Abstract
Background
One of the many ascribed functions of CCCTC-binding factor (CTCF) in vertebrates is insulation of genes via enhancer-blocking. Insulation allows genes to be shielded from "cross-talk" with neighboring regulatory elements. As such, endogenous insulator sequences would be valuable elements to enable stable transgene expression. Recently, CTCF joined Su(Hw), Zw5, BEAF32 and GAGA factor as a protein associated with insulator activity in the fruitfly, Drosophila melanogaster. To date, no known insulators have been described in mosquitoes.
Results
We have identified and characterized putative CTCF homologs in the medically-important mosquitoes, Aedes aegypti and Anopheles gambiae. These genes encode polypeptides with eleven C2H2 zinc fingers that show significant similarity to those of vertebrate CTCFs, despite at least 500 million years of divergence. The mosquito CTCFs are constitutively expressed and are upregulated in early embryos and in the ovaries of blood-fed females. We have uncovered significant bioinformatics evidence that CTCF is widespread, at least among Drosophila species. Finally, we show that the An. gambiae CTCF binds two known insulator sequences.
Conclusion
Mosquito CTCFs are likely orthologous to the widely-characterized vertebrate CTCFs and potentially also serve an insulating function. As such, CTCF may provide a powerful tool for improving transgene expression in these mosquitoes through the identification of endogenous binding sites.
Similar content being viewed by others
Background
CTCF (CCCTC-binding factor) was originally identified as a transcriptional repressor in studies of the chicken lysozyme silencer [1] and the regulation of the chicken c-myc gene [2]. Since that time, CTCF has been extensively characterized in vertebrates as a ubiquitously-expressed, highly-conserved, multivalent transcription factor that utilizes different zinc finger (ZF) combinations to specifically bind diverse nucleotide sequences, resulting in the repression or activation of target genes, creation of hormone-responsive silencers and the formation of enhancer-blocking boundary elements (reviewed in [3]). Multiple, independent studies have established vertebrate CTCF as a central player in the regulation of gene expression via its association with every known vertebrate insulator [3–5]. Further characterization of these proteins revealed their insulator function to be central in three contexts: (a) constitutive insulation of the chicken β-globin gene at the 5'HS4 site [6, 7] and the human apolipoprotein B gene at the 5' boundary [8], (b) imprinted insulation via methylation-sensitive binding to the Igfr2-H19 control locus [6, 9–14], the DM1 locus [5] and the DLK1/GTL2 locus [15], and (c) as part of a more complex, multipartite insulator regulated by ligand binding [16]. Most recently, CTCF-dependent insulators have been identified in transitional chromatin, with high levels of H3 acetylation and essentially no CpG methylation, between escape and inactivated genes on both mouse and human inactivated X chromosomes [17]. Finally, Tsix and CTCF have been proposed to comprise a regulated epigenetic switch for X-inactivation in mammals [18]. Clearly, CTCF plays a pivotal role at multiple levels of gene regulation and genome organization in vertebrate organisms.
Long thought to be exclusive to vertebrates, a CTCF orthologue was recently characterized in Drosophila melanogaster with domain structure, binding site specificity and transcriptional repressor activity similar to that of vertebrate CTCF [19]. Significantly, these researchers also demonstrated that a known Drosophila insulator, Fab8, mediates enhancer-blocking via CTCF in both Drosophila and vertebrate cell lines. We have cloned and characterized two mosquito CTCF-like cDNAs encoding polypeptides with significant similarity and insulator binding properties to both the vertebrate and Drosophila CTCFs. Analysis of available genome sequence from numerous invertebrate species yields promising candidates for additional CTCF orthologues. Clearly, this versatile protein has much more ancient roots than once thought.
Results
Cloning of Ae. aegypti and An. gambiae CTCF-like cDNAs
A BLAST search using the human CTCF protein sequence [20] as a query uncovered a cDNA from D. melanogaster [AAL78208], subsequently characterized by Moon et al. [19] as an orthologous CTCF factor. This sequence was then used to query the An. gambiae genome assembly at the Ensembl database [21], resulting in a highly significant hit of the predicted novel gene ENSANGG00000015222 (e-139). These two dipteran sequences were aligned with known vertebrate CTCF sequences from Gallus gallus [22], Mus musculus and Homo sapiens [20], Rattus norvegicus [NP_114012.1] and Xenopus laevis [23] using the ClustalW algorithm (Vector NTI™ Suite 8, InforMax, Inc., 1999). This multiple sequence alignment was used for degenerate PCR primer design. Degenerate PCR amplification, using Ae. aegypti larval cDNA as a template, yielded a single PCR product of 504 base pairs, corresponding to a 168 amino acid polypeptide containing six of the eleven predicted zinc-finger domains. PCR amplification was initially performed with an An. gambiae larval cDNA template and primers corresponding to the 5' and 3' ends of the predicted novel coding sequence. This yielded a single product of 2040 base pairs, corresponding to a translated polypeptide of 680 amino acid residues. Subsequent 5' and 3' RACE (r apid a mplification of c DNA e nds) in both species yielded putative full-length cDNAs of 2616 and 4544 base pairs for Ae. aegypti (AY935523) and An. gambiae (AY939827), respectively. Alignment of the corresponding polypeptide sequences with both the D. melanogaster and H. sapiens CTCFs revealed significant differences in the N-terminal and C-terminal regions of the protein, however there was 38% identity and 56% similarity across all eleven zinc finger domains (Fig. 1). Furthermore, 68% of the critical binding residues were conserved, despite at least 500 million years of divergence between invertebrate and vertebrate species [24].
The zinc-finger (ZF) domain is highly conserved between humans and the dipteran insects, Ae. aegypti, An. gambiae and D. melanogaster . Each of the eleven ZFs were aligned using the ClustalW algorithm. Identical and highly conserved residues are highlighted in gray. Weakly conserved residues, the zinc-coordinating residues, and the amino acids with identical binding site recognition properties are indicated in gray, red and blue font respectively.
CTCF appears widespread in Drosophila species
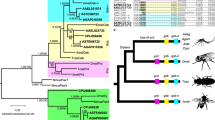
Available genome sequence for multiple drosophilid species was queried at Flybase [25] using the An. gambiae amino acid sequence and the tBLASTx algorithm. All species searched produced single hits of very high significance, ≤ e-126. Each of these was submitted as a BLASTp query of the non-redundant database at NCBI [26] and confirmed to be a significant match to known CTCFs. Sequences with complete zinc finger regions were trimmed to the zinc-finger region plus five flanking amino acid residues and aligned with the corresponding region of CTCFs from H. sapiens, G. gallus, X. laevis, Danio rerio [NP_001001844], Tetraodon nigroviridis [CAF99566], and Fugu rubripes (Ensembl novel gene SINFRUG00000147322). The corresponding region of zinc finger protein 2 from Caenorhabditis elegans [NP_500033], a protein that contains 11 C2H2 zinc finger domains, a coil-coil region and predicted nuclear localization sequence, was also included in the alignment and used as an outgroup in the subsequent phylogenetic analysis. Two consensus distance-based trees, Neighbor-Joining [27] (Fig. 2) and Fitch-Margoliash [28] (data not shown), were generated with 5000 bootstrap replicates using the Phylip software package [29, 30]. Additionally, a maximum-likelihood tree generated by 200,000 iterations of Tree-Puzzle [31] (data not shown) and a Bayesian analysis tree generated by 200,000 cycles of BAMBE [32] with 20,000 cycles of burn-in (data not shown), yielded identical branch topologies.
Phylogenetic analysis of CTCF-like candidates in multiple species. Dendrogram of a neighbor-joining consensus tree of 5000 bootstrap replicates for an alignment of the 11 ZF region of known and predicted CTCFs. The tree topology is consistent with the taxonomic classification of all Drosophila species.
Mosquito CTCF is expressed constitutively in all developmental stages and is upregulated in early embryos and the ovaries of blood-fed females
Reverse-transcriptase (RT)-PCR amplifications of RNA isolated from embryos, ovaries, larvae, pupae and adults shows CTCF expression across all stages of development and in the ovarian tissues of both Ae. aegypti and D. melanogaster (Fig. 3). Early Ae. aegypti embryos and ovarian tissues from both species clearly show increased expression levels.
Developmental expression profile of CTCF protein in Ae. aegypti and D. melanogaster. The expression of CTCF was analyzed using RNA isolated from multiple individuals at each of the indicated stages: E1 and E24 (embryos ≤ 1 hr and 24 hrs post-oviposition respectively), Lv (larvae), Pf (female pupae), Pm (male pupae), Pu (pupae), Af (adult females), Am (adult males), Ov- and Ov+ (ovaries from non-blood-fed and blood-fed females respectively). -RT, no reverse-transcriptase. A. and B.) Ae. aegypti CTCF, 20 cycles and 30 cycles respectively. C.) Ae. aegypti S17, 25 cycles. D.) D. melanogaster CTCF, 25 cycles. E.) D. melanogaster Rp49, 20 cycles.
Polyclonal antisera raised against An. gambiae CTCF recognizes a single protein band in lysates from An. gambiae Sua4 cultured cells
Immunoblotting of total cell lysate from An. gambiae Sua4 cultured cells with rabbit antisera raised against a c-terminal fragment of An. gambiae CTCF results in identification of a single band migrating at ~84 kD (Fig. 4).
An. gambiae CTCF polyclonal antisera recognizes a distinct band migrating ~84 kD in SDS-PAGE. Lysates from An. gambiae Sua4 cultured cells were separated by 8% SDS-PAGE and immunoblotted with CTCF rabbit antisera. The arrow indicates the position of the 81.1 kD marker. The bounding box marks the edges of the gel.
Mosquito CTCF binds in-vitro to both the chicken 5'HS4 and the Drosophila Fab8 insulators
As we were unable to express the full-length mosquito CTCF protein in bacteria, whole cell lysates were prepared from the An. gambiae Sua4 [33] cell line and used in an electrophoretic mobility shift assay (EMSA) to assess whether mosquito CTCF could bind known CTCF-associated insulator sequences (Fig. 5). The intensity of the shifted bands increased with application of greater amounts of protein lysate. The detectable complex was competed by cold, unlabeled probe, indicating that binding was indeed specific. In addition, all reactions contained a 1200-fold excess of cold, non-specific C/G-rich sequences, further illustrating specificity. Finally, the complex could be partially shifted by polyclonal anti-sera generated against the C-terminal region of the An. gambiae CTCF protein.
An. gambiae CTCF specifically binds the chicken 5'HS4 and Drosophila Fab8 insulator sequences. Sua4 cells were lysed and increasing amounts of total cell protein (1.5, 7.5, 15 μg represented as solid triangle) were incubated with radiolabeled insulator sequences as follows: A.) Drosophila Fab8 insulator sequence [19]; B.) chicken β-globin FII insulator sequence [6]. The complex was competed (Cp) with ~150-fold excess of cold, unlabeled probe DNA and supershifted (Ab) with polyclonal antibody sera raised against the C-terminal fragment of An. gambiae CTCF. The probe only lane is indicated by P.
Discussion
Vertebrate CTCFs, from fish to human, are ≥ 98% identical across the entire zinc finger core of the protein. Comparison of the three dipteran CTCFs reveals 54% identity and 68% similarity within this same region. In addition, amino acid residues considered critical for DNA binding [34] are 89% conserved among these three insect species. This apparent discrepancy can be partially addressed by investigating the molecular substitution rate heterogeneity among vertebrates and invertebrates. Recent maximum likelihood analysis of a set of 50 nuclear genes for vertebrates and dipterans, with Arabidopsis as an outgroup, suggests that the rate of vertebrate molecular evolution slowed considerably with respect to that of dipterans, prior to the origin of the crown-group, Osteichthyes [24]. The much shorter generation times of dipterans have undoubtedly facilitated significant differences in their genome sizes (ranging from 179 Mb in D. melanogaster [35] to 813 Mb in Ae. aegypti [36]) and gene organization patterns, attributable primarily to the number and distribution of repetitive sequences [37]. This would perhaps result in predictions of even greater sequence divergence than is observed in the CTCF genes. It seems likely that at least some of the many attributed vertebrate functions of CTCF are ancestral.
Each of the species examined yielded a single, extremely significant match followed by numerous matches of lesser significance, suggesting a single copy locus. Significant divergence in available N-terminal or C-terminal sequence supports the earlier observation that dipteran genomes have evolved very quickly, and thus these regions may not be critical to the conserved ancestral function(s) of this gene. Additionally, these regions may be more directly involved in protein-protein interactions with other proteins having likewise undergone evolutionary adaptation. High bootstrap support and essentially identical trees generated by four independent methods establishes the tree presented in Fig. 2 as representative of the evolution of this gene sequence. Less bootstrap support in the vertebrate clade is more indicative of the homogeneity of the sequence, rather than uncertainty as to where these species should be located in the tree. Clearly, CTCF is present in vertebrates from fish through mammals and is highly conserved. Of interest is its consistent presence in all Drosophila species queried. The relatedness of the protein sequences mirror the accepted taxonomic relationships among these species as presented at FlyBase [25], likely indicative of a conserved critical function. Significant EST evidence from the flour beetle, Tribolium castaneum, the honey bee, Apis mellifera, and the silkworm moth, Bombyx mori, suggests the presence of CTCF-like genes in multiple insect orders.
The RT-PCR data from both mosquito and fly are consistent with one another, repeatable, and in agreement with both in-situ hybridization data [38] posted for the fly at the Berkeley Drosophila Genome Project website [39] and fly microarray data summarized at Yale University's Drosophila Developmental Gene Expression Timecourse website [40]. In-situ hybridization shows high-levels of Drosophila CTCF transcript ubiquitously distributed throughout stage 1–3 embryos. mRNA levels then decrease until approximately stage 9 where they then increase primarily in the developing nervous and sensory tissues. The neural-specific expression pattern also corresponds to findings in X. laevis where in-situ hybridization with staged embryos revealed weak homogeneous staining prior to stage 14, with subsequent upregulation in neural tissues and the sensory organs of the head [23]. Furthermore, over-expression of CTCF in mice during early embryogenesis resulted in decreased expression of the highly conserved homeobox gene Pax6, causing ocular defects [34]. Microarray data analysis clusters fly CTCF (CG8591) with genes exhibiting a single peak in expression during development, those showing significant expression increases in early embryogenesis, genes with expression changes of at least four-fold across development, and those expressed in the female germline [41]. Taken together, these expression data and the corresponding functional data from vertebrates suggest that CTCF may indeed also be multi-functional in insects. Some possible roles include the regulation of homeobox genes like Pax6, the facilitation of chromatin organization during early development and the establishment and/or maintenance of heterochromatic and euchromatic regions.
The EMSA data support a role for CTCF in endogenous mosquito insulator function and confirm recent findings that the insulator function of CTCF is conserved from invertebrate to vertebrate species [19]. Currently, position effect and position-effect variegation complicate efforts to establish stable transgenic lines in Ae. aegypti and other mosquitoes. Particularly problematic is the highly repetitive nature of much of the intergenic sequence, as well as the compact nature of the genome, which places regulatory elements from neighboring genes in close proximity to one another, where they may inappropriately impact the transgene of interest. The ability to flank transgenes with short, conserved endogenous insulator sequences could significantly improve observed expression levels, and possibly increase the frequency of recovery of transgenic individuals.
Conclusion
We have cloned the cDNAs for two putative mosquito CTCF proteins. We have presented bioinformatics evidence that CTCF is likely present in many arthropod species and that the ancestral portion of the protein is clearly the zinc-finger region. Constitutively expressed in all life stages, mosquito CTCFs are highly upregulated in early embryos and in the ovarian tissues of blood-fed female mosquitoes. Finally, mosquito CTCF specifically binds both the chicken 5'HS4 β-globin and the fly Fab8 insulator sequences. Further characterization of these CTCFs and their binding sites will provide a promising avenue for insulating transgenes in these medically-important mosquito species.
Methods
Isolation of RNA and preparation of cDNA by reverse-transcription
Total RNA was isolated from ~30 mg each of Ae. aegypti and An. gambiae larvae using the RNeasy® Mini Kit (Qiagen, Valencia, CA) followed by DNase I-treatment with DNA-free™ (Ambion, Austin, TX) and was used to synthesize first strand cDNA using the SuperScript II™ reverse transcriptase (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. In order to increase the efficiency of the reverse-transcription reaction, 150 ng/μL of T4 Gene 32 Protein [42] was added to the 1st strand buffer.
Isolation of Ae. aegypti CTCF by degenerate PCR amplification
The amino acid sequences of all known and predicted CTCFs [EAA11339.1, AAL78208, AAG40852, NP_031820, NP_114012, P49711 and Q08705] were identified using the BLAST search algorithm at the National Center for Biotechnology Information (NCBI) website http://www.ncbi.nlm.nih.gov and aligned using the ClustalW algorithm in the Vector NTI™ Suite (InforMax, Inc., 1999). Two completely nested and degenerate PCR primer pairs were designed to a highly-conserved 168 amino acid region using CODEHOP [43, 44]. A 504 base pair nested PCR product was obtained from Ae. aegypti larval cDNA using G-1F and K-1R primers in the first PCR reaction, followed by a nested reaction with primers G-2F and J-1R. Each reaction was performed with 2 mM MgCl2, 0.2 μM each primer, 10 mM dNTPs, 0.5 μL cDNA or 1st reaction product and 2.5 units of Taq polymerase (Continental Lab Products, San Diego, CA). The following touchdown PCR conditions were used: 96°C for 4'; 2 cycles of 96°C for 20", 72°C for 1'; 11 cycles of 96°C for 20", 71°C -1.0°C/cycle for 15", 72°C for 45"; 25 cycles of 96°C for 15", 59°C for 15", 72°C for 45"; final extension at 72°C for 2'. Degenerate primers were as follows: G-1F 5' cattccgaggacccgccncayaartg 3', G-2F 5' ggccgctgcagaaccacctiaayacncaya 3', J-1R 5' cgcactgctcgcacctgwancayttytc 3', K-1R 5' ccaggtccagcagctgcykytgickraa 3'.
PCR-amplification and cloning of An. gambiae CTCF
The predicted ORF of An. gambiae CTCF was PCR amplified from ~100 ng of cDNA with 0.2 μM of each primer and 2.5 units of Herculase® Hotstart DNA Polymerase (Stratagene, La Jolla, CA) per the manufacturer's instructions using the following conditions: 95°C for 2'; 5 cycles of 95°C for 30", 55°C for 30", 72°C for 2'45"; 25 cycles of 95°C for 30", 65°C for 30", 72°C for 2'45"; final extension at 72°C for 5'. The primer sequences were: Anopheles CTCFforw 5' caaacgccatatggaggacgtggagctgatat 3' and Anopheles CTCFrev 5' attacctcttgcggccgcttccgtggagaggataaact 3'.
Rapid amplification of cDNA ends (RACE) in Ae. aegypti and An. gambiae
Total RNA was prepared from freshly collected and snap-frozen larvae using the RNeasy® Mini Kit (Qiagen) and immediately DNase I-treated with DNA-free™ (Ambion) according to the manufacturers' instructions. The BD SMART™ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA) was then used to prepare first-strand cDNA and to amplify 5' and 3' RACE products according to the manufacturer's instructions. The gene-specific primers (GSPs) used for each species were: Aedes GSP1 5' gtctgtcttgcgcccacatgttg 3', Aedes GSP2 5' cgaaagcacgtttacaacttctgg 3', Anopheles GSP1 5' ccacaggtcgtcgggcagagtttgca 3', Anopheles GSP2 5' caatcggagtaagattgtccgaagaaaggtct 3'. GSP1 indicates the primer used for 5' RACE reactions while GSP2 indicates the primer used for 3' RACE reactions. Reaction conditions were as follows: 94°C for 5'; 5 cycles of 94°C for 10", 72°C for 3'; 5 cycles of 94°C for 10", 70°C for 10", 72°C for 3'; 25 cycles of 94°C for 10", 68°C for 10", 72°C for 3'; final extension at 72°C for 8'.
Cloning and sequencing of PCR and RACE products
Products were visualized on a 1% agarose gel, gel purified, cloned into pGEM-T (Promega, Madison, WI) and had their DNA sequence determined using an ABI 3100 capillary sequencer with M13 (-20) and M13 Reverse primers followed by primer walking. At least 3 different clones were analyzed for each PCR or RACE product. The resulting sequences have been deposited in the NCBI GenBank database and have the following accession numbers: [AY935523] (Ae. aegypti) and [AY939827] (An. gambiae).
Phylogenetic analysis
Sequences were trimmed to the 11 ZF region plus five flanking amino acid residues and aligned using MultAlin [45] with the Blosum62 model, a gap opening penalty of 35, a gap extension penalty of 0.5 and no end gap penalty. The resulting alignment was analyzed using the Phylip software package [29]: bootstrapped (5000 replicates) with Seqboot, a distance matrix computed using Protdist (5000 datasets), the matrix submitted to Neighbor or Fitch (5000 trees), a consensus tree determined using Consense and the tree drawn using Drawgram. The MultAlin alignment was also submitted to Tree-Puzzle [31] with 200,000 replicates and to BAMBE [32] with 200,000 cycles and 20,000 burn-in.
Reverse-Transcriptase (RT)-PCR analysis/developmental profile
Total RNA was prepared from freshly collected and snap-frozen samples, DNase treated and the reverse-transcription reaction performed as described above. PCR reactions were assembled with 100 ng cDNA template, 10X buffer, 1.5 μL 10 mM dNTPs, 0.2 μM each primer (Table 1) and 1 μL Advantage2 Taq Polymerase (Clontech) in a total volume of 50 μL. Reaction conditions were as follows: 95°C for 5'; 20, 25 or 30 cycles (see Fig. 3) of 95°C for 15", 55°C for 15", 72°C for 30"; final extension at 72°C for 2'. Products were electrophoresed on a 2% agarose gel, stained with ethidium bromide, destained with ddH2O and imaged. The constitutively expressed D. melanogaster Rp49 gene (153 bp product) and Ae. aegypti S17 gene (200 bp product) were used as controls. Primers were as follows: Aedes RT-Forw 5' gtgtttcattgcgagctttgcc 3', Aedes RT-Rev 5' tgtctcgatcctccggaatg 3', S17 RT-Forw 5' cgaagcccctgcgcaacaagat 3', S17 RT-Rev 5' cagctgcttcaacatctccttg 3', Drosophila RT-Forw 5' atggagactcacgatgattcgg 3', Drosophila RT-Rev 5' ctcgtcgccattaaccagct 3', Rp49 RT-Forw 5' gcgcaccaaggacttcatc 3', Rp49 RT-Rev 5' gaccgactctgttgtcgatacc 3'.
Generation of polyclonal antisera against An. gambiae CTCF
The coding sequence for a C-terminal region (amino acid residues 444–680) was PCR amplified and cloned into the pET-30 plasmid (Novagen, VWR International, Bristol, CT), expressed in E. coli (BL21-DE3) and His-tag purified on a Ni-NTA column (Novagen). The purified protein was used to immunize two New Zealand white rabbits following standard procedures.
Immunoblotting
Sua4 cells were lysed in ice-cold lysis buffer (50 mM Tris, pH 7.8; 150 mM NaCl; 1% IGEPAL CA360 (Sigma, St. Louis, MO)) with Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN) and 1 mM PMSF. Total cell lysate protein was quantitated using the BCA Protein Assay (Pierce, Rockford, IL), aliquoted and frozen at -20°C. Total cell lysate was separated on 8% SDS-PAGE gel and electroblotted to a PVDF membrane in 1X Towbin buffer according to standard protocols. Upon completion of the protein transfer, the gel was washed twice for 10 minutes in 1X TBS buffer (10 mM Tris-HCl, pH 7.5; 150 mM NaCl). It was then blocked in blocking buffer (1.5% non-fat dry milk (NFDM), 1.5% fraction V Bovine Serum Albumin (BSA), 1X TBS, 0.05% Tween-20) with 20% 5X casein (Novagen), in a sealed bag overnight at 4°C. The blot was then washed twice for 10 minutes in 1X TBSTT and once for 10 minutes in 1X TBS and was incubated for 1 hour at room temperature on an orbital shaker with CTCF polyclonal antisera diluted 1:250 in blocking buffer without casein. After antibody binding, the blot was washed twice in 1X TBSTT (1X TBS, 0.05% Tween-20, 0.2% Triton X-100) for 10 minutes and once in 1X TBS for 10 minutes. Anti-Rabbit IgG (Fc) AP conjugate (Promega, Madison, WI 53711) was diluted 1:7500 in blocking buffer without casein and incubated with the blot for 1 hour at room temperature on an orbital shaker. The blot was then washed for 10 minutes five times in 1X TBSTT. Finally, it was developed for 1–10 minutes in Sigma-FAST™ (Sigma Aldrich Chemical Company, St. Louis, MO 63178) according to the manufacturer's instructions.
Electromobility Shift Assay (EMSA)
Sua4 cell lysates were prepared and the total protein quantitated as described above. Probes for EMSA were amplified and simultaneously labelled with α-32P (Amersham) by PCR using the following primers: 5'HS4Forw 5' gagctcacggggacagcccccc 3', 5'HS4Rev 5' aagctttttccccgtatccccc 3', Fab8Forw 5' ggcacaatcaagttaatgttgg 3', Fab8Rev 5' gcaagcgaagagttccattc 3'. The chicken 5'HS4 fragment (250 bp) was amplified from pJC13-1 [46] and the Drosophila Fab8 fragment (309 bp) was amplified from Drosophila genomic DNA. The binding reaction protocol was adapted from Filippova et al. [20]. Approximately 10 fmol of labelled probe was incubated for 15 minutes on ice with 0, 1.5, 7.5 or 15 μg of total cell protein in binding buffer (1X PBS with 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM DTT, 0.1% IGEPAL CA360 (Sigma), 10% glycerol) in the presence of a mixture of non-specific, cold, double-stranded competitor DNAs (500 ng polydI· polydC, 500 ng polydG· polydC, 500 ng SpI oligos, 500 ng Egr1 oligos). The SpI and Egr1 ds oligos contain strong, C/G-rich binding sites for the zinc-finger proteins SpI and Egr1 respectively. Sample 5 contained 150-fold excess unlabeled specific competitor. For the supershift, anti-sera against the An. gambiae CTCF was then added and the reactions incubated an additional 15 minutes on ice. Complexes were separated from the free probe on a 5% native PAGE gel in 0.5X TBE. The gel was run for 3.5 hours at 4°C at 10 V/cm.
References
Baniahmad A, Steiner C, Kohne AC, Renkawitz R: Modular structure of a chicken lysozyme silencer: Involvement of an unusual thyroid hormone receptor binding site. Cell. 1990, 61: 505-514. 10.1016/0092-8674(90)90532-J
Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH: A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene. 1990, 5: 1743-1753.
Ohlsson R, Renkawitz R, Lobanenkov V: CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001, 17: 520-527. 10.1016/S0168-9525(01)02366-6
West AG, Gaszner M, Felsenfeld G: Insulators: many functions, many mechanisms. Genes Dev. 2002, 16: 271-288. 10.1101/gad.954702
Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ: CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001, 28: 335-343. 10.1038/ng570
Bell AC, West AG, Felsenfeld G: The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999, 98: 387-396. 10.1016/S0092-8674(00)81967-4
Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G: Position-effect protection and enhancer blocking by the chicken b-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002, 99: 6883-6888. 10.1073/pnas.102179399
Antes TJ, Namciu SJ, Fournier REK, Levy-Wilson B: The 5' Boundary of the Human Apolipoprotein B Chromatin Domain in Intestinal Cells. Biochem. 2001, 40: 6731-6742. 10.1021/bi0100743. 10.1021/bi0100743
Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM: CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000, 405: 486-489. 10.1038/35013106
Ishihara K, Sasaki H: An evolutionarily conserved putative insulator element near the 3' boundary of the imprinted Igf2/H19 domain. Human Mol Genet. 2002, 11: 1627-1636. 10.1093/hmg/11.14.1627. 10.1093/hmg/11.14.1627
Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS: Transgenic RNAi Reveals Essential Function for CTCF in H19 Gene Imprinting. Science. 2003, 303: 238-240. 10.1126/science.1090934. 10.1126/science.1090934
Du M, Beatty LG, Winjing Z, Lew J, Schoenherr C, Weksberg R, Sadowski PD: Insulator and silencer sequences in the imprinted region of human chromosome 11p15.5. Human Mol Genet. 2003, 12: 1927-1939. 10.1093/hmg/ddg194. 10.1093/hmg/ddg194
Schoenherr CJ, Levorse JM, Tilghman SM: CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet. 2003, 33: 66-69. 10.1038/ng1057
Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, Ohlsson R: The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 2003, 17: 586-590. 10.1101/gad.254903
Wylie AA, Murphy SK, Orton TC, Jirtle RL: Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000, 10: 1711-1718. 10.1101/gr.161600
Lutz M, Burke LJ, LeFevre P, Myers FA, Thorne AW, Crane-Robinson C, Bonifer C, Filippova GN, Lobanenkov V, Renkawitz R: Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. Embo J. 2003, 22: 1579-1587. 10.1093/emboj/cdg147
Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM: Boundaries between Chromosomal Domains of X Inactivation and Escape Bind CTCF and Lack CpG Methylation during Early Development. Devel Cell. 2005, 8: 31-42. 10.1016/j.devcel.2004.10.018. 10.1016/j.devcel.2004.10.018
Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT: CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002, 295: 345-347. 10.1126/science.1065982
Moon H, Filippova GN, Loukinov DI, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke LJ, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V: CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Reports. 2005, 6: 165-170. 10.1038/sj.embor.7400334
Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV: An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996, 16: 2802-2813.
Ensembl Mosquito Genome Browser. http://www.ensembl.org/Anopheles_gambiae/
Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV: CTCF, a conserved nuclear factor required for chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993, 13: 7612-7624.
Burke LJ, Hollemann T, Pieler T, Renkawitz R: Molecular cloning and expression of the chromatin insulator protein CTCF in Xenopus laevis. Mech Dev. 2002, 113: 95-98. 10.1016/S0925-4773(02)00005-9
Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA: Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A. 2004, 101: 6536-6541. 10.1073/pnas.0401670101
National Center for Biotechnology Information (NCBI). http://www.ncbi.nlm.nih.gov/
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987, 4: 406-425.
Fitch WM, Margoliash E: Construction of phylogenetic trees. Science. 1967, 155: 279-284.
Felsenstein J: PHYLIP-Phylogeny Interference Package (Version 3.2). Cladistics. 1989, 5: 164-166.
Felsenstein J: Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996, 266: 418-427.
Strimmer K, von Haeseler A: Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996, 13: 964-969.
Larget B, Simon D: Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetics trees. Mol Biol Evol. 1999, 16: 750-759.
Muller JM, Dimopoulos G, Blass C, Kafatos FC: A Hemocyte-like Cell Line Established from the Malaria Vector Anopheles gambiae Expresses Six Prophenoloxidase Genes. J Biol Chem. 1999, 274: 11727-11735. 10.1074/jbc.274.17.11727
Suzuki M, Gerstein M, Yagi N: Sterochemical basis of DNA recognition by Zn fingers. Nucl Acids Res. 1994, 22: 3397-3405.
Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R: The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002, 298: 129-149. 10.1126/science.1076181
Warren AM, Crampton JM: The Aedes aegypti genome: complexity and organization. Genet Res. 1991, 58: 225-232.
Severson DW, Knudson DL, Soares MB, Loftus BJ: Aedes aegypti genomics. Insect Biochem Mol Biol. 2004, 34: 715-721. 10.1016/j.ibmb.2004.03.024
Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu SQ, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM: Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002, 3: research0088.1-88.14. 10.1186/gb-2002-3-12-research0088. 10.1186/gb-2002-3-12-research0088
Berkeley Drosophila Genome Project. http://www.fruitfly.org
Drosophila Developmental Gene Expression Timecourse. http://genome.med.yale.edu/Lifecycle/query_gen.php?input1=FBgn0035769
Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP: Gene Expression During the Life Cycle of Drosophila melanogaster. Science. 2002, 297: 2270-2275. 10.1126/science.1072152
Villalva C, Touriol C, Seurat P, Trempat P, Delsol G, Brousset P: Increased Yield of PCR Products by Addition of T4 Gene 32 Protein to the SMART™ PCR cDNA Synthesis System. Biotechniques. 2001, 31: 81-86.
Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S: Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucl Acids Res. 1998, 26: 1628-1635. 10.1093/nar/26.7.1628
Rose T, Henikoff J, Henikoff S: CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucl Acids Res. 2003, 31: 3763-3766. 10.1093/nar/gkg524
Corpet F: Multiple sequence alignment with hierarchical clustering. Nucl Acids Res. 1988, 16: 10881-10890.
Chung JH, Whiteley M, Felsenfeld G: A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against postion effect in Drosophila. Cell. 1993, 74: 505-514. 10.1016/0092-8674(93)80052-G
Acknowledgements
We gratefully acknowledge Hans-Michael Muller for the Sua4 cell line, Gary Felsenfeld for the plasmid containing 5'HS4, and Andrea Taylor at the LARR facility at TAMU for antibody generation. This work was supported by NIH grant RO1 AI46432 to CJC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
CEG carried out the studies described in this paper and drafted the manuscript. CJC participated in the design and planning of this study and edited the manuscript. Both authors conceived of the study and have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Gray, C.E., Coates, C.J. Cloning and characterization of cDNAs encoding putative CTCFs in the mosquitoes, Aedes aegypti and Anopheles gambiae. BMC Molecular Biol 6, 16 (2005). https://doi.org/10.1186/1471-2199-6-16
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2199-6-16