with probes coxI revealed that all of the cell lines analyzed contained B. scorzonerifolium sequences and cell lines B9 and C10 had donor bands and novel bands (Figure 5A). The chloroplast type of the hybrid cell lines was determined using rbcL as a hybridization probe. Hybridizations of HindIII digests with rbcL show that all of the cell lines analyzed contained B. scorzonerifolium fragments and 1-2 novel fragments, and cell lines B9 and B24 contained S. mussotii fragments (Figure 5B). Thus, some recombination within the mitochondrial and chloroplast genome of both parents also occurred in some hybrids.
RFLP of profiles of mitochondrial and chloroplast DNA of S. mussotii, B. scorzonerifolium , and the hybrid cell lines from combinations B and C between S. mussotii and B. scorzonerifolium. M, labeled λDNA digested by HindIII+EcoRI; Bs, B. scorzonerifolium; Sm, S. mussotii; B9, B24, C10 and C64, hybrid cell lines of S. mussotii -B. scorzonerifolium. Arrows indicate bands of the S. mussotii and B. scorzonerifolium; arrowheads indicate new bands. A, HindIII-digested genomic DNA probed with the mitochondrial-specific probes coxI. B, HindIII-digested genomic DNA probed with the chloroplast-specific probe rbcL.
Content of medicinally active compounds
The HPLC-based analysis of 74 of the hybrid clones determined that none accumulated gentiopicroside (Figure 6). Clones B24, B27, B132, C18, C26, C47 and C124 contained 7.4-81.2 μg/g swertiamarin, while clones B6, B40, B56, C10 and C121 contained 86.3-816.8 μg/g mangiferin. Notably, the mangiferin content of clones B6, B56 and C10 was higher than that of the callus derived from the S. mussotii biparent (Table 4).
HPLC analysis of hybrid clones. A, Standard preparations of swertiamarin, gentiopicroside and mangiferin; B, S. mussotii; C, B. scorzonerifolium; D, Hybrid clone B24; E, Hybrid clone C18. UV spectrum of swertiamarin and mangiferin from samples were indicated in D and E. 1, S. mussotii; 2, Standard preparations; 3, Hybrid clone B24; 4, Hybrid clone C18.
Volatile compounds in the biparent and hybrid clones
The volatile compound content of the hybrid clones, as assessed by GC-MS, largely resembled that of the B. scorzonerifolium biparent (Additional file 4). Nevertheless, a few donor compounds, in particular coumaron and linoleic acid, were detectable in some of the hybrid clones, along with a small number of compounds (e.g., cyclohexanol and dodecanoyl) which were not detected in either biparent (Table 5).
The introgression of P450 genes
Degenerate PCR analysis was used to detect the P450 genes in clones A6, A67, B24, B27, B132, C18, C26, C47 and C124, with various contents of swertiamarin and mangiferin (Figure 7 and Additional file 5). Only amplicon of primer CYP76 was distinguished among the bipatents and hybrid A6 (Figure 7). Each cDNA template amplified a single fragment in the size range 1100~1500 bp in hybrids above and the bipatents using primer CYP76. Sequencing identified 11 distinct fragments. An analysis of the set of polypeptides predicted from these nucleotide sequences identified their homology to the G10H gene of Catharanthus roseus (geraniol 10-hydroxylase gene, GenBank accession number AJ251269). A full length SmG10H sequence of 1488 bp (Genebank accession GU168041) was obtained from S. mussotii. The G10H sequences present in clones B24, B27, B132, C18, C26, C47 and C124 were identical to that of SmG10H (Additional file 6). In two clones (A6 and A67), the G10H sequence shared 53.1% homology with SmG10H (Additional file 7).
Up-regulation of SmG10H is correlated with the accumulation of swertiamarin
Semi-quantitative RT-PCR suggested that the expression SmG10H varied among the clones (S. mussotii > B24 > B132 > C47 > A6, see Figure 8 and Table 4). The swertiamarin content of S. mussotii (933 μg/g) was substantially higher than that in the hybrid clones (0-81.2 μg/g), while clones B24 and B132 produced more than clone C47; neither swertiamarin nor SmG10H expression were detected in clone A6. These results suggest that up-regulation of SmG10H is correlated with the accumulation of swertiamarin.
The expression of SmG10H and accumulation of swertiamarin in hybrid clones. A, Variation for level of SmG10H expression. B, Swertiamarin content. Sm, S. mussotii; Bs, B. scorzonerifolium; B24 and B132, hybrid clones from combination B; C47, hybrid clone from combination C; A6, hybrid clone from combine A. Bars represent the standard error of the mean; t test, *, P < 0.05.
Discussion
Hybrid clones experience both chromosome elimination and introgression
Across a range of hybrid combinations, the regeneration of viable plants has proven to be the main bottleneck in the somatic hybridization process [1, 3, 4]. Much of the problem appears to be related to the hybrid incompatibility of the biparents. This hybrid incompatibility can be alleviated if sufficient of the donor biparent's chromosomes are either completely eliminated, or at least are broken down so that sub-chromosomal segments become fused with the recipient biparent's chromosome [2, 17, 18]. The somatic chromosome number of successful regenerants has been shown to be close to or just slightly lower than that of the recipient biparent [19, 20]. Here, only three of the population of the 194 somatic B. scorzonerifolium / S. mussotii fusion nuclei proved to be regenerable. Both the genetic and cytological analyses showed that the constitution of the regenerable hybrid calli was close to that of the recipient parent B. scorzonerifolium, which suggested that large-scale chromosome elimination is necessary to restore the somatic hybrids' ability to regenerate.
UV irradiation of the donor biparent's protoplasts prior to fusion has been shown to encourage chromosomal elimination [21–23]. The hybrid cell lines B24 and C10 both retained 11-13 B. scorzonerifolium chromosomes, none entire S. mussotii, but the former retained 1-3 introgression chromosomes, while the latter retained more (5-9) introgression chromosomes (Figure 4). This result is consistent with the pattern whereby raising the UV dosage decreases the number of intact donor chromosomes but increases the frequency of donor introgression [20, 23].
Characteristics of the hybrid cytoplasmic genome
Earlier investigations showed that recombination and (or) coexistence mitochondria DNA from both parents is common in somatic hybrids [23]. In contrast, chloroplast DNA often had random and equal segregation [24, 25]. Mixed populations and recombination of chloroplast DNA have only rarely been detected [26]. In our previous studies, mixed and recombined mitochondrial DNA was also seen in wheat somatic hybrids [23, 27, 28]. In this study, the mitochondrial and chloroplast DNA of both parents also coexisted in most hybrid cell lines. Novel DNA segments appeared in some hybrids, which may have been the result of recombination of mitochondria and chloroplast DNA (Figure 5). We conclude that in the inter-familial hybridization between B. scorzonerifolium and S. mussotii, it is possible to transfer donor mitochondrial and chloroplast genes.
The engineering of medicinally active compounds
Somatic hybrids could increase the content of the efficacious compounds from traditional Chinese medicinal materials. However, only a few cases succeeded [29]. The content of swertiamarin, mangiferin and gentiopicroside varied markedly among the hybrid clones (Table 4). With respect to swertiamarin, no clone accumulated close to the level which was achieved by the donor calli (Figure 6). However, with respect to mangiferin, five clones (B6, B40, B56, C10 and C121) outperformed the donor, two accumulated markedly less (B40 and C121), and two (A6 and B24) produced no detectable level (Table 4). The accumulation by the hybrid clones of a number of volatiles associated with the donor species is also indicative of the transfer of whole synthetic pathways from S. mussotii to a genotype which is largely B. scorzonerifolium.
The clones best able to accumulate mangiferin tended to have retained the most introgressed chromosomes (Figure 4). Similarly the RAPD fingerprinting showed that these clones also inherited the most donor DNA (Table 2 and Figure 2). Presumably maximizing the yield of the donor's medicinally active compounds in a somatic hybrid clone requires transferring as much donor DNA as possible.
The relationship between SmG10Hexpression and swertiamarin content
According to RT-PCR at least, the expression of SmG10H varied among the hybrid clones. Nevertheless, its level was largely correlated with the accumulation of swertiamarin (Figure 8), implying that the transfer of SmG10H alone cannot be expected to be sufficient to guarantee heterologous expression. The implied requirement for other genes in the synthetic pathway underlines the difficulty that a more reductive, tansgenic strategy would face in obtaining the successful production of swertiamarin in a heterologous situation. The identification of hybrid clones able to accumulate this medicinally significant compound therefore confirms the potential of somatic hybridization as a viable route for engineering the production of such molecules in plants.
Conclusions
In conclusion, somatic hybridization provides a new way to introgression secondary metabolites and related genes in phylogenetic distant species. Here we have managed to obtain somatic hybrids of B. scorzonerifolium / S. mussotii with an appreciable content of swertiamarin. The nuclear and cytoplasmic genes from donor were transferred into the genome DNA of hybrid clones. The introgression of SmG10H was necessary for the accumulation of swertiamarin. Therefore, the potential of somatic hybridization is a viable route for engineering the production of such molecules in plants.
Methods
Origin of biparental protoplasts
Immature seed of Swertia mussotii Franch was collected from Yushu county, Qinghai province, China. Voucher specimens had been deposited at Qinghai Normal University. The seed was surface-sterilized by immersion first in 70% (v/v) ethanol for 30 s and then in 0.1% w/v aqueous mercuric chloride for 10 min. The seeds were plated on Murashige and Skoog [30] basal medium (MS) containing 1 mg/l 2, 4-dichlorophenoxyacetic acid (2, 4-D) using the method described by Xiang et al. [31] to induce the production of callus, the source of donor protoplasts. The callus was subcultured on MB medium (MS medium supplemented by B5 vitamins [23], 2 mg/l glycine, 146 mg/l glutamine, 300 mg/l casein hydrolysate, 1 mg/l 2, 4-D, 30 g/l sucrose and 7.5 g/l agar at pH 5.8) at interval of 15 days. The recipient protoplasts were obtained from Bupleurum scorzonerifolium Willd calli induced in the same way; these have been kept in culture for 12 years under 18-20 μmol m-2 s-1 cool white light in MB medium. Protoplasts were isolated from all calli following established methods [32].
Protoplast fusion and post fusion culture
Preparations of both protoplast types were washed in 0.6 M mannitol, 5 mM CaCl2, then transferred to a 3.5 cm petridish to form a thin layer. Donor protoplasts were UV irradiated at 380 μW/cm2 for either 0 s (S1), 30 s (S2), 1 min (S3), 2 min (S4) or 3 min (S5), after which they were mixed with the recipient protoplasts at a ratio of 1:1. The fusion protocol followed the PEG method described by Xia and Chen [32]. Fusion products, which combinations A-E were corresponding donor protoplasts S1-S5, were cultured on P5 medium [32]. Once calli had reached a diameter of 1.5-2.0 mm, they were transferred to MB2 medium and sub-cultured every two weeks for 1-2 months. At this stage, the calli were removed to a MB3 solid medium (MB medium supplemented with 1 mg/l 6-benzylaminopurine and 1 mg/l indoleacetic acid). Regenerated plantlets were transferred to a seedling-strengthening medium [32].
Callus genotying
Esterase isozymes were extracted from calli and assayed as described elsewhere [33]. DNA was extracted from selected calli according to Doyle and Doyle [34], and used as template for RAPD reactions based on 88 decamer primers (Promega Inc., Madison, Wis.). The PCRs were conducted according to Xia et al. [35], and the amplicons were electrophoresed through 1.5% agarose gels before staining in 0.5 μg/ml ethidium bromide. These RAPD experiments were repeated at least 3 times and only the repeatable bands were record.
Mitotic chromosome analysis
Mitotic chromosome spreads from callus and root tip cells were prepared as described by [20]. For GISH analysis, total genomic S. mussotii DNA was used as the probe, and the procedure described by Xiang et al. [20] was applied.
RFLP analyis
Genomic DNA of the putative hybrid cell lines and their parents was extracted as described previously [34]. For the analysis of chloroplast (cp) and mitochondrial (mt) DNA, 15-20 μg of total DNA was digested with HindIII and electrophoresed through 0.8-1% agarose gels in TBE buffer. The DNA was transferred onto a nylon membrane (Hybond N+, Amersham-Phamarcia, UK) using 0.4 M NaOH. Probe labelling, hybridization, and washing were carried out with the ECL Random Labeling and detection system (Amersham-Phamarcia, UK) according to the manufacturer's instructions. Plasmids containing mtDNA fragments (coxI) from maize (Zea mays L.) and a cpDNA fragment (rbcL) from spinach (Spinacia oleracea L.) were kindly provided by Dr. G. Spangenberg (Institute for Plant Sciences, Swiss Federal Institute of Technologies, CH-8092 Zürich, Switzerland). The inserts were cut out of a gel and labelled.
Semi-quantitative RT-PCR analysis
Semi-quantitative RT-PCR was conducted on total RNA isolated from hybrid and two parents calli using the TriZOL reagent (Invitrogen, USA). First strand cDNA was synthesized using Superscript II reverse transcriptase M-MLV (TakaRa, Japan), following the manufacturer's directions. Degenerate primers targeting the gene from Arabidopsis thaliana encoding cytochrome P450 monooxygenase (Additional file 2) were applied to the cDNA templates. The amplicons derived from degenerate primers targeting the two S. mussotii actin genes SmAct1 and SmAct2 were used to normalize the RT-PCR signal. Each PCR comprised 19-25 cycles of 94°C/30 s, 53°C/30 s, 72°C/90 s, and was completed with a 10 min extension at 72°C. Each PCR was replicated at least three times, based on independent biological samples.
HPLC analysis
The calli were shade-dried for seven days and ground to powder. After the addition of 20 ml methanol to 1 g powdered callus, the resulting suspension was sonicated for 1 h at room temperature, then filtered through a 0.45 μm membrane filter. A 10 μl aliquot of filtrate was injected into a LC-10AD HPLC system (Shimadzu Co., Japan) equipped with a C18 column (Phenomenex Luna, 4.6 × 250 mm i.d., 5 μm). The mobile phase was water:methanol (75:25), and the outflow (0.8 ml/min) was scanned at 259 nm [14]. Standards for swertiamarin, gentiopicroside and mangiferin were provided by the National Institute for the Control of Pharmaceutic and Biological Products (Beijing, China). All above experiments were carried out four times. In all statistical tests, values of P lower than 0.05 were interpreted as indicating statistically significant differences. Results were analysed with SAS statistical package (Version5.1, SAS Institute Inc., Cary, NC).
Capillary gas chromatography/mass spectrometry (GC-MS) analysis
Methanol extracts of the calli were prepared according to Liu et al. [36], and then subjected to GC/MS, using a Micromass GCT gas chromatograph-mass spectrometer (England) fitted with a DB-5 ms column (0.25 mm × 30 m, 0.25 μm film thickness) (J W Scientific, Folsom, CA), with a helium flow rate of 1 ml/min, and operating at 70 eV ionization voltage with a scan range of 20-600 Da. The column temperature was set at 200°C for 2 min, then elevated to 300°C at 15°C/min and held at 300°C for 7 min.
References
Dudits D, Maroy E, Praznovszky T, Olah Z, Gyorgyey J, Cella R: Transfer of resistance traits from carrot into tobacco by asymmetric somatic hybridization: regeneration of fertile plants. Proc Natl Acad Sci USA. 1987, 84: 8434-8438. 10.1073/pnas.84.23.8434.
Kisaka H, Lee H, Kisaka M, Kanno A, Kang K, Kameya T: Production and analysis of asymmetric hybrid plants between monocotyledon (Oryza sativa L.) and dicotyledon (Daucus carota L.). Theor Appl Genet. 1994, 89: 365-371.
Wang M, Xia G, Peng Z: High UV-tolerance with introgression hybrid formation of Bupleurum scorzonerifolium Willd. Plant Sci. 2005, 168: 593-600. 10.1016/j.plantsci.2004.09.017.
Zhou S, Brown DC: High efficiency plant production of North American ginseng via somatic embryogenesis from cotyledon explants. Plant Cell Rep. 2006, 25: 166-173. 10.1007/s00299-005-0043-z.
Yamahara J, Konoshima T, Sawada T, Fujimura H: Biologically active principles of crude drugs: pharmacological actions of Swertia japonica extracts, swertiamarin and gentianine. Yakugaku Zasshi. 1978, 98: 1446-1451.
Kikuzaki H, Kawasaki Y, Kitamura S, Nakatani N: Secoiridoid glucosides from Swertia mileensis. Planta Med. 1996, 62: 35-38. 10.1055/s-2006-957792.
Brahmachari G, Mondal S, Gangopadhyay A, Gorai D, Mukhopadhyay B, Saha S, Brahmachari AK: Swertia (Gentianaceae): chemical and pharmacological aspects. Chem Biodivers. 2004, 1: 1627-1651. 10.1002/cbdv.200490123.
Dewick PM: Medicinal Natural Products: A Biosynthetic Approach. 2nd edition. Chichester UK, John Wiley and Sons Press, 2002.
Oudin A, Courtois M, Rideau M, Clastre M: The iridoid pathway in Catharanthus roseus alkaloid biosynthesis. Phytochem Rev. 2007, 6: 259-276. 10.1007/s11101-006-9054-9.
Mansuy D: The great diversity of reactions catalyzed by cytochrome P450. Comp Biochem Physiol Part C. 1998, 121: 5-14.
Werck-Reichhart D, Feyereisen R: Cytochromes P450: a success story. Genome Biol. 2000, 1: REVIEWS3003
Kahn R, Durst F: Function and evolution of plant cytochrome P450. Recent Adv Phytochem. 2000, 34: 151-189.
Collu G, Unver N, Peltenburg-Looman AM, van der Heijden R, Verpoorte R, Memelink J: Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett. 2001, 508: 215-220. 10.1016/S0014-5793(01)03045-9.
Wang JF, Liu YL, Cai YF, Zhang FF, Xia GM, Xiang FN: Cloning and functional analysis of geraniol 10-hydroxylase, a cytochrome P450 from Swertia mussotii Franch. Biosci Biotechnol Biochem. 2010, 74: 100175-1-8.
Chen Y, Tan LL, Cai X, Hu ZH: Advances on the studies of chemical constituents of Bupleurum L. Chinese Wild Plant Resources. 2006, 2: 8-11.
Xia GM, Li ZY, Chen HM: Plant regeneration from protoplast of Bupleurum scorzonerifolium. Plant Cell Reports. 1992, 11: 155-158.
Kisaka H, Kameya T: Production of somatic hybrids between Daucus carota and Nicotiana tabacum. Theol Appl Genet. 1994, 88: 75-80.
Kisaka H, Kisaka M, Kanno A, Kameya T: Intergeneric somatic hybridization of rice (Oryza sativa L.) and barley (Hordeum vulgare L.) by protoplast fusion. Plant Cell Reports. 1998, 17: 362-367. 10.1007/s002990050407.
Vlahova M, Hinnisdaels S, Frulleux F: UV irradiation as a tool for obtaining asymmetric somatic hybrids between Nicotiana plumbaginifolia and Lycopersicon esculentum. Theor Appl Genet. 1997, 94: 184-191. 10.1007/s001220050398.
Xiang FN, Xia GM, Chen HM: Effect of UV dosage on somatic hybridization between common wheat (Triticum aestivum L.) and Avena sativa L. Plant Sci. 2003, 164: 697-707. 10.1016/S0168-9452(03)00021-9.
Hall RD, Rouwendal GJA, Krens FA: Asymmetrc somatic cell hybridization in plants: The early effects of (sub) lethal doses of UV gamma radiation on the cell physiology and DNA intergrity of culture sugarbeet (Beta vulgaris L.) protoplasts. Mol Gen Genet. 1992, 234: 234-314.
Forsberg J, Dixelius C, Lagercrantz U, Glimelius K: UV dose dependent DNA elimination in asymmetric somatic hybrids between Brassica napus and Arabidopsis thaliana. Plant Sci. 1998, 131: 65-76. 10.1016/S0168-9452(97)00242-2.
Xiang F, Xia G, Zhi D, Wang J, Nie H, Chen H: Regeneration of somatic hybrids in relation to the nuclear and cytoplasmic genomes of wheat and Setaria italica. Genome. 2004, 47: 680-8. 10.1139/g04-023.
Escalante A, Imanishi S, Hossain M, Ohmido N, Fukui K: RFLP analysis and genomic in situ hybridization (GISH) in somatic hybrids and their progeny between Lycopersicon esculentum and Solanum lycopersicoides. Theor Appl Genet. 1998, 96: 719-726. 10.1007/s001220050794.
Li Y, Sink C: Cell type determines plastid transmission in tomato intergeneric somatic hybrids. Curr Genet. 1992, 22: 167-171. 10.1007/BF00351478.
Cardi T, Bastia T, Monti L, Earle ED: Organelle DNA and male fertility variation in Solanum sp., and interspecific somatic hybrids. Theor Appl Genet. 1999, 99: 819-828. 10.1007/s001220051301.
Xu CH, Xia GM, Zhi DY, Xiang FN, Chen HM: Integration of maize nuclear and mitochondrial DNA into the wheat genome through somatic hybridization. Plant Sci. 2003, 165: 1001-1008. 10.1016/S0168-9452(03)00287-5.
Cai Y, Xiang F, Zhi D, Liu H, Xia G: Genotyping of somatic hybrids between Festuca arundinacea Schreb. and Triticum aestivum L. Plant Cell Rep. 2007, 26: 1809-1819. 10.1007/s00299-007-0397-5.
Han L, Zhou CN, Shi J, Zhi DY, Xia GM: Ginsenoside Rb1 in asymmetric somatic hybrid calli of Daucus carota with Panax quinquefolius. Plant Cell Rep. 2009, 28: 627-638. 10.1007/s00299-009-0674-6.
Murashige T, Skoog F: A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962, 15: 473-497. 10.1111/j.1399-3054.1962.tb08052.x.
Xiang FN, Li JM, Ma JX, Wu XM: Tissue culture of calli of Swertia musstii Franch. Chinese Traditional and Herbal Drugs. 1996, 27: 492-495.
Xia GM, Chen HM: Plant regeneration from intergeneric somatic hybridization between Triticum aestivum and Leymus chinensis. Plant Sci. 1996, 120: 197-203. 10.1016/S0168-9452(96)04492-5.
Hu NS, Wan XG: Application of isozyme technique. Changsha Hunan, Hunan Science and Technology Press, In Chinese 1985.
Doyle JJ, Doyle JI: Isolation of plant DNA from fresh tissue. Focus. 1990, 12: 13-15.
Xia GM, Li ZY, Wang SL, Xiang FN, Liu JY, Chen PD, Liu DJ: Asymmetric somatic hybridization between haploid wheat and UV irradiated Haynaldia villosa. Plant Sci. 1998, 37: 217-223.
Liu YL, Cai YF, Zhao ZJ, Wang JF, Li J, Xin W, Xia GM, Xiang FN: Cloning and functional analysis of a β-amyrin synthase gene associated with oleanolic acid biosynthesis in Gentiana straminea. Biol Pharm Bul. 2009, 32: 818-824. 10.1248/bpb.32.818.
Acknowledgements
This research was made possible by financial support from the Chinese 'National Special Science Research Program' (grant no. 2007CB948203), 'Natural Education Ministry Doctor Station Foundation Fellowship' (grant no. 913111006) and 'National Natural Science Foundation' (grants no. 30970243 and 30771116), and Excellent Youth Foundation of Shandong Province of China (grant no. JQ200810), and 'Science &Technology Plan of Shandong Province' (grant no. 2009GG10002001). We acknowledge the linguistic help given by http://www.smartenglish.co.uk in English editing this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Authors' contributions
JFW conducted most of the experiments and helped in the writing of the ms; CZZ contributed to the GC/MS experiment and participated in the drafting of the manuscript; GMX was responsible for the design and coordination of the study; FNX conceived the study and was responsible for the final version of the ms. The final manuscript were read and approved by all authors.
Electronic supplementary material
12870_2010_853_MOESM1_ESM.JPEG
Additional file 1: Esterase analysis of calli. Sm, S. mussotii; Bs, B. scorzonerifolium. ►, Isozymes not present in either the donor or the recipient; →, Distinctive isozymes inherited from the donor or recipient. *, important calli. (JPEG 497 KB)
12870_2010_853_MOESM2_ESM.DOC
Additional file 2: RAPD analysis of biparental and hybrid calli. Sm: Fragments inherited from S. mussotii; Bs: Fragments inherited from B. scorzonerifolium; T, total of the parents and new bands; N: fragments not present in either biparental profile. (DOC 244 KB)
12870_2010_853_MOESM3_ESM.DOC
Additional file 3: Frequency in the hybrid clones of donor fragments, and fragments absent from both biparental profiles. (DOC 30 KB)
12870_2010_853_MOESM6_ESM.JPEG
Additional file 6: Alignment of G10H nucleotide sequences. Smg10 h, g10 h from S. mussotii; B24 g10 h, g10 h from hybrid B24; C26 g10 h, g10 h from hybrid C26. (JPEG 1 MB)
12870_2010_853_MOESM7_ESM.JPEG
Additional file 7: Alignment of G10H peptide sequences. The red line indicates the conserved domain within the sequence. SmG10H-P, G10H from S. mussotii; B24G10H-P, G10H from hybrid B24; A6G10H-P, G10H from hybrid A6; BsG10H-P, G10H from B. scorzonerifolium. (JPEG 701 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wang, J., Zhao, C., Liu, C. et al. Introgression of Swertia mussotii gene into Bupleurum scorzonerifoliumvia somatic hybridization. BMC Plant Biol 11, 71 (2011). https://doi.org/10.1186/1471-2229-11-71
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-11-71