Abstract
Background
Chloroplasts are the green plastids where photosynthesis takes place. The biogenesis of chloroplasts requires the coordinate expression of both nuclear and chloroplast genes and is regulated by developmental and environmental signals. Despite extensive studies of this process, the genetic basis and the regulatory control of chloroplast biogenesis and development remain to be elucidated.
Results
Green cauliflower mutant causes ectopic development of chloroplasts in the curd tissue of the plant, turning the otherwise white curd green. To investigate the transcriptional control of chloroplast development, we compared gene expression between green and white curds using the RNA-seq approach. Deep sequencing produced over 15 million reads with lengths of 86 base pairs from each cDNA library. A total of 7,155 genes were found to exhibit at least 3-fold changes in expression between green and white curds. These included light-regulated genes, genes encoding chloroplast constituents, and genes involved in chlorophyll biosynthesis. Moreover, we discovered that the cauliflower ELONGATED HYPOCOTYL5 (BoHY5) was expressed higher in green curds than white curds and that 2616 HY5-targeted genes, including 1600 up-regulated genes and 1016 down-regulated genes, were differently expressed in green in comparison to white curd tissue. All these 1600 up-regulated genes were HY5-targeted genes in the light.
Conclusions
The genome-wide profiling of gene expression by RNA-seq in green curds led to the identification of large numbers of genes associated with chloroplast development, and suggested the role of regulatory genes in the high hierarchy of light signaling pathways in mediating the ectopic chloroplast development in the green curd cauliflower mutant.
Similar content being viewed by others
Background
Chloroplast biogenesis from proplastids requires coordinate expression of nuclear and chloroplast genes [1], and is largely regulated by developmental and environmental cues such as light. Approximately 3000 proteins in chloroplasts are encoded by the nucleus [2]. They participate in a large number of functional processes that are required for chloroplast biogenesis. These processes include import of nuclear encoded proteins through the Toc/Tic complexes, protein assembly and disassembly with chaperone proteins, thylakoid formation, pigment synthesis, plastid divisions, and retrograde signaling [3, 4]. In addition, a great number of proteins localized outside chloroplasts, such as photoreceptors, light-signaling transducers, and transcription factors, have been shown to be involved in chloroplast development [3, 4]. On the one hand, most genes belonging to these two classes are essential for chloroplast development since suppression of their expressions leads to impaired chloroplasts. On the other hand, some light signaling pathway genes, such as constitutive photomorphogenic 1 (COP1), COP10, COP11, De-etiolated 1 (DET1) and Phytochrome-interacting transcription factor 3 (PIF3), function as suppressors of light-regulated gene expression and loss-of-function mutations of these genes result in ectopic chloroplast development [5–7]. In contrast, Elongated Hypocotyl 5 (HY5) that acts downstream of multiple families of photoreceptors [8–10] has been genetically characterized as a positive regulator of photomorphogenesis under a broad spectrum of light and affects chloroplast development [4, 11]. Overexpression of HY5-ΔN77 has been shown to result in precocious development of chloroplasts in the hypocotyls [12]. Determining how these genes are coordinately expressed during chloroplast development requires a genome-wide examination of gene expression during the transition from non-colored plastids into chloroplasts.
Mutations in model and other plant species are important resources for functional genomics studies. Analyses of some plastid development mutants identify important regulatory genes of plastid development. For example, ARC6, the first gene discovered to have a global effect on plastid differentiation in higher plants, was identified from an Arabidopsis mutant arc6 [13]. The Orange (Or) gene that encodes a zinc-finger DnaJ cysteine rich domain containing protein is isolated from the orange curd cauliflower mutant and has been proven to be responsible for the conversion of leucoplasts into chromoplasts [14]. The green curd cauliflower mutant is a spontaneous mutation with an abnormal pattern of chloroplast development in curds. Compared with other mutants in which chloroplast development is impaired, the green curd mutant is unique in turning otherwise non-photosynthetic white tissue into green color with the ectopic development of chloroplasts in the inflorescence meristematic cells. The mutation in the green curd cauliflower could involve the gene(s) sufficient for chloroplast development, although there is possibility that the white curd cauliflower carries a genetic mechanism for the suppression of chloroplast development, which the green curd mutation would suppress.
In the present study, we profiled gene expression in green and white curds on the genome scale using the RNA-seq approach. We assembled 118,000 unigenes with an average length of 406 bp from cDNA libraries of green and white curds and detected 7155 differentially expressed genes with a change in expression of at least 3-fold. Among them are a large number of genes associated with chloroplast development. We also observed that BoHY5 was expressed at higher level in green curds than in white curds and that 2616 HY5-targeted genes were expressed differentially. Among these HY5-targeted genes, all the 1600 up-regulated genes were found to be HY5-targeted genes in the light in Arabidopsis, suggesting a role of BoHY5 with the ectopic chloroplast development in the green curd cauliflower mutant.
Results
Cauliflower mutant with green curds
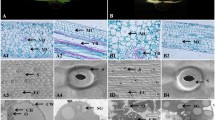
Cauliflower curd is composed of inflorescence meristems that normally contain proplastids and leucoplasts and is therefore white [15]. In the commercially available green cauliflower mutant, chloroplasts are developed in the curd, turning the otherwise white tissue green (Figure 1a and 1b). While the mutant plants produced green curds under normal growth conditions in greenhouse and in field, the intensity of green hue in the curd tissues was affected by light intensity. Under field growth conditions, the curd tissues exposed to direct sunlight showed dark green color and those grown in shade exhibited less green hue. Autofluorescence of chlorophyll in chloroplasts was clearly observed in the green curd cells under the confocal microscope (Figure 1c and 1d).
Phenotype and chlorophyll content of green curd cauliflower mutant. (a) and (b) Field grown curds from white cauliflower variety (Stovepipe) and green curd line (ACX800), respectively. (c) and (d) Autoflorescence of chloroplasts in green curds. Scale bar in (c) = 20 μm and in (d) = 10 μm. (e) Chlorophyll a and b content in young leaves and curds of Stovepipe (WT) and ACX800 (Green) cauliflower. The numbers above bars show the ratio of chlorophyll a/b. Error bars represent ± SD (n = 3).
To investigate chloroplast development in the green curd mutant, we first measured chlorophyll content in young leaf and curd tissues. Higher level of total chlorophyll was detected in leaf tissue of green cauliflower plants than that of the white control. The concentration of chlorophyll in green curd cauliflower leaves was 1780.4 μg/g fresh weights (FW), while that in the white curd leaves was 1056.6 μg/g FW. Although different levels of total chlorophyll were observed between the two samples, the ratio of chlorophyll a/b for leaves in white and green mutant was similar at 2.70:1 and 2.75:1, respectively. In comparison to leaf tissue, the chlorophyll level in the curd of green cauliflower was lower at 344.4 μg/g FW. The chlorophyll a/b ratio was 3.43:1, showing that the accumulation of chlorophyll a was much greater than that of chlorophyll b in green curds (Figure 1e). As expected, no chlorophyll accumulation was detected in the white curd tissue. The green curd cauliflower mutant serves as an excellent model system for investigating the genetic basis of chloroplast biogenesis in plants.
Comparative analysis of gene expression between green and white curd cauliflower
To investigate the transcriptional control of chloroplast development, RNA-seq was employed to monitor differences in gene expression between the green curd mutant and the white cauliflower. A single lane of an Illumina GAII run was utilized for each library and a total of more than 15 million 86-bp reads from each lane were produced. Since currently there is no full genome sequence available for cauliflower (Brassica oleracea) and the genomics resources from other Brassica species are not applicable due to the short length of RNA-seq reads, we developed a novel analysis strategy for our RNA-seq data as described in the Methods section. A total of 118,000 unigenes (including alternative spliced isoforms) with an average length of 406 bp were obtained. Statistical analysis identified 7155 unigenes that were differentially expressed between green curd mutant and white curd control. Among them, 4436 genes (3.76%) were expressed at least 3-fold higher (Additional file 1) and 2719 genes (2.3%) were expressed at least 3-fold lower in green curd than in white curd (Additional file 2). Functional categorization revealed that these genes were largely involved in cellular process (1317), response to stress (980), metabolic process (810), response to abiotic stimulus (654), and biosynthetic process (574). Yet, a large group of genes (3602) remained unclassified (Figure 2).
Verification of gene expression by quantitative RT-PCR
In order to verify the expression profiles obtained from the RNA-seq approach, qRT-PCR was utilized to analyze the expression of 14 selected genes. These genes encode light signal transducers (FAR1, CRY2, PHOT2, LSH7, HY5, CIP1), photosystem II component (LHCB5), chloroplast constituents (GUN5, LHCB1.5, Toc159, HSC70-1, ACP), ATP-dependent peptidase (FtsH8) and chlorophyll synthetase (G4). Among them were 11 up-regulated genes (FAR1, CRY2, PHOT2, HY5, G4, Toc159, LHCB5, GUN5, LHCB1.5, FtsH8, and ACP) and 3 down-regulated genes (HSC70-1, CIP1, LSH7). The trends of the observed expression patterns of these genes from qRT-PCR were consistent with that determined by the RNA-seq approach (Table 1). However, there were differences at the fold level as reported in other studies [16].
Metabolic pathway changes
To identify the metabolic pathways that were affected in the green curd mutant, a cauliflower metabolic pathway database was created based on annotation of the assembled cauliflower unigenes. The significantly affected pathways were identified by using the Plant MetGenMAP analysis system http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi[17]. A total of 198 specific metabolic pathways were significantly changed in green curd mutant (p < 0.01) (Additional file 3). As expected, many metabolic pathways involved in chloroplast biogenesis and function were significantly altered. These included those associated with chlorophyll biosynthesis, such as chlorophyllide a biosynthesis I, chlorophyll a biosynthesis I, chlorophyll a biosynthesis II, chlorophyll a degradation, and chlorophyll cycle, as well as with carotenoid biosynthesis (Additional file 3). In addition, those pathways associated with photosynthesis, such as oxygenic photosynthesis, Calvin cycle, and photorespiration, and with other metabolic processes that take place in chloroplasts, such as amino acid biosynthesis and starch biosynthesis, were also significantly changed (Additional file 3).
Genes involved in chloroplast formation
Chlorophylls and carotenoids compose the photosynthetic pigments that play key roles in photosynthesis. Many genes involved in chlorophyll biosynthesis were found to be expressed highly in green curd in comparison with white (Figure 3). The upregulated genes included Mg-chelatase that plays a key regulatory role in chlorophyll biosynthesis. Genomes uncoupled 4 (GUN4, PP005347) and Genomes uncoupled 5 (GUN5, PP031929) involved in chlorophyll biosynthesis were also expressed at higher levels in green curds. These two genes are among those that produce plastid-to-nuclear retrograde signaling molecules [18, 19]. The upregulation of many genes in chlorophyll biosynthesis resulted in the accumulation of chlorophyll a and b in chloroplasts. Concomitantly, a number of genes involved in carotenoid biosynthesis were also up-regulated (Table 2), suggesting an increased capacity for the synthesis of photosynthetic pigments. Consistent with the accumulation of chlorophyll a and b in green curds, genes encoding chlorophyll binding proteins were also up-regulated (Table 2). Moreover, genes encoding photosystem I and photosystem II proteins were among the up-regulated genes (Table 2), indicating the development of chloroplast structures in the green curd tissue.
Alignment of the associated differentially expressed genes with chlorophyll biosynthesis and degradation pathways. (a) Simplified chlorophyll a biosynthesis and degradation pathway. (b) Chlorophyll cycle. The pathway maps were generated using the Plant MetGenMAP system [17].
In addition to the enhanced biosynthesis of photosynthetic apparatus, genes involved in a number of other chloroplast biogenesis processes were also differentially expressed in green curd mutant. TRANSLOCON AT THE OUTER ENVELOPE MEMBRANE OF CHLOROPLASTS 34 (Toc34) and Toc159 are important parts of the Toc/Tic complexes mediating protein import from cytosol [1]. High levels of Toc34 (PP019500 and PP051864) and Toc159 (PP013646 and PP007289) transcripts were observed in the green curds. Proteins imported into chloroplasts need to be properly assembled and folded, a process that is mediated by a group of chaperone proteins, such as HSP70 and Cpn60, and protein disulfide isomerase [3, 20, 21]. Accordingly, chaperone HSP70 (PP031462, PP020739, and PP094292) and protein disulfide isomerase (PP012584, PP000051, and PP028760) were found to be significantly upregulated in green curds (Additional file 1).
Chlorophyllase catalyzes degradation of chlorophyll a to yield chlorophyllide and phytol [22]. Chlorophyllase (PP095462) was expressed lower in green curds than in white curds, which could account for the accumulation of chlorophyll a in green curds (Figure 3).
Signaling genes for chloroplast biogenesis
The large number of differentially expressed genes between the green curd mutant and white curd cauliflower suggests that genes at high hierarchy in the signal transduction cascade could be involved. COP/DET/FUS are a group of evolutionarily conserved proteins that represent central repressors of photomorphogenesis including chloroplast development [11]. No changes were detected in the expression of COP1, COP10, COP11, and DET1. COP9 complex acts as a suppressor of chloroplast development [5, 23]. Unexpectedly, we found that COP9 (PP010178) and FUSCA 12 (FUS12)/COP9 signalosome complex subunit 2 (PP014936) were expressed at higher levels in green curds than in white curds. Such higher expression could be a result of a negative feedback as the case of SPA1, a partner of COP1, which is frequently found to be light induced [24, 25]. PIFs are another group of regulators that repress photomorphogenesis. No changes were observed for the expression of PIF3 and PIF4 in the green vs. white curds. Interestingly, the transcript of PIL2 (PP058986) was increased in the green curd mutant.
In contrast to those photomorphogenesis repressors, HY5 is a key regulator that promotes photomorphogenic development in all light conditions and directly regulates the light-responsive gene expression [8, 9, 26–28]. Here, we found that BoHY5 (PP014970 and PP017071) and BoHY5-HOMOLOG (PP001428) were expressed at higher levels in green curds than white curds (Table 1 and Figure 4c). A recent study on genome-wide mapping of the Hy5-mediated gene networks in Arabidopsis reveals that HY5 could potentially bind to 11,797 genes with 2770 and 2191 being light and dark regulated genes, respectively [26]. Sequence comparison with the HY5-targeted genes in Arabidopsis revealed that a total of 2616 cauliflower HY5-targeted homolog genes were differentially expressed in green curds (Figure 4a). Among them included 1600 up-regulated genes and 1016 down-regulated genes (Additional file 4 and 5). All of the 1600 up-regulated genes were found to be HY5-targeted genes in the light, while 48 down-regulated genes were HY5-targeted genes in the dark (Figure 4b). Among the 1600 up-regulated HY5-targeted genes were 98 transcription factors, including ARABIDOPSIS THALIANA HOMEOBOX 1 (ATHB-1, PP003454), PHYTOCHROME-ASSOCIATED PROTEIN 1 (PP002569), PHYTOCHROME INTERACTING FACTOR 3-LIKE 2 (PP058986) and INDOLE-3-ACETIC ACID INDUCIBLE (IAA1, PP013005). Forty-four transcription factors including RAP2.2 (PP072648), APETALA1 (PP029050), AUXIN RESPONSE FACTOR 6 (PP006891), and SHORT HYPOCOTYL 2 (PP011787) were down-regulated in green curds. The significant alteration of a large number of transcription factors could cause profound effects on chloroplast biogenesis and/or other processes.
Comparison of the numbers of Hy5 -mediated genes in Arabidopsis [26]with differentially expressed genes in green curds of cauliflower and a simplified model of light signaling pathway for chloroplast development based on [11]. (a) Venn diagram showing the number of common genes between HY5-targeted genes in Arabidopsis and total differentially expressed genes in green curds of cauliflower. (b) Venn diagrams showing the numbers of common genes between HY5-regulated genes in Arabidopsis under light and dark and the up- and down-expressed genes among the 2616 homologs of HY5-trageted genes in green curds of cauliflower. (c) Alignment of the associated differentially expressed genes with light signaling pathway for chloroplast development. Abbreviations are as follows: CIP1, COP1-INTERACTIVE PROTEIN 1; COP1, CONSTITUTIVE PHOTOMORPHOGENIC 1; COP9, CONSTITUTIVE PHOTOMORPHOGENIC 9; COP9-sub2, COP9 SIGNALOSOME COMPLEX SUBUNIT 2; HY5, ELONGATED HYPOCOTYL 5; HYH, HY5-HOMOLOG.
Discussion
Large-scale transcriptome sequencing by next generation sequencing platforms such as the Illumina GA sequencing system has been proven to be a powerful and efficient approach for gene expression analysis at the genome level and offers several advantages over microarray technologies [29]. Since the RNA-seq approach provides digital representation of the gene abundance and the statistics are well modeled by the Poisson distribution, even a single replication has been shown to be adequate [30]. Currently, the RNA-seq approach has been widely used to investigate transcriptomes of plants and animals, especially for those having whole genome sequences [31]. A number of tools to map RNA-seq data to reference genomes and to quantify the expression of transcripts have been developed [32]. However, relatively fewer reports have shown studies on using the RNA-seq approach for organisms without reference genomes. In this report we employed the RNA-seq approach to investigate the gene expression changes in a green curd mutant in order to elucidate the genetic basis of chloroplast biogenesis and development. RNA-seq reads along with publicly available ESTs of cauliflower were assembled de novo using a novel assembly strategy as described in the Methods section. A total of 118, 000 unigenes were obtained and 7155 genes showed at least 3-fold changes in expression in green curd mutant. Among them, a large number of genes involved in photomorphogenesis including chloroplast development were revealed, demonstrating a successful use of the RNA-seq approach to profile gene expression in a species without a fully sequenced genome.
Chloroplast biogenesis and development proceed with the coordinated action of many processes [3, 4]. Both environmental signals and plastidic/nuclear factors affect these processes. Light regulation of chloroplast development has been well-documented [3, 4, 33]. The light signaling pathways are composed of phytochromes, transcription factors and numerous intermediates which control photomorphogenesis including chloroplast development. The COP/DET/FUS proteins are suggested to have a function in suppressing chloroplast development in non-photosynthetic tissues [4]. Loss of function mutation of these regulators, such as cop1 and det1, has been shown to result in ectopic chloroplast development, leading to greening in Arabidopsis roots [5, 6]. The fact that the transcripts of COP1 and DET1 remained unchanged and a large number of light-responsive genes were altered in green curds of cauliflower suggests that other regulatory genes in the hierarchy of photomorphogenic regulation are responsible for chloroplast development in the green curd.
In the light signaling cascade, HY5 plays an important role in light signaling and chloroplast development. HY5 receives upstream signals and activates a large number of genes by directly binding to the G-box in the promoters of these genes [9, 26, 27]. Here, we observed higher level of HY5 transcript in the green curd mutant. Furthermore, 2616 cauliflower homologs of HY5-targeted genes were differentially expressed in green curds. Noticeably, among the 2616 genes, 1600 were up-regulated genes in green curd cauliflower. The fact that all 1600 up-regulated genes were the HY5-targeted genes in the light suggests an important role of elevated expression of BoHY5 in mediating chloroplast development in green curd cauliflower mutant. Furthermore, it is known that COP1 negatively controls HY5 activity [12]. Although COP1 was expressed at the same level between green curds and white curds, we found that CIP1 was significantly reduced in green curds (Figure 4c). Arabidopsis CIP1 is associated with the cytoskeleton and has been hypothesized to affect partitioning of COP1 in the nucleus and cytoplasm [34]. It is possible that COP1 activity in the nucleus might be affected by low level of CIP1, causing ectopic chloroplast development in green curds. Thus, BoHY5 and/or the other genes at the high hierarchy in the signal transduction cascade could be responsible or work in concert to regulate chloroplast biogenesis and development in otherwise white tissue to give rise to the striking green curd mutant phenotype.
Ultimately, the development of chloroplasts requires the coordinated action of a number of processes, including the biosynthesis of photosynthetic complexes, transportation of nuclear encoded proteins into chloroplasts, processing of the imported proteins, and assembly of the photosynthetic apparatus [3, 4]. Indeed, many genes involved in photosynthetic pigment biosynthesis along with pigment-binding proteins such as chlorophyll a/b binding proteins were discovered to be upregulated in our genome-wide profiling of green curd cauliflower. The majority of chloroplast proteins are nucleus-encoded and enter the chloroplasts via the Toc/Tic translocon complexes [1]. The increased expression of Toc genes in the green curd mutant supports an enhanced activity of chloroplast-targeted protein import. The imported proteins are folded and processed to form functional proteins. Molecular chaperones HSP70 and Cpn60 have long been known to be involved in this process [3, 20]. A recent study shows that a protein disulfide isomerase is also required for protein folding [21]. Consistent with the increased activity of protein import, genes associated with protein folding and assembling were expressed highly in the green curd mutant for chloroplast development.
Conclusions
In the present study, we compared gene expression on a genome-wide scale by using RNA-seq in a species without a reference genome. This study identified a great number of genes associated with chloroplast development and suggested the potential role of elevated expression of BoHY5 and/or other regulatory genes in the high hierarchy of light signaling pathways for the ectopic chloroplast development in green curd cauliflower. Our results indicate that RNA-seq as a powerful tool in a genomic era could accelerate the functional identification of genes and aid in dissecting the genetic basis of naturally-occurring variations in crops.
Methods
Plant materials
White curd cauliflower cultivar Stovepipe (Brassica oleracea L. var. botrytis) and the green curd mutant line ACX800 were used in this study. Cauliflower plants were grown either in a greenhouse under 14-h-light/10-h-dark cycle at 23°C or in a field. In the greenhouse, the natural daylight was supplied by full-spectrum lamps with the light intensity at 400 μmol photons m- 2 s-1. Fresh curd tissues were harvested, immediately frozen in liquid nitrogen, and stored at -80°C for RNA extraction and chlorophyll extraction.
RNA extraction and construction of cDNA library for sequencing
Total RNA was extracted from pooled curd tissue using the TRIzol reagent according to the manufacturer's instruction (Invitrogen, Carlsbad, CA), and was further purified with the RNeasy®Plant Mini Kit (Qiagen, Valencia, CA). The cDNA libraries of green and white cauliflower from five micrograms of total RNA were constructed using the mRNA Sequencing Sample Preparation Kit following the manufacturer's instruction (Illumina, San Diego, CA, USA). Sequencing was carried out on an Illumina/Solexa Genome Analyzer II system at the Cornell University Life Sciences Core Laboratories Center.
RNA-seq data processing and analysis
The raw Illumina RNA-seq reads were first processed to remove low quality regions and adaptor sequences using an in-house perl script. To eliminate rRNA sequence contamination, the reads were then aligned to cauliflower ribosomal RNA (rRNA) sequences using Bowtie [35], allowing up to two mismatches. A total of ~60,000 cauliflower Sanger ESTs were collected from GenBank in June, 2010. These ESTs were screened against the NCBI UniVec database, the Escherichia coli genome, and cauliflower rRNA sequences, to remove those contaminant sequences. The resulting high quality ESTs were assembled into unigenes using iAssembler http://bioinfo.bti.cornell.edu/tool/iAssembler. The processed Illumina reads were then aligned to the cauliflower EST-unigenes using Bowtie [35], allowing up to two mismatches. A de novo assembly of the unaligned reads was then performed using ABySS [36]. The unigenes assembled from ESTs and unaligned Illumina reads, respectively, were further assembled using iAssembler. Following mapping to EST-unigenes and de novo assembly, transcript count information for sequences corresponding to each unigene were compared to obtain relative expression levels following normalization to RPKM (reads per kilobase of exon model per million mapped reads) [37]. The significance of differential gene expression between the green and white curds was determined using the R statistical method described by Stekel et al. [38] and raw p-values were adjusted for multiple tests using the false discovery rate [39]. Genes with a false discovery rate ≤ 0.01 and a fold change no less than 3 were identified as differentially expressed genes between green and white curds.
To identify biological processes affected in the green curd mutant, the differentially expressed genes were annotated by assigning gene ontology (GO) terms. Potential roles of differentially expressed genes in some specific biological processes were identified. In addition, we created a metabolic pathway database based on the annotation information of the assembled cauliflower unigenes using the Pathway Tools [40]. The pathway database was then integrated into the Plant MetGenMAP system [17] to identify the significantly affected pathways.
Verification of RNA-Seq by quantitative RT-PCR
The cDNA was synthesized using oligo-dT primers and Superscript® reverse transcriptase III (Invitrogen, Carlsbad, CA). qRT-PCR was conducted by using the SYBR Green PCR master mix (Applied Biosystems, CA). The cycling conditions involved denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The dissociation curves were analyzed to verify the specificity of RT-PCR. The relative expression of selected genes was normalized to a cauliflower actin gene [14]. Values reported represent the average of two biological repeats with three independent trials. Gene-specific primers used are listed in Additional file 6.
Chlorophyll determination
Fifty milligrams of curds were ground in liquid nitrogen, and 1 mL of 80% acetone was added to extract chlorophyll. After centrifugation at 12,000 g for 5 min, the supernatant was transferred into the new tube and measured at OD645 and OD663. Chlorophyll concentrations were calculated by using MacKinney's coefficients in the following equations: Chlorophyll a = 12.7*(OD663)-2.69*(OD645) and Chlorophyll b = 22.9*(OD645)-4.48*(OD663) [41].
Confocal analysis of chloroplasts in green curds
Fresh green curd cauliflower tissue was hand-sectioned and examined under Leica TCS SP5 Laser Scanning Confocal Microscope (Leica Microsystems, Exon, PA USA) to detect the autofluorescence of chlorophyll with argon laser excitation at 488 nm and emission filter at 680 nm.
Abbreviations
- COP1:
-
CONSTITUTIVE PHOTOMORPHOGENIC 1
- DET1:
-
DE-ETIOLATED 1
- HY5:
-
ELONGATED HYPOCOTYL 5
- PHYA:
-
PHYTOCHROME A
- PIF3:
-
PHYTOCHROME-INTERACTING TRANSCRIPTION FACTOR 3
- FAR1:
-
FAR-RED-IMPAIRED RESPONSE 1
- CRY2:
-
CRYPTOCHROME 2
- PHOT2:
-
PHOTOTROPIN 2
- LSH7:
-
LIGHT SENSITIVE HYPOCOTYLS 7
- CIP1:
-
COP1-INTERACTIVE PROTEIN 1
- LHCB5:
-
LIGHT HARVESTING COMPLEX OF PHOTOSYSTEM II 5
- GUN5:
-
GENOMES UNCOUPLED 5
- LHCB1.5:
-
PHOTOSYSTEM II LIGHT HARVESTING COMPLEX GENE 1.5
- Toc159:
-
TRANSLOCON AT THE OUTER ENVELOPE MEMBRANE OF CHLOROPLASTS 159
- HSC70-1:
-
CHLOROPLAST HEAT SHOCK PROTEIN 70-1
- FtsH8:
-
ATP-dependent peptidase/ATPase/metallopeptidase
- G4:
-
chlorophyll synthetase.
References
Kessler F, Schnell D: Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr Opin Cell Biol. 2009, 21: 494-500.
Leister D: Chloroplast research in the genomic age. Trends Genet. 2003, 19: 47-56.
Pogson BJ, Albrecht V: Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 2011
Waters MT, Langdale JA: The making of a chloroplast. EMBO J. 2009, 28: 2861-2873.
Wei N, Deng XW: The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 1996, 112: 871-878.
Chory J, Peto CA: Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc Natl Acad Sci USA. 1990, 87: 8776-8780.
Stephenson PG, Fankhauser C, Terry MJ: PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA. 2009, 106: 7654-7659.
Oyama T, Shimura Y, Okada K: The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997, 11: 2983-2995.
Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N: Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998, 10: 673-683.
Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW: The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003, 17: 2642-2647.
Lau OS, Deng XW: Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol. 2010, 13 (5): 571-577.
Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW: Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998, 1: 213-222.
Robertson EJ, Pyke KA, Leech RM: arc6, an extreme chloroplast division mutant of Arabidopsis also alters proplastid proliferation and morphology in shoot and root apices. J Cell Sci. 1995, 108: 2937-2944.
Lu S, Van EJ, Zhou X, Lopez AB, O'Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al: The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell. 2006, 18: 3594-3605.
Paolillo DJ, Garvin DF, Parthasarathy MV: The chromoplasts of Or mutants of cauliflower (Brassica oleracea L. var. botrytis). Protoplasma. 2004, 224: 245-253.
Wu JA, Zhang YL, Zhang HQ, Huang H, Folta KM, Lu JA: Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol. 2010, 10:
Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei Z: Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol. 2009, 151: 1758-1768.
Larkin RM, Alonso JM, Ecker JR, Chory J: GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003, 299: 902-906.
Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J: Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001, 98: 2053-2058.
Latijnhouwers M, Xu XM, Moller SG: Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta. 2010, 232: 567-578.
Shimada H, Mochizuki M, Ogura K, Froehlich JE, Osteryoung KW, Shirano Y, Shibata D, Masuda S, Mori K, Takamiya KI: Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell. 2007, 19: 3157-3169.
Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya Ki: Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA. 1999, 96: 15362-15367.
Wei N, Chamovitz DA, Deng XW: Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994, 78: 117-124.
Hoecker U, Quail PH: The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem. 2001, 276: 38173-38178.
Hoecker U, Tepperman JM, Quail PH: SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999, 284: 496-499.
Zhang HY, He H, Wang XC, Wang XF, Yang XZ, Li L, Deng XW: Genome-wide mapping of the HY5-mediated genenetworks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011, 65: 346-358.
Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao HY, Lee I, Deng X: Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007, 19: 731-749.
Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, et al: Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome a signaling. Plant Cell. 2010, 22: 3634-3649.
Hurd PJ, Nelson CJ: Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief Funct Genomics. 2009, 8: 174-183.
Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al: Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotech. 2000, 18: 630-634.
Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al: The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010, 42: 1060-1067.
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al: Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010, 42: 961-967.
Mullet JE: Chloroplast development and gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1988, 39: 475-502.
Matsui M, Stoop CD, von Arnim AG, Wei N, Deng XW: Arabidopsis COP1 protein specifically interacts in vitro with a cytoskeleton-associated protein, CIP1. Proc Natl Acad Sci USA. 1995, 92: 4239-4243.
Langmead B, Trapnell C, Pop M, Salzberg SL: Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10:
Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE, et al: De novo transcriptome assembly with ABySS. Bioinformatics. 2009, 25: 2872-2877.
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B: Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Med. 2008, 5: 621-628.
Stekel DJ, Git Y, Falciani F: The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000, 10: 2055-2061.
Benjamini Y, Hochberg Y: Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995, 57: 289-300.
Karp PD, Paley S, Romero P: The pathway tools software. Bioinformatics. 2002, 18: S225-S232.
Arnon DI: Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24: 1-15.
Acknowledgements
We thank Yong-Qiang Wang for helping with the analysis of the BoHY5-targeted genes. We thank Lei Li and Huiyong Zhang from University of Virginia for sharing the list of HY5-targeted genes in Arabidopsis. We thank D. Reed for assistance in growing cauliflower in the field. This work was supported by the USDA-ARS base fund and by the CAS/SAFEA International Partnership Program for Creative Research Teams 20090491019. USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
XZ designed the research, conducted molecular and biochemical analyses, and wrote the manuscript. ZF performed the bioinformatics data analysis. TWT participated in the initial design and discussion of the project, and editing of the manuscript. LL conceived the research and participated in the writing of the manuscript. All authors read and approved the final version of the manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zhou, X., Fei, Z., Thannhauser, T.W. et al. Transcriptome analysis of ectopic chloroplast development in green curd cauliflower (Brassica oleracea L. var. botrytis). BMC Plant Biol 11, 169 (2011). https://doi.org/10.1186/1471-2229-11-169
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-11-169