Abstract
Background
During the development of the central nervous system, oligodendrocytes generate large amounts of myelin, a multilayered insulating membrane that ensheathes axons, thereby allowing the fast conduction of the action potential and maintaining axonal integrity. Differentiation of oligodendrocytes to myelin-forming cells requires the downregulation of RhoA GTPase activity.
Results
To gain insights into the molecular mechanisms of oligodendrocyte differentiation, we performed microarray expression profiling of the oligodendroglial cell line, Oli-neu, treated with the Rho kinase (ROCK) inhibitor, Y-27632 or with conditioned neuronal medium. This resulted in the identification of the transmembrane protein 10 (Tmem10/Opalin), a novel type I transmembrane protein enriched in differentiating oligodendrocytes. In primary cultures, Tmem10 was abundantly expressed in O4-positive oligodendrocytes, but not in oligodendroglial precursor cells, astrocytes, microglia or neurons. In mature oligodendrocytes Tmem10 was enriched in the rims and processes of the cells and was only found to a lesser extent in the membrane sheets.
Conclusion
Together, our results demonstrate that Tmem10 is a novel marker for in vitro generated oligodendrocytes.
Similar content being viewed by others
Background
Oligodendrocytes are specialized cells of the central nervous system that produce myelin, a multilayered membrane spirally ensheathing axons and facilitating rapid nerve conduction [1, 2]. The development of oligodendrocytes is a gradual process, in which each step of the differentiation process is characterized by stage specific markers [3–6]. Oligodendrocytes originate from oligodendrocyte precursor cells that arise from multiple foci along the neuronal tube and migrate into the future white matter of the brain. After reaching their final position, they develop into mature post-mitotic cells that produce the myelin sheaths [7]. During the progression through the oligodendroglial lineage, the cells loose their migratory and proliferative capacities and undergo dramatic changes in their morphology by the formation of a highly branched network of processes. This transformation is accompanied by the expression of a number of gene products that are highly enriched or even specific to oligodendrocytes such as the myelin basic protein (MBP), proteolipid proteins (PLP/DM20), myelin-associated glycoprotein (MAG), cyclic nucleotide phosphodiesterase (CNP) and the glycolipids, galactosylceramide and sulfatide. The capacity of oligodendrocyte precursor cells to differentiate into oligodendrocytes that express these different gene products is intrinsic to the lineage and occurs even in the absence of neurons [4, 8]. Oligodendrocytes need to provide specific sorting and transport mechanisms to enable the synthesis of an extensive amount of myelin membrane in a very short time [3, 9]. Since oligodendrocytes must produce myelin at the appropriate time of neuronal development, a number of reciprocal signalling systems are likely to operate to coordinate the organisation of axonal domains and the biogenesis of myelin [10–15]. A number of recent studies have shown that neuronal-derived signalling molecules control the development of myelin-forming glial cells [16–21]. We have recently shown that neurons regulate membrane trafficking in oligodendrocytes [19]. In the absence of neurons, the major myelin protein, PLP, is internalized and stored in late endosomes. After receiving an unknown soluble signal from neurons, oligodendrocytes reduce the rate of endocytosis and increase the retrograde transport of PLP from late endosomes to the plasma membrane. A fraction of PLP is released in association with exosomes [22, 23]. Our previous work shows that changes in Rho GTPase activity were responsible for switching between these two modes of transport [24]. Inactivation of Rho GTPase activity reduced the transport of cargo to late endosomes and at the same time increased the mobilization of membrane from late endosomes. We found that a neuronal soluble factor was responsible for the downregulation of RhoA GTPase activity in the oligodendroglial cell line, Oli-neu [24]. The downregulation of RhoA function during morphological differentiation of oligodendrocytes is supported by a number of other studies [18, 25]. In this study, we were interested in the transcriptional changes that occur after differentiation of Oli-neu cells by conditioned neuronal medium or by inactivation of Rho GTPase function. This effort led to the identification of the transmembrane protein 10 (Tmem10/Opalin) as a novel marker for oligodendrocytes. The transmembrane protein 10 is known as Tmem10/TMEM10 in mice, rats and humans, with the synonyms TMP10 or HTMP10. Recently four Tmem10 homologs of prosimian species (Eulemur macaco, Lemur catta, Microcebus murinus and Otolemur garnetti) have been named Opalin [26]. In this work the human, rat and mouse transmembrane protein 10 will be referred to as Tmem10.
Results and Discussion
As a cellular model for oligodendrocyte differentiation we use the oligodendroglial cell line, Oli-neu. The advantage of this system is that morphological differentiation of a pure oligodendroglial culture can be triggered synchronously by adding conditioned medium from primary neuronal cultures to the cells. To characterize the gene changes that occur after incubation of Oli-neu cells with conditioned neuronal medium, we used Affymetrix microarrays. Oli-neu cells were incubated for 16 hours with conditioned neuronal medium and compared to untreated parallel cultures. Cell fractions were used to purify mRNA for microarray analysis (the data is available at NCBI Geo accession number GSE10291). Using a 1.7 fold cut-off, we found that 716 genes were up-regulated, whereas 386 genes were down-regulated by conditioned neuronal medium as compared to the control. The eighty most highly induced genes are shown in Table 1. We have previously shown that incubation of Oli-neu cells with conditioned neuronal medium leads to RhoA GTPase inactivation. We, therefore, performed gene expression profiling analysis after treating cells with the Rho kinase (ROCK) inhibitor, Y27632 for 16 hours and compared the transcriptional changes to the ones obtained after incubation with conditioned neuronal medium. Interestingly, 70% of the genes that were controlled by both conditioned neuronal medium and by treatment with Y27632 were regulated into the same direction, indicating that these treatments affected the fate of the cells into a common path (Table 2). Some of these genes such as the UDP galactosyltransferase 8a and CNP are known to be upregulated during the progression through the oligodendroglial lineage [4], whereas known housekeeping genes such as the ATP synthase served as internal controls and were not differentially expressed after incubation with conditioned neuronal medium or Y27632 (data not shown). One so far not characterized, but highly upregulated gene is the brain-specifically expressed, Tmem10 [27]. For our further study, we decided to focus on Tmem10 for a number of reasons. Tmem10 was the strongest up-regulated gene in our analysis of transcriptional changes induced by conditioned neuronal medium and as well highly up-regulated after treatment with Y27632. The analysis of the average fold up-regulation in both conditions shows that Tmem10 is the most up-regulated gene together with Cyp2c39 (cytochrome P450, family2, polypeptide 39) (Table 3). In addition, a previous microarray analysis identified a 23.75 fold up-regulation of Tmem10 during oligodendrocyte differentiation [28] and in situ expression data of Tmem10 in the Allen Brain Atlas http://www.brain-map.org suggested enrichment in the white matter of the brain.
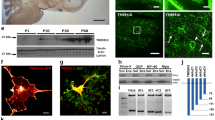
To begin our characterization of Tmem10 we raised an antibody against the C-terminal part of the protein and performed immunofluorescence analysis on primary cultures of oligodendrocytes. We did not detect Tmem10 on NG2-positive oligodendrocyte precursor cells, whereas pre-oligodendrocytes that were still NG2-positive, but also contained O4 started to express Tmem10 (Fig 1A, B). Higher expression of Tmem10 was identified on NG2-negative and O4-positive or O1-positive oligodendrocytes (Fig 1A, C). Low levels of Tmem10 expression can be detected in A2B5-positive oligodendrocyte progenitors (Fig 1D). Colocalization studies of Tmem10 with MBP indicated that Tmem10 was present in mature oligodendrocytes where it was enriched in the rims and processes of the cells and was found only to a lesser extent in the membrane sheets (Fig. 1E). Comparisons of Tmem10 expression in O4-positive/MBP-negative and MBP-positive cells shows that Tmem10 is redistributed to the rims of the membrane sheets, but the expression level does not change significantly compared to O4-positive oligodendrocytes (Fig 1B).
Tmem10 is expressed during oligodendrocyte differentiation. (A) Primary oligodendrocytes were cultured for 2–3 days and levels of Tmem10 (green) expression were determined by immunofluorescence. The developmental stage of the cells was determined by costaining for NG2 (red) or O4 (blue). (B) Quantitative analysis of the Tmem10 immunofluorescence intensities. Values represent the mean ± SEM (n > 20 cells, ***p < 0,001). (C) Tmem10 (green) is expressed in O1 (red) positive oligodendrocytes (day 2) and (D) Tmem10 (green) is weakly expressed in A2B5 (red) positive cells (day 2). (E) Tmem10 (green) localizes to processes and rims of mature oligodendrocytes, expressing MBP (red) (day 5). Scale bars, 10 μm.
Interestingly, double labelling of Tmem10 with GFAP or neuron specific βIII Tubulin showed that Tmem10 could not be detected on GFAP-positive astrocytes or neurons, which are present as a minor cell population in the same cultures, indicating a specific expression of Tmem10 in differentiating oligodendrocytes (Fig. 2A, B). Additionally, we analysed primary cultures of astrocytes and microglia cultures, but could not detect Tmem10 on either of these cell types (Fig. 2C, D).
Tmem10 is not detected on neurons, astrocytes and microglia. (A) Tmem10 (red) is specifically expressed in oligodendrocytes, but absent form GFAP-positive astrocytes (GFAP, green). (B) Tmem10 (green) is enriched in oligodendrocytes, labelled with O4 (blue) compared to neurons stained for neuronal βIII Tubulin (red). Tmem10 (green) is not detectable on (C) astrocytes (GFAP, red) and (D) microglia (stained with isolectin IB4 conjugated to Alexa Fluor 568, red) in cultures enriched for these cells types. Scale bars, 10 μm.
The analysis of the primary amino acid sequence of Tmem10 for protein domains and functional sites with InterProScan [29] revealed a predicted signal peptide (amino acid 1–15 in the mouse sequence) and a putative transmembrane domain (amino acid 31–51 in the mouse sequence) (Fig. 3A). To further characterize the protein structure and the membrane orientation of Tmem10, we used N-terminal ECFP- and C-terminal EYFP-fusion proteins of Tmem10. Oli-neu cells were transfected with either of the fusion proteins and live staining was performed at 4°C with anti-GFP antibody to specifically label the proteins at the cell surface. We found that only the N-terminal ECFP-fusion protein was detectable by surface staining, whereas premeabilization of the cells uncovered both fusion proteins (Fig. 3B). These results show that Tmem10, as predicted from its primary amino acid sequence, is a type I membrane protein.
Tmem10 is a type I transmembrane protein. (A) Alignment of mouse, rat and human Tmem10 amino acid sequence. Black/grey box indicates the predicted position of the transmembrane domain. (B) Oli-neu cells were transfected to express either N-terminal ECFP-tagged or C-terminal EYFP-tagged Tmem10. Subsequent staining with anti-GFP antibody was performed either on living, unpermeabilized (surface anti-GFP) or on fixed, permeabilized (after permeabilization anti-GFP) cells. Scale bars, 10 μm.
After ectopic expression of both Tmem10 fusion proteins and an untagged expression construct, we found that it was mainly localized at the plasma membrane with only very little intracellular staining. Tmem10 appeared to be enriched in actin-rich membrane ruffles at the cell surface as shown by its colocalization with rhodamine-phalloidin (Fig 4A). Additionally, comparison of Tmem10 transfected Oli-neu with untransfected control cells shows that our anti-Tmem10 antibody specifically recognizes Tmem10 (Fig 4A).
Tmem10 colocalizes with actin in processes and membrane ruffles. (A) Oli-neu cells were transfected with a plasmid encoding for rat Tmem10 and subsequently stained for Tmem10 (green) and Actin (red) with rhodamin-labelled phalloidin. Note the colocalization of Tmem10 with Actin in processes and membrane ruffles. The absence of Tmem10 labelling in untransfected cells shows the specificity of the generated anti-Tmem10 antiserum. (B, C) Oli-neu cells were transfected with Tmem10-EYFP and treated with 2 μM latrunculin A for 30 min 16 h after transfection. (B) FRAP was measured by bleaching a squared region of interest within the cell body and fluorescence recovery in this regions was examined. Average FRAP tracings for 15 cells form 2 independent experiments are shown. (C) Tmem10-EYFP (green) accumulates in intracellular sites (arrow head) after disruption of the F-actin (red) cytoskeleton with latrunculin A. Scale bar, 10 μm.
To investigate the role of the actin cytoskeleton on the lateral mobility of Tmem10 we performed fluorescence recovery after photobleaching (FRAP) experiments in the presence or absence of the F-actin disrupting drug, latrunculin A. The mobile fraction and the half time of fluorescence recovery did not change after treatment with latrunculin A (Fig 4B), indicating that the lateral mobility of Tmem10 within the plasma membrane is not dependent on a functional cortical actin cytoskeleton. Interestingly, we did observe a redistribution of a fraction of plasma membrane Tmem10 into intracellular sites after treatment with latrunculin A (Fig 4C), suggesting a role of the actin cytoskeleton in keeping Tmem10 at the cell surface.
In summary, we have applied an expression profiling approach to identify genes upregulated during Oli-neu cell differentiation. We used two different experimental approaches – incubation with conditioned neuronal medium or treatment with the Rho kinase (ROCK) inhibitor, Y-27632. This procedure led to the identification of Tmem10, a protein that appears during oligodendrocyte differentiation. In cultured oligodendrocytes, Tmem10 was absent from bipolar precursor cells and started to be expressed after the cells had acquired the O4 epitope.
A previous comparative genome analysis suggested that Tmem10 is a mammalian-specific gene [26]. Interestingly, the comparison of the genome structure of the Tmem10 gene and its flanking region identified an evolutionary conserved region within the first intron that functions as an oligodendrocyte-specific enhancer. This domain contains binding sites for Myt1 and cAMP-response element binding protein (CREB) and the treatment of Oli-neu cells with cAMP enhanced the expression of Tmem10 [26]. Previous studies have already shown that cAMP regulates the expression of several other oligodendroglial-specific genes [30], suggesting a general role for cAMP dependent signalling in the differentiation of oligodendrocytes into myelin-forming cells. Another factor that appears to regulate Tmem10 gene expression was the leukaemia inhibitory factor (LIF) [26], which seems to be released by astrocytes in response to ATP secreted by neurons and to promote myelination by mature oligodendrocytes [31]. Together, these data suggest a function for Tmem10 in an oligodendroglial specific process. Our finding that Tmem10 colocalizes with F-actin in plasma membrane ruffles and in F-actin-rich processes, points to a role in the regulation of the oligodendroglial actin cytoskeleton. This is reminiscent to another oligodendroglial-specific protein, Ermin, which has been implicated in the regulation of cell morphology by modulating the actin cytoskeleton [32]. The localization of Tmem10 to the leading edge of myelin sheets in mature oligodendrocytes suggests a role for Tmem10 in myelin membrane sheet extension. As Tmem10 localizes to the growing tip of the myelin sheet it could also be involved in the process of recognition or adhesion to potential axonal targets. Further analysis will be required to elucidate these issues.
Conclusion
During the development of the nervous system oligodendrocytes form a highly branched network of processes and several oligodendroglial-specific genes such as Ermin, CNP and Tmem10 are expressed during this process. Gene profiling using microarrays is a useful starting point to identify genes relevant to oligodendrocyte differentiation and myelination [28, 33, 34]. Functional analysis of these proteins, as performed for CNP [35], will be required to elucidate their exact biological function in the generation of myelin-forming processes.
Methods
Cell culture, transfections and immunofluorescence
Primary cultures of mouse oligodendrocytes were prepared as described previously [36]. In brief, cells were plated in MEM containing B27 supplement, 1% horse serum, L-thyroxine, tri-iodo-thyronine, glucose, glutamine, gentamycine, pyruvate, and bicarbonate on poly-L-lysine coated glass-coverslips after shaking. The minor population of neurons and astrocytes which arise together with the oligodendrocytes in the mixed brain cultures were used to assess Tmem10 expression in other cell types. Primary cultures of microglia and astrocytes were prepared as described previously [37]. In brief, microglial cells were shaken off, centrifuged and plated on poly-L-lysine coated coverslips in D-MEM containing 10% FCS, glutamine, penicillin and streptomycin. The remaining astrocytes were trypsinized, centrifuged and plated on poly-L-lysine coated coverslips in D-MEM containing 10% FCS, glutamine, penicillin and streptomycin. The oligodendroglial cell line Oli-neu was cultured as described previously [38]. For microarray experiments we used Oli-neu cells stably expressing PLP-EGFP [19]. Cells were treated for 16 h with conditioned neuronal medium or Y27632 (Calbiochem). Conditioned neuronal medium was obtained from primary cultures enriched in neurons after culturing for 2 weeks and used directly as described previously [24]. Transient transfections were performed using FuGENE transfection reagent (Roche Diagnostics, Basel, Switzerland) according to the manufacturer's protocol. Immunofluorescence was performed as described previously [19]. For surface labelling of ECFP-Tmem10 or Tmem10-EYFP transfected, living cells were incubated with anti-GFP antibody in medium for 10 min at 4°C, washed, fixed and labelled with secondary antibody. Disruption of the actin cytoskeleton was done 16 h after transfection with 2 μM latrunculin A for 30 min at 37°C.
Antibodies and plasmids
The following plasmids were used: human Tmem10 cDNA C-terminally fused with EYFP or N-terminally fused with ECFP [39] generated from modified pECFP-C1 or pEYFP-N1 expression vectors (Clonetech, Heidelberg, Germany), rat Tmem10 cDNA subcloned in pExpress-1 expression vector purchased form RZPD (Deutsches Ressourcenzentrum für Genomforschung GmbH, Berlin, Germany).
Anti-Tmem10 antiserum was induced in rabbits against the C-terminal Tmem10 sequence LERRRGLWWLVPSLSLE and the affinity purified IgG fraction was used. Peptid synthesis, immunization of the rabbit and affinity purification was carried out by Davids Biotechnology (Regensburg, Germany). Further the following primary antibodies were used: A2B5 (mouse monoclonal IgM, Chemicon (Millipore)), GFAP (mouse monoclonal IgG1; Vision BioSystems Novocastra, New Castle upon Tyne, UK), GFP (rabbit polyclonal IgG; Abcam, Cambridge, UK), MBP (mouse monoclonal IgG1; Sternberger Inc., Lutherville, MD), NG2 (rat IgG) [40], O1 (monoclonal IgM) [41], O4 (monoclonal IgM) [41], Tmem10 (rabbit polyclonal IgG fraction), neuron specific βIII Tubulin (mouse monoclonal IgG1; Promega, Madison, WI). Microglial cells were stained with isolectin IB4 conjugated to Alexa Fluor 568 from Molecular Probes (Invitrogen, Carlsbad, CA). Secondary antibodies were purchased from Dianova (Hamburg, Germany) and rhodamin-labelled phalloidin from Molecular Probes (Invitrogen, Carlsbad, CA).
RNA isolation
RNA isolation was performed using the Trizol (Invitrogen, Carlsbad, CA) method according to the manufacturer's recommendations and stored at -80°C. Afterwards, the samples were DNAse I treated in order to remove genomic DNA contaminations. RNA quality was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies) microfluidic electrophoresis. Only sample pairs with comparable RNA integrity numbers were selected for microarray analysis.
Experimental design and sample preparation for 2 colour-microarrays
For gene expression profiling, a two-colour 1 × 2 design including a dye swap using 6 arrays was applied, comparing Oli-neu cells stably expressing PLP-EGFP treated for 16 h with conditioned neuronal medium or Y27632, respectively, to untreated parallel cultures.
The samples for hybridization were prepared from total RNA according to the Atlas SMART Fluorescent Probe Amplification Kit (Clonetech-Takara Bio Europe) protocol, except, that the RNA template was hydrolyzed under alkaline conditions before cDNA purification, and the PCR amplification process was monitored and stopped in the exponential phase. Quantity and Cy-dye incorporation rates of the generated target material were assessed using a NanoDrop ND-100. Cy3- and Cy5-labelled cDNA fragments, respectively, were hybridized to Agilent Technologies 44 K Mouse Whole Genome Microarrays (G4112A) for 17 h at 65°C. Post-processing washes were done according to the Agilent Technologies SSPE protocol (v2.1), replacing wash solution 3 by acetonitril, followed by immediate scanning using an Agilent G2505B scanner. Intensity data were extracted using the software 'Automatic Image processing for Microarrays'.
Statistical analysis
Normalization of the raw intensity data was done with a non-linear loess regression [42].
Uni- and multivariate designs
Differentially expressed genes were identified by an ANOVA fixed effects model [43]. The resulting P-values were adjusted with the Benjamini-Hochberg method to control the False-Discovery-Rate [44]. Normalization and statistical computation was done for two independent datasets derived from a high gain and a low gain scan, allowing replacement of saturated features in the high gain scan with data from the low gain measurement.
Sequence analysis
The prediction of the transmembrane domain of Tmem10 was done using the InterProScan algorithm provided at EMBL-EBI [29].
Microscopy and analysis
Fluorescence images were acquired on a confocal laser scanning microscope (TCS SP equipped with AOBS, Leica) with a 40× or 63× oil plan-apochromat objective (Leica). Image processing and analysis was performed using Meta Imaging Series 6.1 software (Universal Imaging Corporation). Quantification of fluorescence intensities was performed as described previously [19]. FRAP experiments were done as described in [20].
References
Sherman DL, Brophy PJ: Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci. 2005, 6: 683-690. 10.1038/nrn1743.
Simons M, Trotter J: Wrapping it up: the cell biology of myelination. Curr Opin Neurobiol. 2007
Pfeiffer SE, Warrington AE, Bansal R: The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993, 3: 191-197. 10.1016/0962-8924(93)90213-K.
Baumann N, Pham-Dinh D: Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001, 81: 871-927.
Miller RH: Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002, 67: 451-467. 10.1016/S0301-0082(02)00058-8.
Baracskay KL, Kidd GJ, Miller RH, Trapp BD: NG2-positive cells generate A2B5-positive oligodendrocyte precursor cells. Glia. 2007, 55: 1001-1010. 10.1002/glia.20519.
Rowitch DH: Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004, 5: 409-419. 10.1038/nrn1389.
Dubois-Dalcq M, Behar T, Hudson L, Lazzarini RA: Emergence of three myelin proteins in oligodendrocytes cultured without neurons. J Cell Biol. 1986, 102: 384-392. 10.1083/jcb.102.2.384.
Kramer EM, Schardt A, Nave KA: Membrane traffic in myelinating oligodendrocytes. Microsc Res Tech. 2001, 52: 656-671. 10.1002/jemt.1050.
Barres BA, Raff MC: Axonal control of oligodendrocyte development. J Cell Biol. 1999, 147: 1123-1128. 10.1083/jcb.147.6.1123.
Salzer JL: Polarized domains of myelinated axons. Neuron. 2003, 40: 297-318. 10.1016/S0896-6273(03)00628-7.
ffrench-Constant C, Colognato H: Integrins: versatile integrators of extracellular signals. Trends Cell Biol. 2004, 14: 678-686. 10.1016/j.tcb.2004.10.005.
Coman I, Barbin G, Charles P, Zalc B, Lubetzki C: Axonal signals in central nervous system myelination, demyelination and remyelination. J Neurol Sci. 2005, 233: 67-71. 10.1016/j.jns.2005.03.029.
Simons M, Trajkovic K: Neuron-glia communication in the control of oligodendrocyte function and myelin biogenesis. J Cell Sci. 2006, 119: 4381-4389. 10.1242/jcs.03242.
Boiko T, Winckler B: Myelin under construction -- teamwork required. J Cell Biol. 2006, 172: 799-801. 10.1083/jcb.200602101.
Stevens B, Porta S, Haak LL, Gallo V, Fields RD: Adenosine: a neuron-glial transmitter promoting myelination in the CNS in response to action potentials. Neuron. 2002, 36: 855-868. 10.1016/S0896-6273(02)01067-X.
Chan JR, Watkins TA, Cosgaya JM, Zhang C, Chen L, Reichardt LF, Shooter EM, Barres BA: NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron. 2004, 43: 183-191. 10.1016/j.neuron.2004.06.024.
Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, Hession C, Sah D, Trapp B, He Z, Jung V, McCoy JM, Pepinsky RB: LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005, 8: 745-751. 10.1038/nn1460.
Trajkovic K, Dhaunchak AS, Goncalves JT, Wenzel D, Schneider A, Bunt G, Nave KA, Simons M: Neuron to glia signaling triggers myelin membrane exocytosis from endosomal storage sites. J Cell Biol. 2006, 172: 937-948. 10.1083/jcb.200509022.
Fitzner D, Schneider A, Kippert A, Mobius W, Willig KI, Hell SW, Bunt G, Gaus K, Simons M: Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. Embo J. 2006, 25: 5037-5048. 10.1038/sj.emboj.7601376.
Nave KA, Salzer JL: Axonal regulation of myelination by neuregulin 1. Curr Opin Neurobiol. 2006, 16: 492-500. 10.1016/j.conb.2006.08.008.
Kramer EM, Bretz N, Tenzer S, Winterstein C, Mobius W, Berger H, Nave K, Schild H, Trotter J: Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons?. Proteomics - Clinical Applications. 2007, 1: 1446-1461. 10.1002/prca.200700522.
Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M: Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science. 2008, 319: 1244-1247. 10.1126/science.1153124.
Kippert A, Trajkovic K, Rajendran L, Ries J, Simons M: Rho regulates membrane transport in the endocytic pathway to control plasma membrane specialization in oligodendroglial cells. J Neurosci. 2007, 27: 3560-3570. 10.1523/JNEUROSCI.4926-06.2007.
Liang X, Draghi NA, Resh MD: Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J Neurosci. 2004, 24: 7140-7149. 10.1523/JNEUROSCI.5319-03.2004.
Aruga J, Yoshikawa F, Nozaki Y, Sakaki Y, Toyoda A, Furuichi T: An oligodendrocyte enhancer in a phylogenetically conserved intron region of the mammalian myelin gene Opalin. J Neurochem. 2007, 102: 1533-1547. 10.1111/j.1471-4159.2007.04583.x.
Nobile C, Hinzmann B, Scannapieco P, Siebert R, Zimbello R, Perez-Tur J, Sarafidou T, Moschonas NK, French L, Deloukas P, Ciccodicola A, Gesk S, Poza JJ, Lo Nigro C, Seri M, Schlegelberger B, Rosenthal A, Valle G, Lopez de Munain A, Tassinari CA, Michelucci R: Identification and characterization of a novel human brain-specific gene, homologous to S. scrofa tmp83.5, in the chromosome 10q24 critical region for temporal lobe epilepsy and spastic paraplegia. Gene. 2002, 282: 87-94. 10.1016/S0378-1119(01)00846-0.
Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA: Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006, 26: 10967-10983. 10.1523/JNEUROSCI.2572-06.2006.
Zdobnov EM, Apweiler R: InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001, 17: 847-848. 10.1093/bioinformatics/17.9.847.
Raible DW, McMorris FA: Cyclic AMP regulates the rate of differentiation of oligodendrocytes without changing the lineage commitment of their progenitors. Dev Biol. 1989, 133: 437-446. 10.1016/0012-1606(89)90047-X.
Ishibashi T, Dakin KA, Stevens B, Lee PR, Kozlov SV, Stewart CL, Fields RD: Astrocytes promote myelination in response to electrical impulses. Neuron. 2006, 49: 823-832. 10.1016/j.neuron.2006.02.006.
Brockschnieder D, Sabanay H, Riethmacher D, Peles E: Ermin, a myelinating oligodendrocyte-specific protein that regulates cell morphology. J Neurosci. 2006, 26: 757-762. 10.1523/JNEUROSCI.4317-05.2006.
Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD: Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J Neurosci. 2006, 26: 9881-9891. 10.1523/JNEUROSCI.2246-06.2006.
Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA: A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008, 28: 264-278. 10.1523/JNEUROSCI.4178-07.2008.
Lee J, Gravel M, Zhang R, Thibault P, Braun PE: Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol. 2005, 170: 661-673. 10.1083/jcb.200411047.
Simons M, Kramer EM, Thiele C, Stoffel W, Trotter J: Assembly of myelin by association of proteolipid protein with cholesterol- and galactosylceramide-rich membrane domains. J Cell Biol. 2000, 151: 143-154. 10.1083/jcb.151.1.143.
Hirrlinger J, Resch A, Gutterer JM, Dringen R: Oligodendroglial cells in culture effectively dispose of exogenous hydrogen peroxide: comparison with cultured neurones, astroglial and microglial cells. J Neurochem. 2002, 82: 635-644. 10.1046/j.1471-4159.2002.00999.x.
Jung M, Kramer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, Khazaie K, Chlichlia K, von Blankenfeld G, Kettenmann H, et al: Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci. 1995, 7: 1245-1265. 10.1111/j.1460-9568.1995.tb01115.x.
Simpson JC, Wellenreuther R, Poustka A, Pepperkok R, Wiemann S: Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000, 1: 287-292. 10.1093/embo-reports/kvd058.
Niehaus A, Stegmuller J, Diers-Fenger M, Trotter J: Cell-surface glycoprotein of oligodendrocyte progenitors involved in migration. J Neurosci. 1999, 19: 4948-4961.
Sommer I, Schachner M: Monoclonal antibodies (O1 to O4) to oligodendrocyte cell surfaces: an immunocytological study in the central nervous system. Dev Biol. 1981, 83: 311-327. 10.1016/0012-1606(81)90477-2.
Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP: Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002, 30: e15-10.1093/nar/30.4.e15.
Landgrebe J, Bretz F, Brunner E: Efficient two-sample designs for microarray experiments with biological replications. In Silico Biol. 2004, 4: 461-470.
Bretz F, Landgrebe J, Brunner E: Multiplicity issues in microarray experiments. Methods Inf Med. 2005, 44: 431-437.
Acknowledgements
We thank S. Bechtel and S. Wiemann (DKFZ, Heidelberg, Germany) for providing us with the ECFP-Tmem10 and Tmem10-EYFP plasmids and J. Trotter (University of Mainz, Germany) for antibodies. The work was supported by the Deutsche Forschungsgemeinschaft (SFB 523).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
AK carried out cell culture, transfections and immunofluorescence, including microscopy and statistical analysis, performed amino acid sequence alignment, created figures and participated in drafting the manuscript. KT performed cell culture, RNA isolation and sample preparation for gene expression profiling experiments. DF carried out FRAP experiments. LO participated in analysis and presentation of gene expression data. MS developed the design of the study, deduced interpretation of the data and wrote the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kippert, A., Trajkovic, K., Fitzner, D. et al. Identification of Tmem10/Opalin as a novel marker for oligodendrocytes using gene expression profiling. BMC Neurosci 9, 40 (2008). https://doi.org/10.1186/1471-2202-9-40
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2202-9-40