Abstract
Background
The mammalian homologue of Seven in Absentia (Siah) can act in the ubiquitin/proteasome pathway. Recent work has shown that Siah can bind group I metabotropic glutamate receptors (mGluRs), but the functional consequences of this interaction are unknown.
Results
The effects of coexpression of Siah on group I mGluR signaling were examined using heterologous expression in rat sympathetic, superior cervical ganglion neurons. Siah1a attenuated heterologously expressed group I mGluR-mediated calcium current inhibition, but was without effect on group II mGluR- or NE-mediated calcium current modulation via heterologously expressed mGluR2 or native a2 adrenergic receptors, respectively, indicating that the effect of Siah was specific for group I mGluRs. Surface expression and subcellular distribution of group I mGluRs were not detectably altered in the presence of Siah1a as assessed by immunoflourescence experiments with epitope tagged receptors and imaging of a GFP/mGluR fusion construct. In addition, an N-terminal Siah deletion construct, which cannot function in the proteolysis pathway, displayed effects similar to the wild type Siah1a. Finally, coexpression of calmodulin, which competes with Siah1a for binding to the C-terminal tail of group I mGluRs, reversed the effect of Siah1a on mGluR-mediated signaling.
Conclusions
These data supported the conclusion that the attenuation of mGluR signaling induced by Siah1a expression was likely a direct consequence of Siah/mGluR association rather than a result of targeting of the receptors to the proteosome. In addition, the data suggest that the binding of CaM and Siah may play an important role in the regulation of group I mGluR function.
Similar content being viewed by others
Background
Metabotropic glutamate receptors (mGluRs) are a family of class 3 G protein-coupled receptors with important functions in the regulation of synaptic transmission, plasticity, neuronal excitability, and the regulation sensory transduction [1–5]. The eight mGluRs (mGluR1-8) and their splice variants have been divided into three groups (I-III) based on sequence homology, pharmacology and G protein coupling profiles [3, 6, 7]. Group II (mGluR2 and 3) and III (mGluR4, and 6 through 8) couple negatively to adenylate cyclase through activation of the Gi/o family of G proteins [8]. Group I mGluRs (mGluR1 and 5) couple to multiple G proteins in native and heterologous systems, and have been observed to couple to Gq/11, Gi/o and/or Gs[9–15]. mGluRs appear to be important mediators of synaptic transmission in the brain. The group I mGluRs typically are located postsynaptically (as well as in other compartments of the cell) [16–18] where they regulate activity of ionotropic receptors and are necessary in some forms of synaptic plasticity [19]. The association of mGluRs with scaffolding proteins such as Homer (group I; [20]) and PICK-1 (group III; [21]) is important in regulating mGluR localization and function [21–24]. In addition to those proteins involved in scaffolding, other proteins have been shown to associate with mGluRs. For example, group I and group III mGluRs are both known to bind Calmodulin (CaM) [25, 26]. In the case of group III mGluRs, the interaction with CaM is important in regulating the efficacy of G protein activation [27, 28]. CaM binding to both group I and group III mGluRs can be inhibited by serine/threonine phosphorylation at specific sites [25, 28]. Therefore, it is of interest in terms of mGluR function and localization to understand the roles of proteins which interact with these receptors.
The Seven in Absentia (SinA) protein was discovered in Drosophila as a mediator of photoreceptor development [29]. Subsequently, SinA was shown to participate in the ubiquitin/proteasome pathway [30, 31]. The presence of a RING finger domain in the N-terminus of the mammalian homologue of SinA, "Siah", suggests it may be involved in targeting specific proteins for degradation in the proteasome as a ubiquitin ligase [32]. Two mammalian genes encoding Siah, Siah1 and 2, are known. These proteins have been implicated in many processes including cancer [33], apoptosis [34] and cell cycle regulation [35]. More recently, an interaction between Siah1a and the C-terminal tail of group I mGluRs was demonstrated [36]. This interaction was specific for group I mGluRs, as it was not observed in the analogous sequences from group II (mGluR2) or group III (mGluR7) receptors. Due to the novelty of the mammalian homolog of Seven in Absentia (Siah), little information is available regarding its tissue distribution or subcellular localization in mammalian cells. To date the functional consequences of Siah1a association with group I mGluRs are unknown.
To assess the role of Siah1a binding, specific group I mGluRs were heterologously expressed in sympathetic neurons from the rat superior cervical ganglion (SCG), which do not natively express mGluRs [37, 38]. The functional role of Siah1a was assessed by examining calcium current inhibition mediated by the expressed mGluRs in the absence and presence of heterologous Siah1a expression. In addition, the surface expression and subcellular distribution of group I mGluRs was examined with and without Siah1a using various fluorescence techniques. Finally, the effect of CaM overexpression on Siah1a function was assessed.
Results and Discussion
Coexpression of Siah1a with group I mGluRs
To examine the role of Siah1a expression on mGluR function, SCG neurons were injected with cDNA encoding several group I mGluRs (mGluR1, 5 and their splice variants) with or without Siah1a expression. Calcium current modulation was then examined upon application of extracellular 100 μM glutamate. The triple pulse voltage protocol [39] was used as the standard test pulse to assess calcium current magnitude and voltage dependence of inhibition [38, 40]. Figure 1A illustrates sample current traces from before ("con") and during inhibition by glutamate ("Glu"), in an mGluR5b-expressing cell. Consistent with previous reports [24, 38], calcium current inhibition via mGluR5b appeared partially voltage dependent. The time course of inhibition by glutamate and 10 μM norepinephrine (NE), acting through endogenous α2 adrenergic receptors [41], in this cell (figure 1B) illustrates that inhibition was rapid and reversible.
Effect of Siah1a expression on mGluR-mediated calcium current modulation. A and C, Sample current traces illustrating calcium current before and during inhibition by 100 μM glutamate in cells expressing mGluR5b (A) and mGluR5b + Siah1a (C). A triple pulse voltage protocol was used wherein cells were sustained at a holding potential of-80 mV and stepped to an initial test pulse to +10 mV for 25 msec, to +80 mV for 50 msec, then following a 10 msec step to -80 mV a second test pulse to +10 mV was applied. The scale bars indicate 20 msec and 0.4 nA. Tail currents were cropped to better illustrate the step currents. B and D, Time course of inhibition by glutamate (Glu) and norepinephrine (NE) in the cells illustrated in A and C, respectively. Closed circles represent measurements taken from the prepulse (the first test pulse to +10 mV). Open circles represent measurements taken from the postpulse (the second test pulse to +10 mV).
In contrast to that in cells expressing mGluR5b alone, calcium current inhibition by glutamate in cells coexpressing mGluR5b and Siah1a was quite small (figure 1C). Average inhibition of calcium currents by glutamate in mGluR5b-expressing cells was 33 ± 5%, (n = 7). Inhibition was significantly reduced to 10 ± 6% (n = 5; p < 0.05) in cells coexpressing mGluR5b and Siah1a (figure 2). This reduction of modulation by glutamate appeared to be specific for mGluRs, as NE-mediated calcium current inhibition was unaltered (figure ID). Calcium current inhibition resulting from NE application in mGluR-expressing cells was 58 ± 6% (n = 12). In cells coexpressing Siah1a, NE produced an inhibition of 54 ± 6% (n = 11; figure 2).
Siah1a effect is specific for group I mGluRs. Bar graph illustrating average (+SEM) calcium current inhibition via the indicated receptor in the absence (filled bars) and presence (open bars) of Siah1a. The number of cells from each group is indicated in parentheses. * indicates a significant difference from control (p < 0.05, ANOVA).
The strong reduction in calcium current modulation by glutamate in the presence of Siah1a was observed with every group I mGluR tested. Cells expressing an extracellularly myc-tagged mGluR5a (mGluR5a-myc) exhibited calcium current inhibition of 42 ± 4% (n = 6) upon glutamate application. This inhibition was significantly reduced to 22 ± 5% (n = 6) in cells coexpressing Siah1a (p < 0.05; figure 2). Similarly, calcium current inhibition in cells expressing a C-terminally GFP-tagged mGluR1a (mGluR1a-GFP) and mGluR1a-GFP + Siah1a was 46 ± 5% (n = 5) and 23 ± 6% (n = 6), respectively. In addition to group I mGluRs, mGluR2 (a group II mGluR) was also examined. However, Siah1a had no detectable effect on mGluR2-mediated calcium current inhibition, consistent with reports demonstrating that Siah1a does not bind to group II mGluRs [36]. Cells expressing mGluR2 alone exhibited 57 ± 5% inhibition of calcium current in response to glutamate (n = 6), while those coexpressing Siah1a were inhibited 61 ± 2% (n = 6; figure 2). Together, these data demonstrate that in the presence of Siah1a, the function of group I mGluRs is selectively inhibited. These data also suggest that Siah1a is acting at the level of the receptor, since much of the calcium current modulation through group I mGluRs proceeds through a Gβγ-mediated pathway similar to that used by α2 adrenergic receptors and mGluR2 [38, 40].
Effect of Siah1a on mGluR surface expression and localization
The reduction in group I mGluR signaling induced by Siah1a expression may be attributed to several possible mechanisms. For example, binding of Siah1a to the C-terminus of group I mGluRs may occlude the region or regions responsible for G protein activation. This is possible since both Siah1a binding and G protein activation have been attributed to areas on the C-terminus of group I mGluRs [36, 42, 43]. Alternatively, Siah1a binding may alter the 'normal' interactions of the receptors with scaffolding proteins or may cause the receptors to be targeted for degradation in the proteasome, effectively reducing the number of receptors that are 'correctly' assembled, or the number of functional receptors at the membrane. The latter hypothesis predicts a reduction in mGluR cell surface expression or perhaps a change in surface distribution in the presence of Siah1a, such as occurs in the presence of Homer proteins [24, 44, 45]. To test whether Siah1a expression altered the surface expression of group I mGluRs, the surface distribution of mGluR5a-myc was examined in the absence and presence of Siah1a. The subcellular distribution of mGluR1a-GFP was also examined with and without Siah1a.
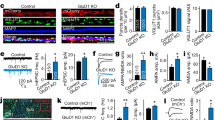
Figure 3A (upper) illustrates a cell injected with cDNA encoding mGluR5a-myc alone (and coexpressing pEGFP as a positive marker for expression). Column 1 ("Phase") and column 2 ("GFP") show a phase contrast image and GFP fluorescence of such a cell. Column 3 ("anti-myc cy3") shows surface labeling of the extracullular myc epitope on mGluR5a-myc using a cy3 fluorescently-conjugated primary antibody. Live cells were exposed to antibody prior to fixation to ensure that only receptors expressed on the cell surface would be detected. Uninjected cells displayed no detectable cy3 labeling (data not shown). In similarly treated cells coexpressing Siah1a, surface expression, measured as total cell fluorescence (figure 3B), was not significantly reduced (p = 0.33) and no receptor clustering was observed (figure 3A, lower). Similar experiments using a GFP-tagged group I mGluR, GFP-mGluR1a, indicated no detectable changes in receptor distribution when coexpressed with Siah1a (figure 3C). These data suggest that neither a reduction in receptor levels nor a change in receptor association with scaffolding proteins appears to be responsible for the diminished group I mGluR-mediated calcium current modulation observed in the presence of Siah1a.
Effect of Siah1a on mGluR surface expression and subcellular distribution. A, phase contrast image, GFP fluorescence, and surface cy3-conjugated anti-myc labeling of mGluR5a-myc from a cell expressing mGluR5a-myc alone (upper) and a cell coexpressing Siah1a (lower). Cy3-conjugated primary anti-myc antibody was applied to live cells before fixation to ensure labeling of only extracellular surface epitopes (see methods). B, Fluorescence measurments for cells expressing mGluR5a-myc (black bar) and mGluR5a-myc + Siah1a (open bar), as in A. The number of cells analyzed for each group is shown in parentheses. Units are arbitrary fluorescence units derived from 8 bit images (see methods). No significant difference was detected between the two groups (T-Test, p > 0.3). C, Phase contrast image and GFP fluorescence from a cell expressing mGluR1a-GFP alone (upper) and mGluR1a-GFP with Siah1a (lower).
Does Siah1a target group I mGluRs for proteolysis?
Siah1a has been shown to be a member of the ubiquitin family of proteins involved in protein degradation through the proteasome [30, 31, 33]. Therefore, group I mGluR degradation was considered a potential mechanism for the decrease in mGluR-mediated calcium current modulation observed in SCG neurons. Although an obvious decrease in receptor levels in the presence of Siah1a was not detected (figure 3), it is possible that introduction of high levels of Siah1a changed the rate of mGluR degradation such that a new equilibrium was reached with lower levels of active receptor than in the absence of Siah1a. Such a change may have been difficult to detect with the fluorescence techniques used here (figure 3). Therefore, a deletion mutant of Siah1a was constructed, GFP-Siah1a-CT, with the N-terminal RING-fmger domain deleted. This RING-fmger domain has been shown to be necessary for the ubiquitination function of Siah1a [46]. As a positive control for expression, this construct was fused to the C-terminus of GFP (figure 5A). Figure 4A illustrates sample currents from a cell expressing mGluR5a alone. As with mGluR5b, mGluR5a produced a rapid and reversible calcium current inhibition when 100 μM glutamate was applied to the bath (figure 4B). Coexpression of GFP-Siah1a-CT caused a significant reduction in the magnitude of calcium current modulation via mGluR5a (figure 4C,D). Glutamate-mediated calcium current inhibition in cells expressing mGluR5a alone was 33 ± 4% (n = 7). In cells expressing mGluR5a and GFP-Siah1a-CT, inhibition was reduced to 14 ± 3 (n = 6; p < 0.05; figure 5B). Therefore, a ubiquitin-deficient Siah1a construct that can bind mGluRs is capable of producing a similar reduction in mGluR-mediated calcium current modulation as the full length Siah1a. This suggests that the effect of Siah1a on group I mGluRs is a result of its physical association with the receptor, rather than a change in receptor levels.
Effect of GFP-Siah1a-CT on mGluR-mediated calcium current modulation. A and C, Sample current traces illustrating calcium current before and during inhibition by 100 μM glutamate in cells expressing mGluR5b (A) and mGluR5b + Siah1a (C). Voltage protocol was as in figure 1. The scale bars indicate 20 msec and 0.5 nA (A) or 1 nA (C). Tail currents were cropped to better illustrate the step currents. B and D, Time course of inhibition by glutamate (Glu) in the cells illustrated in A and C, respectively. Closed circles represent measurements taken from the prepulse (the first test pulse to +10 mV). Open circles represent measurements taken from the postpulse (the second test pulse to +10mV).
Summary of GFP-Siah1a-CT effect. A, Image showing GFP flourescence from a cell expressing GFP-Siah1a-CT. B, Bar graph illustrating average (+SEM) calcium current via the indicated receptor in the absence (filled bar) and presence (open bar) of GFP-Siah1a-CT. The number of cells from each group is indicated in parentheses. * indicates a significant difference from control (p < 0.05, ANOVA).
The role of Calmodulin
Calmodulin (CaM), a calcium binding protein intricately involved in intracellular signaling, interacts with the C-terminal tail of group I mGluRs [25]. This interaction appears to be dynamically regulated. Phosphorylation of the receptors at specific sites prevents CaM binding to mGluRs [25]. Association of Siah1a with group I mGluRs can also prevent the interaction of CaM and the receptor [36]. This competition for binding was predicted due to the overlap in binding sites on the C-terminal tail for CaM and Siah1a [36]. Although CaM has been known to associate with group I mGluRs for some time, the functional role of this interaction is unknown. However, CaM can also interact with group III mGluRs [26, 28]. Recent data suggests that the association of CaM with group III mGluRs is necessary for normal G protein activation [27]. By analogy to group III mGluRs, the ability of Siah1a to compete with CaM for binding to group I mGluRs was considered a potential mechanism for the actions of Siah1a on group I mGluR-mediated calcium current inhibition. Therefore, overexpression of CaM was tested for its ability to reverse the effect of Siah1a.
Calcium current inhibition in cells expressing the group I mGluR, mGluR5b was 29 ± 2% (figure 6; n = 21), similar to that through other group I mGluRs tested (see figure 2). Coexpression of Siah1a caused a reduction in calcium current modulation to 12 ± 3%, as observed previously (n = 14). Next, when CaM was coexpressed with mGluR5b and Siah1a, calcium current modulation was significantly increased (over that in cells expressing mGluR5b and Siah1a) to 25 ± 5% (n = 12; figure 6). Finally, CaM binding to the mGluRs did not appear to be Ca2+-dependent. When results obtained in the presence of 11 mM EGTA, a relatively high level of Ca2+ buffering, were compared to those in low Ca2+ buffering (with 0.1 mM intracellular BAPTA), no difference was detected. Under low Ca2+-buffering conditions, calcium current inhibition in control (mGluR5b alone) was 25 ± 4% (n = 9). With Siah1a and Siah1a + CaM inhibition was 7 ± 6% (n = 3) and 30 ± 18% (n = 3), respectively. Since the results were indistinguishable with high or low Ca2+ buffering, the data from both sets were pooled in the final results (figure 6). It should also be noted that coexpression of CaM with mGluR5a in the absence of Siah1a yields no changes in calcium current modulation (data not shown). These results indicate that the presence of high levels of CaM can reverse the effect of Siah1a on calcium current modulation, presumably by competing for binding on the C-terminal tail of the receptor.
CaM reverses the effect of Siah1a on group I mGluR-mediated calcium current inhibition. Bar graph illustrating average (+SEM) calcium current via the indicated receptor in cells expressing mGluR5b alone ("mGluR5b"), mGluR5b plus Siah1a ("+Siah1a") and mGluR5b + Siah1a and CaM ("+CaM"). The number of cells from each group is indicated in parentheses. * indicates a significant difference from control (p < 0.05, ANOVA). ** indicates a significant difference from "+Siah1a".
The data imply two possible mechanisms for the effect of Siah1a on mGluR signaling: First, CaM association with group I mGluRs increases G protein coupling (or perhaps is required, as may be the case with group III mGluRs). This model predicts that Siah1a association produces its effect by disrupting the interaction of CaM with the group I mGluRs. In this case, it must be assumed that under control conditions (cells heterologously expressing only the receptors), mGluRs associate with natively-expressed CaM. CaM expression levels would presumably be much lower natively than in cells injected with CaM cDNA. When Siah1a is caused to express heterologously (at presumably high levels) it expresses at levels high enough to out-compete native CaM for mGluR binding.
Alternatively, the presence of Siah1a itself may occlude G protein coupling, with the CaM interaction being incidental. Since a portion of the C-terminal tail of group I mGluRs is involved in G protein activation [43], this model is plausible. However, only the proximal ~20 amino acids of the C-terminal tail appear to be involved in G protein coupling. The site of Siah1a interaction is ~50 residues distal to that site [36]. Although this does not rule out this model, it suggests that it may be less likely than the former model, which implicates CaM as the pivotal interacting protein. However, until a more detailed picture of these interactions is available, it will be difficult to answer this question definitively. Nonetheless, the data presented here suggest a novel role for Siah1a and CaM in regulating the function of group I mGluRs.
The notion that Siah, a putative ubiquitin ligase, would have other roles in cell signaling is not surprising. Many RING-finger-containing proteins that may act as ubiquitin ligases under certain conditions have otherwise diverse structures and functions [32]. It is clear that Siah can act in the ubiquitin pathway, targeting some proteins for proteolysis, under certain circumstances. In addition, the data presented here do not rule out the possibility that Siah may target group I mGluRs to the proteasome should they become damaged, misfolded or should other conditions in the cell warrant mGluR proteolysis. At this time however, there is no evidence to support this claim. Still, it appears likely that Siah1a may play an important role in mGluR signaling in addition to any putative function regarding the proteasome pathway.
Further studies will be needed to determine if group I mGluRs and Siah1a interact physiologically, and whether this interaction has functional consequences. For example, are these proteins coexpressed in certain regions of the brain? Do they co-localize when expressed in a particular cell type? If the results presented here can be exprapolated to physiological situations that involve group I mGluRs, Siah1a expression in the mammalian nervous system could potentially play a role in cell excitability by influencing mGluR-mediated potassium current modulation (of M-type currents and perhaps of GIRK currents), and synaptic plasticity due to the role that group I mGluRs play in these processes [2, 5, 38].
Conclusions
The findings described here demonstrate that the mammalian homologue of seven in absentia, Siah1a regulates the function of group I metabotropic glutamate receptors. Sympathetic neurons from the rat SCG that were made to express Siah1a with a group I mGluR attenuated mGluR-mediated calcium current modulation. This attenuation did not appear to be accompanied by a reduction in cell surface expression levels of receptor nor by an obvious change in receptor distribution in the cells. Further, a mutant of Siah1a was constructed with the RING finger domain deleted, such that the protein was unable to act in the ubiquitin/proteasome pathway [46]. This deletion construct was as effective in attenuating group I mGluR function as the wild type Siah1a, providing supporting evidence that the effect of Siah1a resulted from its physical association with the receptors, rather than through a resulting reduction in receptor expression levels. Finally, coexpression of CaM reversed the effect of Siah1a on mGluR5b, indicating that the interaction of CaM and Siah1a in binding to group I mGluRs plays a novel and important role in mGluR function.
Materials and Methods
Cell preparation and microinjection
Detailed descriptions of the isolation and injection procedures have been reported previously [47]. Briefly, the superior cervical ganglia were dissected from adult rats and incubated in Earle's balanced salt solution (Life Technologies, Rochelle, MD) containing 0.45 mg ml-1 trypsin (Worthington, Freehold, NJ), 0.6 mg ml-1 collagenase D (Boehringer Mannheim, Indianapolis, IN) and 0.05 mg ml-1 DNAse I (Sigma, St. Louis, MO) for 1 hour at 35°C. Cells were centrifuged twice, transferred to minimum essential medium (Fisher Scientific, Pittsburgh, PA), plated and incubated at 37°C for 4 hours before cDNA injection. Injection was performed with an Eppendorf 5246 microinjector and 5171 micromanipulator (Madison, WI) 4–6 hours following cell isolation. Plasmids were stored at -20 °C as a 1 μg μl-1 stock solution in TE buffer (10 mM TRIS, 1 mM EDTA, pH 8). The mGluR5b insert (from J.-P. Pin) was cloned in the pRK5 vector (Genentech, South San Francisco, CA). The rat calmodulin (CaM) construct was amplified from an EST clone (accession #AA165448) and subcloned into the pCI vector (Promega, Madison, WI). The mGluR5a-myc (pRK5; from P. F. Worley) has been described previously [24]. mGluR1a-GFP was constructed by inserting the mGluR1a cDNA into the pEGFPN1 vector (Clontech Laboratories, Palo Alto, CA). Siah1a (from D. Bowtell) was in the pcDNA3 vector (In Vitrogen, Carlsbad, CA). The GFP-Siah1a-CT construct was made by subcloning the 3' end of Siah1a (from base 228) into pEGFPC1 in frame. All constructs were injected at 0.1 μg μl-1, unless otherwise noted. Constructs were sequence verified before injection. All neurons except those injected with the mGluR1a-GFP or GFP-Siah1a-CT constructs, were co-injected with "enhanced" green fluorescent protein cDNA (0.005 μg μl-1; pEGFP-N1; Clontech) to facilitate later identification of successfully injected cells. Following injection, cells were incubated overnight at 37°C and patch clamp experiments were performed the following day.
Electrophysiology and data analysis
Patch clamp recordings were made using 7052 glass (Gamer Glass, Claremont, CA). Pipette resistances ranged from 1–3 MΩ, yielding uncompensated series resistances of 2–7 MΩ. Series resistance compensation of 80% was used in all recordings. Data were recorded using an Axopatch 200A patch clamp amplifier from Axon Instruments (Foster City, CA). Voltage protocol generation and data acquisition were performed using custom data acquisition software on a Macintosh Quadra series computer with a MacAdios II data acquisition board (G. W. Instruments, Somerville, MA). Currents were sampled at 0.5–5 kHz low-pass filtered at 5 kHz using the 4-pole Bessel filter in the patch clamp amplifier, digitized, and stored on the computer for later analysis. Experiments were preformed at 21–24°C (room temperature). Data analysis was performed using Igor software (WaveMetrics, Lake Oswego, OR). Statistical analyses were performed using Statview software (SAS Institute, Cary, NC).
In all recordings, the external (bath) solution contained (in mM): 145 tetraethylammonium (TEA) methanesulfonate (MS), 10 4-(2-Hydroxy ethyl)-1-piperazineethanesulfonic acid (HEPES), 15 glucose, 10 CaCl2, and 300 nM tetrodotoxin, pH 7.4, osmolality ~320 mOsm kg-1. The internal (pipette) solution contained: 120 N-methyl-D-glucamine (NMG) MS, 20 TEA, 11 EGTA, 10 HEPES, 10 sucrose, 1 CaCl2, 4 MgATP, 0.1 Na2GTP, and 14 tris creatine phosphate, pH 7.2, osmolality ~300 mOsm kg-1. L-glutamate (Fisher Scientific, Pittsburgh, PA) was applied at a concentration of 100 μM in all experiments.
Immuno fluorescence and imaging
To detect surface expression of the mGluR5a-myc construct, a cy3-conjugated, mouse anti-myc antibody (Sigma) was applied to live cells at room temperature for 20 minutes at a 1:150 dilution. Cells were subsequently fixed in a 4% formaldehyde (Fisher) and 4% sucrose (Fisher) solution in phosphate buffered saline (PBS) for 20 minutes, washed with PBS and imaged. Images were acquired using a Spot RT Monochrome, cooled CCD camera (Diagnostic Instruments, Inc.) mounted onto a Nikon Eclipse TE300 inverted microscope with a 40X objective (Nikon). Images were obtained using Spot software (Diagnostic Instruments) and figures were constructed using Adobe Photoshop and Canvas (Deneba Systems) software. Image analysis was performed using NIH Image software. All images analyzed in figure 3B were taken using a 3 second exposure, chosen to put the majority of cells' fluorescence near the center of the camera's dynamic range. Images of 3 cells (out of 38) were discarded for having large areas of saturation. Using 8 bit images, total fluorescence of a small area encompassing the entire cell was measured and the fluorescence of an identically sized area of background was subtracted to yield a measure of total fluorescence for each cell. Since there was no detectable change in cell size accompanying Siah1a expression measured electrophysiologically (data not shown), the fluorescence measurments were not normalized to total area.
References
Bhave G, Karim F, Carlton SM, Gereau RW: Peripheral group I metabotropic glutamate receptors modulate nociception in mice. Nature Neuroscience. 2001, 4: 417-423. 10.1038/86075.
Bortolotto ZA, Bashir ZI, Davies CH, Collmgridge GL: A molecular switch activated by metabotropic glutamate receptors regulates induction of long-term potentiation. Nature. 1994, 368: 740-743. 10.1038/368740a0.
Conn PJ, Pin JP: Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997, 37: 205-237. 10.1146/annurev.pharmtox.37.1.205.
Dingledine R, Conn PJ: Peripheral glutamate receptors: molecular biology and role in taste sensation. J Nutr. 2000, 130: 1039S-1042S.
Wilsch VW, Behnisch T, Jager T, Reymann KG, Balschun D: When are class I metabotropic glutamate receptors necessary for long-term potentiation?. J Neurosci. 1998, 18: 6071-6080.
Nakanishi S: Metabotropic glutamate receptors: synaptic transmission, modulation and plasticity. Neuron. 1994, 13: 1031-1037. 10.1016/0896-6273(94)90043-4.
Pin J-P, Duvoisin R: Neurotransmitter receptors I. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995, 34: 1-26. 10.1016/0028-3908(94)00129-G.
De Blasi A, Conn PJ, Pin JP, Nicoletti F: Molecular determinants of metabotropic glutamate receptor signaling. Trends Pharmacol Sci. 2001, 22: 114-120. 10.1016/S0165-6147(00)01635-7.
Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S: Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2+ signal transduction. J. Biol. Chem. 1992, 267: 13361-13368.
Aramori I, Nakanishi S: Signal transduction and pharmacological characteristics of a metabotropic glutamate receptor, mGluR1, in transfected CHO cells. Neuron. 1992, 8: 757-765.
Choi S, Lovinger DM: Metabotropic glutamate receptor modulation of voltage-gated Ca2+ channels involves multiple receptor subtypes in cortical neurons. J Neurosci. 1996, 16: 36-45.
Hay M, Kunze DL: Glutamate metabotropic receptor inhibition of voltage-gated calcium currents in visceral sensory neurons. J. Neurophysiol. 1994, 72: 421-430.
Joly C, Gomeza J, Brabet I, Curry K, Bockaert J, Pin JP: Molecular, functional, and pharmacological characterization of the metabotropic glutamate receptor type 5 splice variants: comparison with mGluR1. J Neurosci. 1995, 15: 3970-3981.
McCool BA, Harpold MM, Stauderman KA, Brust PF, Lovinger DM: Relative contributions of G protein, channel, and receptor to voltage-dependent inhibition of neuronal N-type and P/Q-type calcium channels in HEK 293 cell lines. Neurosci Lett. 1997, 239: 89-92. 10.1016/S0304-3940(97)00893-8.
Pin J-P, Waeber C, Prezeau L, Bockaert J, Heinemann SF: Alternative splicing generates metabotropic glutamate receptors inducing different patterns of calcium release in Xenopus oocytes. Proc. Natl. Acad. Sci. USA. 1992, 89: 10331-10335.
Lujan R, Roberts JD, Shigemoto R, Ohishi H, Somogyi P: Differential plasma membrane distribution of metabotropic glutamate receptors mGluR1 alpha, mGluR2 and mGluR5, relative to neurotransmitter release sites. J Chem Neuroanat. 1997, 13: 219-241. 10.1016/S0891-0618(97)00051-3.
Rouse ST, Marino MJ, Bradley SR, Awad H, Wittmann M, Conn PJ: Distribution and roles of metabotropic glutamate receptors in the basal ganglia motor circuit: implications for treatment of Parkinson's disease and related disorders. Pharmacol Ther. 2000, 88: 427-435. 10.1016/S0163-7258(00)00098-X.
Hubert GW, Paquet M, Smith Y: Differential subcellular localization of mGluR1a and mGluR5 in the rat and monkey Substantia nigra. J Neurosci. 2001, 21: 1838-1847.
Bortolotto ZA, Fitzjohn SM, Collingridge GL: Roles of metabotropic glutamate receptors in LTP and LTD in the hippocampus. Curr Opin Neurobiol. 1999, 9: 299-304. 10.1016/S0959-4388(99)80044-0.
Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF: Homer: a protein that selectively binds metabotropic glutamate receptors [see comments]. Nature. 1997, 386: 284-288. 10.1038/386284a0.
Boudin H, Doan A, Xia J, Shigemoto R, Huganir RL, Worley P, Craig AM: Presynaptic clustering of mGluR7a requires the PICK1 PDZ domain binding site. Neuron. 2000, 28: 485-497. 10.1016/S0896-6273(00)00127-6.
Boudin H, Craig AM: Molecular determinants for PICK1 synaptic aggregation and mGluR7a receptor coclustering: role of the PDZ, coiled coil, and acidic domains. J Biol Chem. 2001, 24: 24.
Dev KK, Nakajima Y, Kitano J, Braithwaite SP, Henley JM, Nakanishi S: PICK1 interacts with and regulates PKC phosphorylation of mGLUR7. J Neurosci. 2000, 20: 7252-7257.
Kammermeier PJ, Xiao B, Tu JC, Worley PF, Ikeda SR: Homer proteins regulate coupling of group I metabotropic glutamate receptors to N-type calcium and M-type potassium channels. J Neurosci. 2000, 20: 7238-7245.
Minakami R, Jinnai N, Sugiyama H: Phosphorylation and calmodulin binding of the metabotropic glutamate receptor subtype 5 (mGluR5) are antagonistic in vitro. J Biol Chem. 1997, 272: 20291-20298. 10.1074/jbc.272.32.20291.
O'Connor V, El Far O, Bofill-Cardona E, Nanoff C, Freissmuth M, Karschin A, Airas JM, Betz H, Boehm S: Calmodulin dependence of presynaptic metabotropic glutamate receptor signaling. Science. 1999, 286: 1180-1184. 10.1126/science.286.5442.1180.
El Far O, Bofill-Cardona E, Airas JM, O'Connor V, Boehm S, Freissmuth M, Nanoff C, Betz H: Mapping of calmodulin and G{betagamma} binding domains within the C-terminal region of the metabotropic glutamate receptor 7A. J Biol Chem. 2001, 6: 6.
Nakajima Y, Yamamoto T, Nakayama T, Nakanishi S: A relationship between protein kinase C phosphorylation and calmodulin binding to the metabotropic glutamate receptor subtype 7. J Biol Chem. 1999, 274: 27573-27577. 10.1074/jbc.274.39.27573.
Carthew RW, Rubin GM: seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990, 63: 561-577. 10.1016/0092-8674(90)90452-K.
Li S, Li Y, Carthew RW, Lai ZC: Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional represser Tramtrack. Cell. 1997, 90: 469-478. 10.1016/S0092-8674(00)80507-3.
Tang AH, Neufeld TP, Kwan E, Rubin GM: PHYL acts to down-regulate TTK88, a transcriptional represser of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997, 90: 459-467. 10.1016/S0092-8674(00)80506-1.
Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM: RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999, 96: 11364-11369. 10.1073/pnas.96.20.11364.
Hu G, Zhang S, Vidal M, Baer JL, Xu T, Fearon ER: Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 1997, 11: 2701-2714.
Roperch JP, Lethrone F, Prieur S, Piouffre L, Israeli D, Tuynder M, Nemani M, Pasturaud P, Gendron MC, Dausset J, Oren M, Amson RB, Telerman A: SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21(Waf1). Proc Natl Acad Sci USA. 1999, 96: 8070-8073. 10.1073/pnas.96.14.8070.
Bruzzoni-Giovanelli H, Faille A, Linares-Cmz G, Nemani M, Le Deist F, Germani A, Chassoux D, Millot G, Roperch JP, Amson R, Telerman A, Calvo F: SIAH-1 inhibits cell growth by altering the mitotic process. Oncogene. 1999, 18: 7101-7109. 10.1038/sj.onc.1203187.
Ishikawa K, Nash SR, Nishimune A, Neki A, Kaneko S, Nakanishi S: Competitive interaction of seven in absentia homolog-1A and Ca2+/calmodulin with the cytoplasmic tail of group 1 metabotropic glutamate receptors. Genes Cells. 1999, 4: 381-390. 10.1046/j.1365-2443.1999.00269.x.
Ikeda SR, Lovinger DM, McCool BA, Lewis DL: Heterologous expression of metabotropic glutamate receptors in adult rat sympathetic neurons: subtype-specific coupling to ion channels. Neuron. 1995, 14: 1029-1038.
Kammermeier PJ, Ikeda SR: Expression of RGS2 alters the coupling of metabotropic glutamate receptor la (mGluR1a) to M-type K+ and N-type Ca2+ channels. Neuron. 1999, 22: 819-829.
Elmslie KS, Zhou W, Jones SW: LHRH and GTP-γ-S modify calcium current activation in bullfrog sympathetic neurons. Neuron. 1990, 5: 75-80. 10.1016/0896-6273(90)90035-E.
Kammermeier PJ, Ruiz-Velasco V, Ikeda SR: A voltage-independent calcium current inhibitory pathway activated by muscarinic agonists in rat sympathetic neurons requires both Gβq/11 and Gβγ. J Neurosci. 2000, 20: 5623-5629.
Schofield GG: Norepinephrine blocks a calcium current of adult rat sympathetic neurons via an α2-adrenoceptor. Eur. J. Pharmacol. 1990, 180: 37-47. 10.1016/0014-2999(90)90590-3.
Francesconi A, Duvoisin RM: Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation. J. Biol. Chem. 1998, 273: 5615-5624. 10.1074/jbc.273.10.5615.
Pin J-P, Joly C, Heinemann SF, Bockaert J: Domains involved in the specificity of G protein activation in phospholipase C-coupled metabotropic glutamate receptor. EMBO J. 1994, 13: 342-348.
Ciruela F, Soloviev MM, McIlhmney RA: Co-expression of metabotropic glutamate receptor type 1 alpha with homer-1a/Vesl-1S increases the cell surface expression of the receptor. Biochem J. 1999, 341: 795-803. 10.1042/0264-6021:3410795.
Tadokoro S, Tachibana T, Imanaka T, Nishida W, Sobue K: Involvement of unique leucine-zipper motif PSD-Zip45 (Homer 1c/vesl-1L) in group 1 metabotropic glutamate receptor clustering. Proc. Natl. Acad. Sci. USA. 1999, 96: 13801-13806. 10.1073/pnas.96.24.13801.
Hu G, Fearon ER: Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999, 19: 724-732.
Acknowledgements
We thank M. King and L. Olmstead for valuable technical assistance, also D. Bowtell, J.-P. Pin, and P. F. Worley for supplying clones. This work was supported by NIH grants NS37615 (to SRI) and NS 10943 (to PJK).
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kammermeier, P.J., Ikeda, S.R. A role for Seven in Absentia Homolog (Siah1a) in metabotropic glutamate receptor signaling. BMC Neurosci 2, 15 (2001). https://doi.org/10.1186/1471-2202-2-15
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2202-2-15