Abstract
Background
Several lines of evidence strongly suggest that agouti-related peptide (AGRP) plays a key role in the regulation of metabolic function but ablation of the AGRP gene has no apparent effect on metabolic function. Since specific pharmacological antagonists of AGRP do not presently exist, we assessed if reduction of hypothalamic AGRP mRNA by RNA interference (RNAI) would influence metabolic function, an outcome suggesting that pharmacological antagonists might constitute useful reagents to treat obesity.
Results
The RNAI protocol specifically reduced hypothalamic expression of AGRP mRNA by 50% and resulted in reduction of AGRP peptide immunoreactivity. Physiologically, the reduction in AGRP levels was associated with increased metabolic rate and reduced body weight without changes in food intake.
Conclusion
AGRP can function to increase body weight and reduce metabolic rate without influencing food intake. The present study demonstrates that RNAI protocols can be used to assess physiological function of neuronal genes in vivo.
Similar content being viewed by others
Background
Agouti-related peptide (AGRP) gene expression is elevated by fasting and in obesity due to leptin insufficiency [1, 2], while reversal of obese phenotypes by adrenalectomy reverses the elevation of AGRP mRNA [3]. Injections of synthetic analogs of AGRP, and of AGRP itself, also stimulate food intake and body weight [4, 5]. Together with the observation that transgenic over-expression of the AGRP gene leads to hyperphagia and obesity [1], these data suggest that antagonism of AGRP may reduce food intake and body weight, thus potentially serving as a therapy for obesity.
RNAI produces sequence-specific reduction of gene expression in various mammalian in vitro systems [6] and most recently, RNAI was shown to suppress the expression and activity of a luciferase transgene in adult mice [7], suggesting that RNAI may become a valuable molecular and possibly clinical tool for use in vivo. A major advantage of RNAI over a similar protocol, using antisense oligonucleotides, is that the use of antisense oligonucleotides to assess chronic effects requires constant or repetitive infusion, is expensive and generally impractical for more than a few days. In contrast, RNAI can be used to study long-term phenotypes (including body weight and memory functions) due to the development of a DNA plasmid-based system to deliver short double-stranded RNA sequences in vivo[8]. While RNAI has been extensively used to assess physiological functions of specific genes in C. elegans and Drosophila [9, 10] and a single recent paper has reported that RNAI can reduce gene expression in vivo (in liver) [7], the applicability of RNAI to assess physiological or neuronal function in mammals has not yet been demonstrated.
Here we show, using both small interfering RNA (siRNA) particles and the plasmid based RNAI delivery system, pSUPER-RNAI, that decreasing endogenous hypothalamic AGRP levels in the central nervous system leads to significant physiological responses including an increase in metabolic rate and a decrease in body weight.
Results and Discussion
Effects of small interfering RNA particles against AGRP
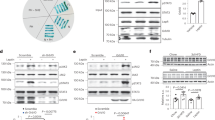
SiRNA particles against both AGRP and GFP were designed based on reports by Elabashir et al. [6] (Fig. 1a). Injection of these siRNA particles into the arcuate nucleus of the hypothalamus reduced AGRP mRNA about 50% 24 hours after injection, compared to the control group injected with siRNA particles targeted to GFP (Fig. 1b and 1c). The 50% decrease in expression level is similar to previously reported effects of RNAI to reduce gene expression from 40–80% both in vitro[6] and in vivo[7] in mammalian systems. Since the construct targeted to AGRP reduced AGRP expression relative to the construct targeted to GFP, this result indicates specific down-regulation by the AGRP RNAI construct.
Small interfering RNA (siRNA) particles decreases AGRP mRNA. (a) Synthetic siRNA particle designed against AGRP mRNA. (b) Representative bands from the Northern blot 24 hours after injection shows decreased levels of AGRP mRNA in the group injected with AGRP-siRNA particle compared to the control GFPi-siRNA injections. (c) Quantification of the Northern blot results yields a statistically significant decrease in expression of 50% with the AGRP-siRNA particle. Data are expressed as a mean percentage ± SEM of GFP-siRNA levels (n = 16; p < 0.05).
The decrease in AGRP mRNA was associated with a striking increase in metabolic rate. Both heat production (Fig. 2a; p < 0.0001) and oxygen consumption, VO2 (Fig. 2b; p < 0.0003) were elevated one day after infusion of the AGRPi construct. Metabolic rate continued to be elevated during day 2 (heat production p < 0.0001; VO2 p < 0.0494). However, by 3 days after infusion, metabolic rate returned to control levels. This result indicates that AGRP normally acts to reduce metabolic rate, consistent with conclusions from indirect evidence based on pair-fed animals treated with AGRP [11].
Decreasing AGRP levels with siRNA increases metabolic rate. (a) Heat production for AGRP-siRNA injected mice was found to be significantly and dramatically elevated one day following surgery compared to the control injected GFP-siRNA mice (n = 16; p < 0.0001). (b) Oxygen consumption, VO2, was also significantly elevated one day after surgery for mice injected with the AGRP-siRNA particles (n = 16; p < 0.0003).
The decrease in AGRP mRNA and increase in metabolic rate was associated with a trend to decrease body weight, but this effect was not significant over the 48 hours after injection (body weight loss of -1.72 ± 0.66 g for the GFPi control mice compared to a loss of -2.85 ± 0.52 g for the AGRPi mice). Surprisingly, decrease in AGRP mRNA did not significantly influence food intake compared to controls (mean intake of 5.0 ± 0.7 g/day for the GFPi control mice compared to 5.3 ± 0.6 g/day for the AGRPi mice). Since both the AGRPi and the control GFPi mice were still recovering from the effects of the surgery and anesthesia at 48 hours, it is not surprising that the effects on body weight were not statistical at this time. Nevertheless the lack of an effect on food intake was unexpected since treatment with synthetic AGRP peptides has profound effects on food intake [4, 5].
Effects of the plasmid based RNAI protocol against AGRP
Since it was anticipated that double-stranded RNAI particles, like antisense oligonucleotides, would produce only very transient reduction in mRNA levels, and indeed the effect on physiological function was only transient, we next assessed effects of AGRP RNAI using a plasmid-based system, pSUPER [8] (Fig. 3a), which leads to the transcription of a short double stranded RNA product with a hair-pin loop in vivo. Using luciferase as a reporter we observed that liposome-mediated transfection of plasmids into brain parenchyma allows significant gene expression for more than one week after infusion [12]. Transfection of neurons in the arcuate nucleus of the hypothalamus led to a decrease in AGRP peptide immunoreactivity, whereas transfection with the control plasmid targeted to GFP had no effect on AGRP immunoreactivity (Fig. 3b,3c). The effect of RNAI to reduce AGRP gene expression was therefore also associated with decreased expression of the AGRP peptide.
pSUPER-AGRPi construct decreases AGRP immunoreactivity. (a) Overall design of the pSUPER-RNAI plasmid construct targeted against AGRP and the predicted transcript with the hairpin loop. (b and c) Immunocytochemistry of fasted mice transfected with the (b) control pSUPER-GFPi vector compared to (c) mice transfected with the pSUPER-AGRPi vector shows decreased AGRP immunoreactivity with the RNAI plasmid construct targeted to AGRP.
The persistent effect of the pSUPER vector to reduce the levels of AGRP peptide was associated with a significant reduction in body weight that persisted over six days (Fig. 4a; p < 0.0001). While the control group recovered body weight initially lost due to surgery, the pSUPER-AGRPi group did not regain the body weight lost. In contrast, as with the double-stranded RNAI construct, infusion of the plasmid-based AGRP RNAI construct did not significantly reduce food intake compared to infusion of the plasmid-based GFP RNAI construct (Fig. 4b; p = 1.6992). Food intake was minimal immediately following surgery, however, both groups of mice recovered to consume four to five grams of food per day. The loss of body weight, without changes in food intake is consistent with increases in metabolic rate observed with the siRNA protocol. This result supports the hypothesis that tonic hypothalamic AGRP reduces metabolic rate independent of food intake, and indeed that metabolic rate may be even more dependent on AGRP tone than is food intake.
Decreasing AGRP levels by pSUPER-AGRPi construct decreases body weight (a) Mice transfected with the pSUPER-AGRPi construct shows persistent loss of body weight after transfection while the mice transfected with the control vector recover body weight over a course of six days (n = 14; p < 0.0001). (b) pSUPER-RNAI targeted to AGRP has no effect on mean daily food intake (n = 14; p = 1.6992).
Conclusions
In summary, the present report represents the first demonstration that RNA interference can be used to reduce expression of an endogenous gene in adult mammals. This protocol has allowed us to assess the physiological function of an endogenous neuronal gene. This study also directly indicates that hypothalamic AGRP regulates metabolic rate. The observation that reduction of AGRP mRNA would produce more profound effects on metabolic rate than on food intake was surprising since infusion of AGRP peptide produces profound effects on food intake [4, 5]. However, since neither AGRP mRNA nor peptide was completely ablated, it seems likely that effects on food intake would become more apparent with a more dramatic manipulation. Although ablation of the AGRP gene was reported to have no effect on metabolic phenotype [13], our results suggest that complete ablation of the gene may have produced metabolic compensation. Thus reducing (but not ablating) AGRP expression in adult mice may have identified a metabolic role for AGRP not apparent from complete ablation of the gene, suggesting that agents antagonizing the effect of AGRP may be useful to treat obesity without producing unacceptable loss of appetite. Consistent with previous findings in mammalian systems [6–8] and unlike other systems such as C. elegans and Drosophila [9, 10], RNAI reduced but did not eliminate expression of AGRP. However, improvements in delivery systems, especially in conjunction with viral-based gene transfer protocols [14], could result in greater and even longer-term reduction of target gene expression suggesting that RNAI will prove to be a useful tool to assess physiological functions of mammalian genes in vivo, especially those expressed in the brain, as RNAI has been so useful in functional mapping of genes in non-mammalian systems.
Methods
Animal and RNAI protocols
The appropriate Institutional Animal Review Board had approved all studies. To study the effects of RNAI, C57Bl/6J mice retired breeders were obtained from The Jackson Laboratory (Bar Harbor, ME). Mice were individually housed with free access to food and water under 12:12 h light-dark cycle (lights on at 07:00 h). Stereotaxic surgery was performed under anesthesia (2.5% 2-2-2-tribromoethanol, 0.015–0.017 ml/g body weight, i.p.). The RNAI constructs were injected bilaterally using a 1 ul Hamilton syringe placed on a stereotaxic frame into the following coordinates from bregma: anterior-posterior -0.12; lateral ± 0.02; vertical -0.6. Double-stranded siRNA particles to reduce expression of AGRP were designed based on Elbashir et al. [6] (Fig. 1a). The siRNA oligonucleotides were designed from the beginning of exon 1 of both AGRP and GFP genes and ordered gel-purified and annealed from Oligos Etc. (Wilsonville, OR). SiRNA particles were re-suspended in Rnase-free water to a concentration of 1 mM and 0.5 ul was injected over 2 minutes by stereotaxic surgery bilaterally. The pSUPER-RNAI plasmid constructs were re-suspended in lipofectin (Invitrogen Corp. Carlsbad, CA) to a concentration of 1 ug/ul, and 1 ul of the mixture was injected bilaterally over five minutes using the same coordinates as for the siRNA particles. Body weight and food intake was monitored daily.
Metabolic rate was monitored continuously by indirect-calorimetry for three days following surgery. Air from each cage was sampled every five minutes, and oxygen and carbon dioxide concentrations from each sample were measured independently. From the change in oxygen, VO2 or oxygen consumption can be calculated after normalizing to body weight. Heat production can then be calculated using the following formula: Heat (cal/hour) = ((4.33+0.67 × (VCO2/VO2)) × VO2 × (Body weight) × 60). Therefore, the parameter VO2 is normalized to body weight, whereas heat production is not normalized to body weight.
For Northern blot analysis, all mice were sacrificed toward the end of the light period (between 17:00 and 19:00 h) by decapitation after a brief exposure to carbon dioxide. Brains were quickly removed and the hypothalamus was dissected out, frozen on dry ice and stored at -70°C until use. For immunocytochemistry, mice were perfused with cold PBS followed by 4% paraformaldehyde. Brains were removed, post-fixed in paraformaldehyde then preserved in 30% sucrose solution for 48 hours.
Northern blot analysis
Total RNA was extracted from tissue using TRIzol (GIBCO BRL, Gaitherburg, MD). Concentration of the samples were measured by UV spectrophotometer and normalized to 1 ug/ul. Uniform concentration and integrity of the samples were verified by agarose gel electrophoresis prior to Northern blot analysis. Six micrograms of total RNA from hypothalamus was subjected to Northern blot analysis, as described previously, to measure AGRP mRNA [15, 16, 2, 3]. Briefly, RNA was denatured by incubating with glyoxal and dimethyl sulfoxide for 1 hour at 50°C and separated on a 1.5% agarose 10 mM NaPO4 (pH = 7.0) gel for 1 hour at 100 V. The RNA was then transferred to an Immobilon S (Millipore) membrane by capillary elution overnight in 20 × standard saline citrate (SSC) buffer. Membranes were briefly washed in 6 × SSC, baked at 80°C and cross-linked by UV light. The membranes were prehybridized in Ultrahyb buffer (Ambion) at 68°C for 2 hours and hydribized overnight with 3.5 × 106 DPM/ml of probe at 42°C. The blots were probed with single-stranded internally labeled DNA probe as described previously [17]. The membranes were washed twice in 1 × SSC/0.1% SDS solution for 15 minutes at room temperature, followed by 0.1 × SSC/0.1% SDS solution for 15 minutes at room temperature twice and then for 3 hours at 42°C. The membranes were then exposed to phosphoimager screen overnight. The total integrated densities of hybridization signals were determined by phosphoimager (STORM 860, Molecular Dynamics, Sunnyvale, CA). To monitor RNA loading, membranes were re-probed and hybridized with a 32P-labeled probe encoding 18S ribosomal RNA.
Immunocytochemistry
Cryopreserved brains were cut to a thickness of 30 um in a cryostat at -20°C and stored in PBS-B buffer (10 mM PO4 in 0.9% NaCl) containing 0.1% sodium azide at 4°C. Immunocytochemistry was performed with a rabbit polyclonal primary antibody to AGRP peptide diluted 1 to 10,000 (Phoenix Pharmaceuticals, Belmont, CA) followed by Vectastain ABC kit (Vector Laboratories, Burlingame, CA). The immunoreactivity was visualized with the Vector VIP kit (Vector Laboratories, Burlingame, CA). Sections were mounted on microscopic slides, air-dried and cover slipped with VectaMount (Vector Laboratories, Burlingame, CA).
Statistical Analysis
Statistical analysis for the Northern blot was performed by student's t-test, using the JMP statistical package implemented on the Macintosh operating system. A p-value of less than 0.05 was considered significant. For the analysis of the metabolic cage data, a two-way ANOVA (time × RNAI construct) was performed using the JMP statistical package. For analysis of food intake and body weight, a two-way ANOVA (time × RNAI construct) followed, where indicated, by student's t-test (p < 0.05 was considered significant) was performed to compare each independent time point.
References
Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS: Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997, 278 (5335): 135-138. 10.1126/science.278.5335.135.
Mizuno TM, Mobbs CV: Hypothalamic agouti-related protein messenger ribonucleic acid is inhibited by leptin and stimulated by fasting. Endocrinology. 1999, 140 (2): 814-817. 10.1210/en.140.2.814.
Makimura H, Mizuno TM, Roberts J, Silverstein J, Beasley J, Mobbs CV: Adrenalectomy reverses obese phenotype and restores hypothalamic melanocortin tone in leptin-deficient ob/ob mice. Diabetes. 2000, 49 (11): 1917-1923.
Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD: Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature. 1997, 385 (6612): 165-168. 10.1038/385165a0.
Rossi M, Kim MS, Morgan DG, Small CJ, Edwards CM, Sunter D, Abusnana S, Goldstone AP, Ruseell SH, Stanley SA, Smith DM, Yagaloff K, Ghatei MA, Bloom SR: A C-terminal fragment of Agouti-related protein increases feeding and antagonizes the effect of alpha-melanocyte stimulating hormone in vivo. Endocrinology. 1998, 139 (10): 4428-4431. 10.1210/en.139.10.4428.
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T: Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001, 411 (6836): 494-498. 10.1038/35078107.
McCaffrey AP, Meuse L, Pham T-TT, Conklin DS, Hannon GJ, Kay MA: RNA Interference in Adult Mice. Nature. 2002, 418 (July): 38-39. 10.1038/418038a.
Brummelkamp TR, Bernards R, Agami R: A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002, 296 (5567): 550-553. 10.1126/science.1068999.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mell CC: Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998, 391 (6669): 806-811. 10.1038/35888.
Misquitta L, Paterson BM: Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc Natl Acad Sci U S A. 1999, 96 (4): 1451-1456. 10.1073/pnas.96.4.1451.
Small CJ, Kim MS, Stanley SA, Mitchell JR, Murphy K, Morgan DG, Ghatei MA, Bloom SR: Effects of chronic central nervous system administration of agouti-related protein in pair-fed animals. Diabetes. 2001, 50 (2): 248-254.
Katsel PL, O'Connell B, Mizuno TM, Mobbs CV, Greenstein RJ: Liposome mediated gene transfer into GH3 cells, and rat brain, liver and gut: comparison of different polar or aliphatic domains. Int J Surg Investig. 2000, 1 (5): 415-429.
Qian S, Chen H, Weingarth D, Trumbauer ME, Novi DE, Guan X, Yu H, Shen Z, Feng Y, Frazier E, Chen A, Camacho RE, Shearman LP, Gopal-Truter S, MacNeil DJ, Van der Ploeg LH, Marsh DJ: Neither agouti-related protein nor neuropeptide Y is critically required for the regulation of energy homeostasis in mice. Mol Cell Biol. 2002, 22 (14): 5027-5035. 10.1128/MCB.22.14.5027-5035.2002.
Kaplitt MG, Makimura H: Defective viral vectors as agents for gene transfer in the nervous system. J Neurosci Methods. 1997, 71 (1): 125-132. 10.1016/S0165-0270(96)00132-X.
Mizuno TM, Kleopoulos SP, Bergen HT, Roberts JL, Priest CA, Mobbs CV: Hypothalamic pro-opiomelanocortin mRNA is reduced by fasting and in ob/ob and db/db mice, but is stimulated by leptin. Diabetes. 1998, 47 (2): 294-297.
Mizuno TM, Makimura H, Silverstein J, Roberts JL, Lopingco T, Mobbs CV: Fasting regulates hypothalamic neuropeptide Y, agouti-related peptide, and proopiomelanocortin in diabetic mice independent of changes in leptin or insulin. Endocrinology. 1999, 140 (10): 4551-4557. 10.1210/en.140.10.4551.
Mizuno TM, Bergen H, Funabashi T, Kleopoulos SP, Zhong YG, Bauman WA, CV Mobbs: Obese gene expression: reduction by fasting and stimulation by insulin and glucose in lean mice, and persistent elevation in acquired (diet-induced) and genetic (yellow agouti) obesity. Proc Natl Acad Sci U S A. 1996, 93 (8): 3434-3438. 10.1073/pnas.93.8.3434.
Acknowledgements
The authors would like to thank I-Wei Shu for his expertise in immunocytochemistry and Chineta L. Pullin for her help in cloning.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
HM participated in the design and coordination of the study, implemented the molecular biological cloning of the RNAI constructs, carried out the animal protocols, processed the tissues, performed Northern blot analysis and immunocytochemistry, performed statistical analysis and drafted the manuscript. TMM participated in the design of the study and the animal protocols. JWM participated in the animal protocols and immunocytochemistry. RA provided the pSUPER construct and provided useful discussion. CVM conceived of the study, participated in the design and coordination of the study and drafted the manuscript.
All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Makimura, H., Mizuno, T.M., Mastaitis, J.W. et al. Reducing hypothalamic AGRP by RNA interference increases metabolic rate and decreases body weight without influencing food intake. BMC Neurosci 3, 18 (2002). https://doi.org/10.1186/1471-2202-3-18
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2202-3-18