Abstract
Background
Wingless gene (Wg) plays a fundamental role in regulating the segment polarity and wing imaginal discs of insects. The rice planthoppers have an obvious wing dimorphism, and the long- and short-winged forms exist normally in natural populations. However, the molecular characteristics and functions of Wg in rice planthoppers are poorly understood, and the relationship between expression level of Wg and wing dimorphism has not been clarified.
Results
In this study, wingless gene (Wg) was cloned from three species of rice planthopper, Sogatella furcifera, Laodelphgax striatellus and Nilaparvata lugens, and its characteristics and role in determining the wing dimorphism of S. furcifera were explored. The results showed that only three different amino acid residuals encoded by Wg were found between S. furcifera and L. striatellus, but more than 10 residuals in N. lugens were different with L. striatellus and S. furcifera. The sequences of amino acids encoded by Wg showed a high degree of identity between these three species of rice planthopper that belong to the same family, Delphacidae. The macropterous and brachypterous lineages of S. furcifera were established by selection experiment. The Wg mRNA expression levels in nymphs were significantly higher in the macropterous lineage than in the brachypterous lineage of S. furcifera. In macropterous adults, the Wg was expressed mainly in wings and legs, and less in body segments. Ingestion of 100 ng/μL double-stranded RNA of Wg from second instar nymphs led to a significant decrease of expression level of Wg during nymphal stage and of body weight of subsequent adults. Moreover, RNAi of Wg resulted in significantly shorter and deformative wings, including shrunken and unfolded wings.
Conclusion
Wg has high degree of identity among three species of rice planthopper. Wg is involved in the development and growth of wings in S. furcifera. Expression level of Wg during the nymphal stage manipulates the size and pattern of wings in S. furcifera.
Similar content being viewed by others
Background
Wingless/Wnt signaling pathway is a complicated protein-protein interaction network that regulates important developmental processes, such as cell proliferation, polarity and fate specification[1]. The wingless gene (Wg) in Drosophila is homologous with the mammalian Wnt-1 gene[2], which manipulates embryonic development[3], as well as the limb formation of adults[4]. Wg encodes a kind of cysteine-rich secreted protein[5], and the secreted location and concentration gradients mediate the composition of the midgut morphology, development of central nervous system and the formation of imaginal discs of wings, eyes and legs[6–8].
Wg protein belongs to a kind of morphogen which acts as a signaling molecule to directly control specific cellular responses[9]. Different concentrations of Wg protein lead to different cell reactions. It can stimulate some specific target genes when its concentration reaches a threshold gradient[10, 11]. It is well known that Wg influences development of Drosophila wing imaginal disc[12–14]. Wg gene is expressed in a narrow stripe along the dorsal/ventral (D/V) boundary during the development of wing disc of Drosophila[15], and it has been considered as one of important signal transduction pathways in the formation of wing pattern[16]. During the formation of wings, Wg regulates the expression of downstream target genes through changing its own concentration, and then results in the morphological differentiation of wing imaginal disc cells[9]. When Wg gene in Tribolium castaneum was knocked down in the late instar of larvae, the wing width of adults decreased[17]. Wg gene of Bombyx mori exhibited the highest expression level in wing primordium of the fifth instar larvae, and its expression level decreased gradually after pupation. When the expression level of Wg was reduced in the fifth instar larvae by RNA interference method, the resulting adults showed a partial or even complete loss of wings[18].
The white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae), is one of the most devastating pests in rice fields in Asia. It sucks phloem sap of rice and causes a decrease of grain weight[19]. Sogatella furcifera has a wing dimorphism which females have either macropterous or brachypterous wings, but males usually are macropterous in China. In their natural environments, the macropterous rice planthoppers can make long-distance migrations to expand their occurrence regions[20]. Although the genetic basis of wing polymorphism in insects is generally not well understood, it has been verified that the wing forms of rice planthopper have a genetic component and are not purely environmentally determined[21]. The titer of juvenile hormone and DNA methylation were also thought to be involved in the determination of wing forms[22–24]. There are some differentially expressed genes between the long- and short-winged brown planthoppers, such as flightin, troponin C4, titin and myosin heavy chain[25, 26]. However, the molecular characteristics, expression and biological function of Wg, an important gene relating to growth and wing imaginal disc development of insects, in rice planthoppers are still not well understood. Moreover, it is unknown whether the Wg plays a key role in determining the development of wings and in manipulating the wing dimorphism in rice planthoppers. In the present study, therefore, the full-length cDNAs encoding Wg were cloned and characterized from the three common species of rice planthopper, S. furcifera, Laodelphgax striatellus and Nilaparvata lugens. And then the expression differences of Wg between the macropterous and brachypterous lineages of S. furcifera which wing forms were selected for more than 20 generations under a constant condition were examined using the quantitative real-time PCR method. Finally, the survival of nymphs, body weight, wing length and wing pattern of adults in the macropterous lineage were measured when the Wg expression was knocked down by ingestion of dsRNA of Wg in nymphs. This study will illustrate the role of Wg in determining the wing dimorphism of rice planthoppers.
Results
Selection response of wing forms of S. furcifera
All the adults from the parents (macropterous female × macropterous male, M♀ × M♂ lineage) were macropterous after seven generations of selection. All the males from the parents (brachypterous female × macropterous male, B♀ × M♂ lineage) were macropterous during all these 40 generations of selection, and more than 95% of the females were brachypterous after 25 generations of selection (Figure 1). The S. furcifera had significant selection response in wing forms. The macropterous pure line had been obtained by seven continuous generations of selection from M♀ × M♂ lineage (Figure 1).
Characteristics of Wg from three species of rice planthoppers
The full-length cDNA clone encoding Wg was isolated from S. furcifera and L. striatellus by the 3′ and 5′ RACE, as well as the open reading frame (ORF) of Wg from N. lugens. The cDNA length of Wg from S. furcifera was 1571 bp which contains 31 bp 5′-untranslated region (UTR), 367 bp 3′-UTR with a consensus polyadenylation sequence and 1173 bp ORF. The deduced protein consisted of 390 amino acid residues. A full-length cDNA of Wg from L. striatellus was 1443 bp containing 34 bp 5′-UTR,1173 bp ORF, and 236 bp 3′-UTR, which also encoded 390 amino acid residues. The cDNA sequence of Wg from N. lugens had a complete 1185 bp ORF encoding 394 amino acid residues. The deduced amino acid sequences encoded by Wg had high level of identity among the three species of rice planthopper. There were only three different amino acid residues between S. furcifera and L. striatellus. Nilaparvata lugens Wg had ten and eleven different amino acid residues compared to S. furcifera and L. striatellus, respectively. The result implied that Wg was highly conservative in rice planthoppers (Figure 2).
Alignment of the deduced amino acid sequences encoded by Wg from three species of rice planthopper, Nilaparvata lugens , Sogatella furcifera and Laodelphax striatellus . Amino acids covered by black, grey, and white bars are 100%, 80%, and below 80% identity among the three species of rice planthoppers, respectively.
Expression of Wg in the macropterous and brachypterous lineages of S. furcifera
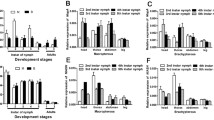
Relative expression levels of Wg mRNA were significantly higher in nymphs than in adults regardless of the macropterous and brachypterous lineages (for the macropterous lineage: F4, 25 = 36.876, P < 0.0001; for the brachypterous lineage: F4, 25 = 51.908, P < 0.0001). Moreover, the expression levels were significantly higher in the macropterous lineage than in the brachypterous lineage during the 3rd instar (t = 4.238, df = 5, P = 0.0082), 4th instar (t = 3.554, df = 5, P = 0.0163), and 5th instar nymphs (t = 5.946, df = 5, P = 0.0019), whereas there were no significant differences between the two lineages during adults (male: t = 0.870, df = 5, P = 0.4243; female: t = 2.018, df = 5, P = 0.0997, Figure 3).
Relative expression levels of Wg mRNA in the macropterous and brachypterous lineages of Sogatella furcifera . Error bars indicate standard error (SE, n = 6). * and ** above the bars indicate significant differences at P < 0.05 and 0.01 level between the macropterous and brachypterous lineages, respectively.
Relative expression levels of Wg mRNA in different parts of the macropterous adults of S. furcifera, such as head, thorax, abdomen, wings and legs, showed that Wg was expressed mainly in wings and legs, and at much lower levels in body segments: head, thorax, and abdomen (males: F4, 10 = 73.556, P < 0.0001; females: F4, 10 = 81.891, P < 0.0001). Relative expression levels between females and males were not significantly different (Figure 4).
Relative expression levels of Wg mRNA in five parts of the macropterous adults of Sogatella furcifera . Error bars indicate standard error (SE, n = 3). Different capital and lowercase letters above the bars indicate significant differences at P < 0.05 level among different parts of male and female adults, respectively.
Survival and body weight of S. furcifera after Wg RNAi
The results showed that mortality rates of S. furcifera nymphs ingested artificial diet with 100 ng/μL dsWg and dsEGFP were 38.81 ± 3.77% and 30.35 ± 6.54%, respectively, and there were no significant differences (t = 1.209, df = 8, P = 0.261). Mortality of nymphs in the control was 6.41 ± 1.73% which was significantly lower than these dsWg and dsEGFP treated nymphs (F2, 10 = 11.041, P = 0.03). Relative expression levels of Wg mRNA in nymphs fed on 100 ng/μL dsWg for seven days significantly decreased (F2, 6 = 8.836, P = 0.018, Figure 5). Body weights of these resulting adults were reduced when they were fed on artificial diet with dsWg during nymphs in comparison with those fed on diet with dsEGFP (males: t = 2.934, P = 0.008; females: t = 5.360, P < 0.0001, Figure 6).
Relative expression levels of Wg mRNA in nymphs of Sogatella furcifera feeding on artificial diet with 100 ng/μ L dsEGFP and dsWg for seven days. Error bars indicate standard error (SE, n = 3). Different lowercase letters above the bars indicate significant differences among the Wg RNAi and controls at P < 0.05 level.
Wings of S. furcifera treated by Wg RNAi
Wg RNAi resulted in seriously deformative wings in S. furcifera. The abnormal rates of wings in the blank (CK) and negative controls (dsEGFP) were 1.90% (n = 105) and 1.82% (n = 110), respectively, however, the value increased to 18.55% (n = 124) when the nymphs were fed on diet with dsWg. There were significant differences among dsWg, dsEGFP and CK in the abnormal rates of wings (x2 = 29.88, df = 2, P < 0.0001). The normal wings of S. furcifera were smooth and folded on the back (Figure 7 A and B), but the adults had shrunken (Figure 7C and D) or unfolded wings on the back (Figure 7E and F) when their nymphs were ingested artificial diet with 100 ng/μL dsWg. The wings from nymphs ingested dsWg had obvious patterning defects in the vines and pigmentation, and the background and veins of wings were depigmented (Figure 7). The lengths of fore- and hind-wing from base to tip became significantly shorter when nymphs were fed with 100 ng/μL dsWg than that fed with 100 ng/μL dsEGFP (female fore-wing: t = 3.68, P < 0.0001; female hind-wing: t = 3.68, P < 0.0001; male fore-wing: t = 4.74, P < 0.0001; male hind-wing: t = 5.88., P < 0.0001, Figure 8). The Wg was involved in the determination of wing length and pattern in S. furcifera.
Discussion
The white-backed planthopper is one of insects with wing-dimorphism which adult was either macropterous or brachypterous. It has been reported that the development of wing was controlled by multiple genes[27–30]. The wing pattern genes are mainly involved in determining three kinds of development of insect wings: proximal-distal, dorsal-ventral and anterior-posterior. The Wg occupies a key position in the genetic determination system of the development of dorsal-ventral axis, and it directly impacts the expression of downstream genes controlling the wing pattern[9]. In this study, we found the wing forms of S. furcifera exhibited a significant selection response, and the macropterous pure line and brachypterous near-pure line were obtained after seven and twenty-five generations of selection, respectively. Additionally, we cloned the full-length cDNA of Wg from S. furcifera and L. striatellus, and the ORF from N. lugens. The amino acid residues encoded by Wg had more than 99% identity between S. furcifera and L. striatellus, whereas the value was 94% between S. furcifera and N. lugens. The Wg is highly conservative within the Delphacidae. Among the three species of rice planthopper, S. furcifera, L. striatellus and N. lugens, the Wg from S. furcifera had higher similarity with L. striatellus, but relatively lower with N. lugens. Peng et al.[17] presumed that the genetic basis of wing forms in S. furcifera was as similar as L. striatellus and both were controlled by two pairs of alleles, one locating on a euchromosome and the other on a sex chromosome, but it was significantly different from N. lugens which was controlled by an allele locating on a euchromosome[28]. The variations of Wg among these three species of rice planthopper supported the opinion of Peng et al.[17].
In this study, we found that the Wg mRNA expressed highly in third, fourth and fifth instar nymphs, but surprisingly, in adult, the expression level of Wg mRNA was significantly lower. The nymphs from third to fifth instar are in the process of development and growth of wings, therefore the Wg mRNA expression shows a high level. In the adult, the wings are fully formed, so the expression level becomes lower. In pea aphid, an insect that also has the wing dimorphism, there is a trend towards higher Wg mRNA expression in the winged morphs than the wingless ones[27]. In the present study, we found that Wg mRNA expression levels in the 3rd-5th instar nymphs of the macropterous pure line of S. furcifera were significantly higher than that in the brachypterous line, but no differences were found in adults between these two lines. Classic Wingless/Wnt signaling pathway in Drosophila is related to wing formation process, including wing growth and morphogenesis[31]. In our study, we found that the higher expression of Wg mRNA in the nymphal stage was beneficial to form the long-winged rice planthoppers. The relative expression of Wg mRNA was more abundant in wings than that in the other parts of adult. These results revealed that the Wg gene indeed played an important role in the wing development and growth of S. furcifera.
Studies had already illustrated that Wg was required for wing cell survival, particularly during the rapid growth phase of wing development. The determination of the final wing size of wings in Drosophila was only a part of functions of Wg[32]. The lower expression level of Wg mRNA in the nymphs of the brachypterous S. furcifera lineage might be resulted from the requirement of the development of short wings or from other functions of Wg.
Nymphs of S. furcifera treated with 100 ng/μL dsWg from the second instar for 7 days had abnormal wings as adults, including shrunken and unfolded wings. In the previous studies, it has been found that the mutation of Wg caused the loss of wings in adults of D. melanogaster[33]. In Bombyx mori, the knocking-down of Wg expression by dsRNA resulted in partially missing or even absent wings[18]. In S. furcifera, we found the wing lengths of the fore- and hind-wing were significantly shortened caused by the ingestion of dsWg. Therefore, the Wg was really involved in the development and growth of wings in rice planthoppers. The β-catenin pathway, one of the Wg signal pathway, is required for growth and cell fate specification[34]. The decrease of body weight and wing length of adults S. furcifera implied that the β-catenin pathway would be disrupted when the nymphs fed on diet with dsWg. The Wg performs the function to determine the growth of wings.
The genetic determination of wing morphs in the rice planthoppers is complex. It has been illustrated that the wing form of rice planthoppers is a quantitative trait controlled by many genes[35–37]. We found here that the wing lengths of female and male adults became significantly shorter when the silencing of the Wg was performed at nymphal stage, however, the wings were still longer than that of the naturally branchypterous ones. The forewing lengths were 1.2-1.8 mm and 3.2-3.8 mm in the brachypterous and macropterous females of S. furcifera, respectively[38]. In this study the wing length from the dsWg nymphs was 2.84 ± 0.07 mm which was an intermediate wing form. We also found that the relative expression level of Wg mRNA in third instar nymphs of the brachypterous lineage was as about 66 percent as the macropterous lineage, and wing length of brachypterous female in natural field was as 47 percent as the macropterous one[38]. However, when the Wg mRNA expression level in the macropterous lineage was interfered 50 percent by dsWg in nymphs, the wing length of female was only shortened 10 percent. The expression levels of Wg mRNA in nymphs were not linearly related with the wing lengths of rice planthoppers. We presumed that the wing forms of rice planthoppers might be a threshold response to Wg. When the expression level of Wg was less than a lower threshold gradient, the adults would be short-winged. Otherwise, when it was more than a higher threshold, the adults would be long-winged. Fortunately, we have attained an adult which wings only covered its abdomen when the nymph was fed with a higher concentration 400 ng/uL of dsWg. Of course, this comprehensive process of Wg manipulating the wing size and pattern of rice planthoppers still needs the further study.
Conclusions
The wingless gene has a high level of identity among the three species of rice planthopper, and was strongly related to the development and growth of wings. The expression levels of Wg mRNA were significantly higher in the macropterous lineage than in the brachypterous lineage of S. furcifera. Interference of Wg resulted in the shorter and abnormal wings in S. furcifera.
Methods
Experimental insects
Three species of rice planthoppers, S. furcifera, N. lugens and L. striatellus, were originally collected from rice fields in Nanjing, Jiangsu province, China, and then reared in laboratory at 25 ± 1°C, relative humidity 75 ± 10%, and 14 h light/10 h dark photoperiod using the rice seedlings (var. Wuyunjing 7).
Selection of the macropterous and brachypterous lineages of S. furcifera
Due to shortage of the brachypterous males in S. furcifera in Jiangsu province, only two mating combinations: macropterous male × macropterous female (M♂ × M♀, called the macropterous lineage) and macropterous male × brachypterous female (M♂ × B♀, called the brachypterous lineage) were established, and reared under a constant condition (25 ± 1°C, RH 75 ± 10%, and 14 h light/10 h dark). In their offspring generations, only the females and males with the same wing form as their parents were chosen to mate and propagate. The detail method for selection experiment of wing forms was described by Peng et al.[28]. Continuous 40 generations of selection were performed, and the macropterous and brachypterous lineages were used in the following experiments.
Cloning of the wingless gene cDNA from three species of rice planthopper
Total RNA was isolated from the homogenization of 10 nymphs of rice planthopper (two individuals from each instar) and four adults (two macropterous males, and one macropterus and one brachypterous female) with Trizol (Invitrogen Co., USA). Total RNA 1 μg was used to synthesize the templates for cloning the Wg cDNA using the BD SMART™ RACE cDNA Amplification Kit (Clontech). The PCR primers were SfGSP1-5′: 5′-AGAAGCCAGGCGACGACTCCAGGTAG-3′, SfNGSP1-5′: 5′-CGTCGAACCTGTCCTTGAGGCTGTC-3′, SfGSP2-3′: 5′-ACTTTGGATTCAAGTTCTCGCGGGAG-3′, and SfNGSP2-3′: 5′-AAGCCCTACAACCCGGAGCACAAGC-3′ for S. furcifera, and LsGSP3-5′:5′-CGTCGAACCTGTCCTTGAGGCTGTC-3′, LsNGSP3-5′:5′-AAGCCCTACAACCCGGAGCACAAGC-3′, LsGSP4-3′:5′-CACAACACATCAGGTCGCAGCCGT-3′, and LsNGSP4-3′:5′-TTGTGCTCCGGGTTGTAGGGCTTCAG-3′ for L. striatellus. PCR reaction system 25 μL was used which contained 1 μL template, 2.5 μL 10 × Taq buffer (Mg2+ free), 2 μL MgCl2 (25 mM), 2 μL dNTP mixture (2.5 mM/each), 1 μL forward and 1 μL reverse primers, 0.25 μL Taq polymerase (Takara Bio.) and 15.25 μL double distilled H2O. PCR was preceded by denaturation at 94°C for 2 min, followed by 5 cycles at 94°C for 30 s and 72°C for 5 min, and by another 5 cycles at 94°C for 30 s, 70°C for 30s, and 72°C for 4 min, and then by 25 cycles at 94°C for 30 s, 68°C for 30 s, 72°C for 4 min and finishing with chain extension at 72°C for 10 min. The amplified product was separated by 1.5% agarose gel, and the target product was recycled and purified with the Wizard DNA Gel Extraction Kit (Unigenes Promega, Madison, Wis., USA), cloned into the EASY-T3 vector (TransGen Biotech, Beijing, China), and transformed into the DH5α competent cells. Positive clones were chosen to sequence. According to the sequences of Wg in S. furcifera and L. striatellus, the end to end primers sfwg-F2 5′-GATGGCAGCGCAATGATG-3′ and Sfwg-R2 5′-CGCTGTGGGTTTGGGTAA-3′ for S. furcifera, Lswg-F2 5′-AGTACAAGCCTGGTGATGG-3′ and Lswg-R2 5′-GGCAATAAAGTGAATTGAATAA-3′ for L. striatellus were designed to check the full-length of Wg. Unfortunately, We did not clone the full-length of Wg from N. lugens, and only cloned its ORF used the end to end primers for S. furcifera. The ORF sequences of Wg gene in the three species of rice planthopper were found using the ORF Finder online service in NCBI (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi).
Expression level of Wg mRNA in the macropterous and brachypterous lineages of S. furcifera
The 3rd, 4th and 5th instar nymphs and adults from the 29th generation of the macropterous lineage and the 30th generation of the brachypterous lineage of S. furcifera were collected and frozen in 300 μL Trizol for 24 hours at -70°C. The total RNA from six individuals was isolated using Trizol. The samples were grinded well by mixing some quartz rocks. The total RNA was adjusted to 1 μg/μL with DEPC-treated H2O, and 2 μg RNA was used for RT-PCR in the 20 μL reaction mixture using the One Step SYBR PrimerScript RT-PCR Kit (Takara) to synthesize the first-strand cDNA. The real-time qPCR was performed on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) to quantify the expression levels of Wg. The first-strand cDNA (2 μL) diluted 50-fold was used as templates in each 20 μL reaction mixture. The qRT-PCR was performed at 95°C for 30 s, and then for 40 cycles at 95°C for 5 s and 60°C for 34 s. The ribosomal 18 s rRNA was used as an internal control. Relative expression level of Wg mRNA was computed based on the internal control gene using the 2-△△Ct method. The special primer sets for quantifying Wg in S. furcifera were the sense primer (SFwg-F) 5′-TCGAATGCCAATTCCAGTTTAG -3′ and the antisense primer (SFwg-R) 5′-CCCCTATCCACAATCTTTCCA -3′. And in the internal control, the primers for S. furcifera 18S rRNA gene were 18S rRNA-F 5′-ACAAGTATCAATTGGAGGGCAAGTCTGG-3′ and 18S rRNA-R 5′-ATGCACACAGTATACAGGCGTGACAAG -3′. The expression levels of Wg mRNA in head, thorax, abdomen, wings and legs were also detected in the macropterous lineage of S. furcifera which was selected for 37 generations. For each sample, we carried out six biological replicates.
Synthesis of the double stranded RNA (dsRNA)
A 246 bp of nucleotide sequence of S. furcifera Wg was cloned using the sense primer dsWg F: 5′- GCTGCCCAACCTGCGCGT-3′ and antisense primer dsWg R: 5′- CCGTGCGTGCCCTGGATG-3′ designed by the coding region sequence of Wg, and inserted into the EASY-T3 vector (Trans Gen Biotech, Beijing, China) to confirm the sequences. The plasmid containing the specific nucleotide sequences was extracted by the MinBEST plasmid Purification Kit (Takara). The diluted plasmid was used as templates for amplification of the target sequence (dsWg) by PCR. The sense primer was T7 Wg F: 5′-TAATACGACTCACTATAGGG GCTGCCCAACCTGCGCGT-3′ and the antisense primer was T7 Wg R: 5′- TAATACGACTCACTATAGGG CCGTGCGTGCCCTGGATG-3′. The PCR procedures for synthesizing dsWg were performed at 94°C for 2 min followed by 35 cycles at 94°C for 30 s, 55°C for 20 s and 72°C for 20 s, and the last cycle was followed by final extension at 72°C for 5 min. The T7 RiboMAX™ Express RNAi System (Promega USA) was used to produce the specific dsRNA. The dsRNA synthesized here was washed twice using 70% ethanol, dried and suspended into an appropriate amount of the nuclease-free water, and then quantified by the Nano Drop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) at 260 nm. The quality and size of the dsRNA products were verified by 1.5% agarose gel electrophoresis. Enhanced green fluorescent protein gene dsRNA (dsEGFP) was used as the negative control. The sense primer was T7 EGFP-R 5′-TAATACGACTCACTATAGGGAAGTTCAGCGTGTCCG-3′ and the antisense primer was T7 EGFP-F 5′-TAATACGACTCACTATAGGGCACCTTGATGCCGTTC-3′ for synthesizing the dsEGFP. The synthesized dsRNA was stored at -70°C.
Nymph RNAi
To determine the function of Wg in S. furcifera, the second instar nymphs were fed on artificial diet with the dsWg[39–41]. Groups of 30 second-instar nymphs were collected from the 37th generation selection of the macropterous lineage of S. furcifera, and placed into a feeding chamber constructed from a 3 cm diameter and 12 cm height cylindrical glass tube with one end covered by insect-proof nylon mesh net (48-micron) and the other end covered by two-layer of Parafilm sandwiched together[42]. Seventy-microliter liquid diets containing 100 ng/ul dsWg were put into the two layers. The nymphs feeding on artificial diet containing 100 ng/ul dsEGFP and water were the negative and blank control, respectively. The artificial diet was replaced daily. After seven days on the diet, the nymphs were counted and collected to examine the expression levels of Wg mRNA by qPCR method. When the nymphs grew into adults, their survival, wing forms, wing lengths, and body weights were measured. Curled wings were unfolded by dipping in absolute ethanol for 48 hours before measurement. The lengths of wings were measured from the base to tip under a stereomicroscope using the CellSens V 1.5 system (Olympus ®). The dsWg and dsEGFP treatments were repeated at least three times.
Data analysis
Wg protein sequences in three species of rice planthopper were aligned in a multiple sequence alignment using CLUSTAL X[43]. The differences between two means, such as the relative expression levels of Wg mRNA between the macropterous and brachypterous lineages, were compared by a student’s t test. The expression levels of Wg mRNA, body weight and wing length among different tissues or RNAi treatments were analyzed by the ANOVA followed by the Tukey test. The proportions of adults with abnormal wings among RNAi treatments by dsWg and dsEGFP and the control were compared by the Chi square analysis using the Crosstabs method. All these statistic analyses were performed using SPSS 13.0 software (SPSS, Chicago, IL, USA).
Abbreviations
- Wg :
-
Wingless gene
- EGFP :
-
Enhanced green fluorescent potein
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Reverse transcriptase PCR
- qRT-PCR:
-
Quantitative real-time PCR
- cDNA:
-
Complementary DNA
- RACE:
-
Rapid-amplification of cDNA ends
- dsRNA:
-
Double-stranded RNA
- RNAi:
-
RNA interference
- ORF:
-
Open reading frame
- UTR:
-
Untranslated region
- SE:
-
Standard error
- ANOVA:
-
Analysis of variance.
References
Seto ES, Bellen HJ: The ins and outs of wingless signaling. Trends Cell Biol. 2004, 14 (1): 45-53. 10.1016/j.tcb.2003.11.004
Rijsewijk F, Schuermann M, Wagenaar E, Parren P, Weigel D, Nusse R: The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell. 1987, 50 (4): 649-657. 10.1016/0092-8674(87)90038-9
Gonsalves FC, DasGupta R: Function of the wingless signaling pathway in Drosophila. Methods Mol Biol. 2008, 469: 115-125. 10.1007/978-1-60327-469-2_10
Schubiger M, Sustar A, Schubiger G: Regeneration and transdetermination: The role of wingless and its regulation. Dev Biol. 2010, 347 (2): 315-324. 10.1016/j.ydbio.2010.08.034
Cadigan KM, Nusse R: Wnt signaling: a common theme in animal development. Genes Dev. 1997, 11 (24): 3286-3305. 10.1101/gad.11.24.3286
Chu-LaGraff Q, Doe CQ: Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science. 1993, 261 (5128): 1594-1597. 10.1126/science.8372355
Siegfried E, Perrimon N: Drosophila wingless: A paradigm for the function and mechanism of Wnt signaling. Bioessays. 1994, 16 (6): 395-404. 10.1002/bies.950160607
Takashima S, Paul M, Aghajanian P, Younossi-Hartenstein A, Hartenstein V: Migration of Drosophila intestinal stem cells across organ boundaries. Development. 2013, 140 (9): 1903-1911. 10.1242/dev.082933
Swarup S, Verheyen EM: Wnt/Wingless signaling in Drosophila. Cold Spring Harb Perspect Biol. 2012, 4: a007930-
Cadigan KM: Regulating morphogen gradients in the Drosophila wing. Semin Cell Dev Biol. 2002, 13 (2): 83-90. 10.1016/S1084-9521(02)00014-9
Tabata T, Takei Y: Morphogens, their identification and regulation. Development. 2004, 131 (4): 703-712. 10.1242/dev.01043
Phillips RG, Whittle JRS: Wingless expression mediates determination of peripheral nervous system elements in late stages of Drosophila wing disc development. Development. 1993, 118 (2): 427-438.
Simcox AA, Roberts IJH, Hersperger E, Gribbin MC, Shearn A, Whittle JRS: Imaginal discs can be recovered from cultured embryos mutant for the segment-polarity genes engrailed naked and patched but not from wingless. Development. 1989, 107 (4): 715-722.
Williams JA, Paddock SW, Carroll SB: Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development. 1993, 117 (2): 571-584.
Campbell G, Weaver T, Tomlinson A: Axis specification in the developing Drosophila appendage: the role of wingless, decapentaplegic, and the homeobox gene aristaless. Cell. 1993, 74 (6): 1113-1123. 10.1016/0092-8674(93)90732-6
Yang L, Meng F, Ma D, Xie W, Fang M: Bridging decapentaplegic and wingless signaling in Drosophila wings through repression of naked cuticle by brinker. Development. 2013, 140 (2): 413-422. 10.1242/dev.082578
Peng YN, Li CJ, Li B, Li ZY: Knocking-down of Wingless/Wnt1 influences the development of Tribolium castaneum. Chin J Biotechnol Mol Biol. 2012, 28 (8): 733-738.
Tong XL: Study on the wing pattern genes in the silkworm, Bombyx mori. PhD thesis. 2008, China: Southwest University
Zhu ZR, Cheng J: Sucking rates of the white-backed planthopper Sogatella furcifera (Horv.) (Homoptera, Delphacidae) and yield loss of rice. J Pest Science. 2002, 75 (5): 113-117.
Otuka A, Matsumura M, Watanabe T: The search for domestic migration of the white-backed planthopper, Sogatella furcifera (Horvath) (Homoptera: Delphacidae), in Japan. Appl Entomol Zool. 2009, 44 (3): 379-386. 10.1303/aez.2009.379. 10.1303/aez.2009.379
Denno RF, Roderick GK: Population biology of planthoppers. Ann Rev Entomol. 1990, 35: 489-520. 10.1146/annurev.en.35.010190.002421. 10.1146/annurev.en.35.010190.002421
Roff DA: The evolution of wing dimorphism in insects. Evolution. 1986, 40 (5): 1009-1020. 10.2307/2408759. 10.2307/2408759
Zhang QX, Sun ZX, Li GH, Wang FH: Effects of three kinds of exogenous hormones on wing dimorphism of Sogatella furcifera (Horvath). Acta Ecol Sin. 2008, 28 (12): 5994-5998.
Zhou X, Chen J, Zhang M, Liang S, Wang F: Differential DNA methylation between two wing phenotypes adults of Sogatella furcifera. Genesis. 2013, 51 (12): 819-826. 10.1002/dvg.22722
Xue JA, Bao YY, Li BL, Cheng YB, Peng ZY, Liu H, Xu HJ, Zhu ZR, Lou YG, Cheng JA, Zhang CX: Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS One. 2010, 5: e14233- 10.1371/journal.pone.0014233
Xue J, Zhang XQ, Xu HJ, Fan HW, Huang HJ, Ma XF, Wang CY, Chen JG, Cheng JA, Zhang CX: Molecular characterization of the flightin gene in the wing-dimorphic planthopper, Nilaparvata lugens, and its evolution in Pancrustacea. Insect Biochem Mol Biol. 2013, 43 (5): 433-443. 10.1016/j.ibmb.2013.02.006
Brisson JA, Ishiawa A, Miura T: Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol Biol. 2010, 19 (Suppl 2): 63-73.
Peng J, Zhang C, An ZF, Yu JL, Liu XD: Genetic analysis of wing-form determination in three species of rice planthoppers (Hemiptera: Delphacidae). Acta Entomol Sin. 2012, 55 (8): 971-980.
Weatherbee SD, Halder G, Kim J, Hudson A, Carroll S: Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 1998, 12 (10): 1474-1482. 10.1101/gad.12.10.1474
Braendle C, Davis GK, Brisson JA, Stern DL: Wing dimorphism in aphids. Heredity. 2006, 97 (3): 192-199. 10.1038/sj.hdy.6800863
Logan CY, Nusse R: The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004, 20: 781-810. 10.1146/annurev.cellbio.20.010403.113126
Johnston LA, Sanders AL: Wingless promotes cell survival but constrains growth during Drosophila wing development. Nat Cell Biol. 2003, 5 (9): 827-833. 10.1038/ncb1041
Sharma RP, Chopra VL: Effect of wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev Biol. 1976, 48 (2): 461-465. 10.1016/0012-1606(76)90108-1
Kohn AD, Moon RT: Wnt and calcium signaling: β-Catenin-independent pathways. Cell Calcium. 2005, 38 (3–4): 439-446.
Mahmud FS: Alary polymorphism in the small brown planthopper Laodelphax striarellus (Homoptera: Delphacidae). Ent Exp Appl. 1980, 28 (1): 47-53. 10.1111/j.1570-7458.1980.tb02986.x. 10.1111/j.1570-7458.1980.tb02986.x
Mori K, Nakasuji F: Genetic analysis of the wing-form determination of the small brown planthopper, Laodelphax striatellus (Hemiptera: Delphacidae). Res Popul Ecol. 1990, 32 (2): 279-287. 10.1007/BF02512563. 10.1007/BF02512563
Liu JN, Gui FR, Li ZY: Factors of influencing the development of wing dimorphism in the rice white-backed planthopper, Sogatella furcifera (Horváth). Acta phytophylacica Sinica. 2010, 37 (6): 511-516.
Cook AG, Perfect TJ: Determining the wing-morph of adult Nilaparvata lugens and Sogatella furcifera from morphometric measurements on the fifth-instar nymphs. Ent Exp Appl. 1982, 31: 159-164. 10.1111/j.1570-7458.1982.tb03128.x. 10.1111/j.1570-7458.1982.tb03128.x
Chen J, Zhang D, Yao Q, Zhang J, Dong X, Tian H, Chen J, Zhang W: Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol Biol. 2010, 19 (6): 777-786. 10.1111/j.1365-2583.2010.01038.x
Fu Q, Zhang ZT, Hu C, Lai FX, Sun ZX: A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Stål) (Homoptera: Delphacidae). Appl Entomol Zool. 2001, 36 (1): 111-116. 10.1303/aez.2001.111. 10.1303/aez.2001.111
Jia S, Wan PJ, Zhou LT, Mu LL, Li GQ: Molecular cloning and RNA interference-mediated functional characterization of a Halloween gene spook in the white-backed planthopper Sogatella furcifera. BMC Mol Biol. 2013, 14: 19- 10.1186/1471-2199-14-19
Yao J, Rotenberg D, Afsharifar A, Barandoc-Alviar K, Whitfield AE: Development of RNAi methods for Peregrinus maidis, the corn planthopper. PLoS One. 2013, 8: e70243- 10.1371/journal.pone.0070243
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ: Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998, 23 (10): 403-405. 10.1016/S0968-0004(98)01285-7
Acknowledgements
This study was supported by the grant from Major Project of Chinese National Programs for Fundamental Research and Development (973 Program, No. 2010CB126201).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JLY and ZFA carried out the experiments. XDL and JLY participated in the data analysis and drafted the manuscript. XDL conceived and supervised the research. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yu, JL., An, ZF. & Liu, XD. Wingless gene cloning and its role in manipulating the wing dimorphism in the white-backed planthopper, Sogatella furcifera. BMC Molecular Biol 15, 20 (2014). https://doi.org/10.1186/1471-2199-15-20
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2199-15-20