Abstract
DNA cruciforms play an important role in the regulation of natural processes involving DNA. These structures are formed by inverted repeats, and their stability is enhanced by DNA supercoiling. Cruciform structures are fundamentally important for a wide range of biological processes, including replication, regulation of gene expression, nucleosome structure and recombination. They also have been implicated in the evolution and development of diseases including cancer, Werner's syndrome and others.
Cruciform structures are targets for many architectural and regulatory proteins, such as histones H1 and H5, topoisomerase IIβ, HMG proteins, HU, p53, the proto-oncogene protein DEK and others. A number of DNA-binding proteins, such as the HMGB-box family members, Rad54, BRCA1 protein, as well as PARP-1 polymerase, possess weak sequence specific DNA binding yet bind preferentially to cruciform structures. Some of these proteins are, in fact, capable of inducing the formation of cruciform structures upon DNA binding. In this article, we review the protein families that are involved in interacting with and regulating cruciform structures, including (a) the junction-resolving enzymes, (b) DNA repair proteins and transcription factors, (c) proteins involved in replication and (d) chromatin-associated proteins. The prevalence of cruciform structures and their roles in protein interactions, epigenetic regulation and the maintenance of cell homeostasis are also discussed.
Similar content being viewed by others
Review
Genome sequencing projects have inundated us with information regarding the genetic basis of life. While this wealth of information provides a foundation for our understanding of biology, it has become clear that the DNA code alone does not hold all the answers. Epigenetic modifications and higher order DNA structures beyond the double helix also contribute to basic biological processes and maintaining cellular stability. Local alternative DNA structures are known to exist in all life forms [1]. The negative supercoiling of DNA can induce local nucleotide sequence-dependent conformational changes that give rise to cruciforms, left-handed DNA, triplexes and quadruplexes [2–4]. The formation of cruciforms is strongly dependent on base sequence and requires perfect or imperfect inverted repeats of 6 or more nucleotides in the DNA sequence [5, 6]. Over-representation of inverted repeats, which occurs nonrandomly in the DNA of all organisms, has been noted in the vicinity of breakpoint junctions, promoter regions, and at sites of replication initiation [3, 7, 8]. Cruciform structures may affect the degree of DNA supercoiling, the positioning of nucleosomes in vivo[9], and the formation of other secondary structures of DNA. Cruciforms contain a number of structural elements that serve as direct protein-DNA targets. Numerous proteins have been shown to interact with cruciforms, recognizing features such as DNA crossovers, four-way junctions, and curved or bent DNA. Structural transitions in chromatin occur concomitantly with DNA replication or transcription and in processes that involve a local separation of DNA strands. Such transitions are believed to facilitate the formation of alternative DNA structures [10, 11]. Transient supercoils are formed in the eukaryotic genome during DNA replication and transcription, and these often involve protein binding [12]. Indeed, active chromatin remodeling is a typical feature for many promoters and is essential for gene transcription [13]. Notably, DNA supercoiling can have a strong impact on gene expression [14]. Using microarrays covering the E. coli genome, it was recently shown that expression of 7% of genes was rapidly and significantly affected by a loss of chromosomal supercoiling [15]. Several complexes that involve extensive DNA-protein interactions, whereby the DNA wraps around the protein, can only occur under conditions of negative DNA supercoiling [10]. Other proteins are reported to interact with the supercoiled DNA (scDNA) at crossing points or on longer segments of the interwound supercoil [16, 17]. Interestingly, the eukaryotic genome has been shown to contain a percentage of unconstrained supercoils, part of which can be attributed to transcriptional regulation [3]. The spontaneous generation of DNA supercoiling is also a requirement for genome organization [18]. Transient supercoils are formed both in front of and behind replication forks as superhelical stress is distributed throughout the entire replicating DNA molecule [19]. A number of additional processes may operate to create transient and localized superhelical stresses in eukaryotic DNA.
The recognition of cruciform DNA seems to be critical not only for the stability of the genome, but also for numerous, basic biological processes. As such, it is not surprising that many proteins have been shown to exhibit cruciform structure-specific binding properties. In this review, we focus on these proteins, many of which are involved in chromatin organization, transcription, replication, DNA repair, and other processes. To organize our review, we have divided cruciform binding proteins into four groups (see Table 1) according to their primary functions: (a) junction-resolving enzymes, (b) transcription factors and DNA repair proteins, (c) replication machinery, and (d) chromatin-associated proteins. For each group, we describe in detail recent examples of research findings. Lastly, we review how dysregulation of cruciform binding proteins is associated with the pathology of certain diseases found in humans.
Formation and presence of c ruciform structures in the genome
Cruciform structures are important regulators of biological processes [3, 5]. Both stem-loops and cruciforms are capable of forming from inverted repeats. Cruciform structures consist of a branch point, a stem and a loop, where the size of the loop is dependent on the length of the gap between inverted repeats (Figure 1). Direct inverted repeats lead to formation of a cruciform with a minimal single-stranded loop. The formation of cruciforms from indirect inverted repeats containing gaps is dependent not only on the length of the gap, but also on the sequence in the gap. In general, the AT-rich gap sequences increase the probability of cruciform formation. It is also possible that the gap sequence can form an alternative DNA structure. The formation of DNA cruciforms has a strong influence on DNA geometry whereupon sequences that are normally distal from one another can be brought into close proximity [20, 21]. The structure of cruciforms has been studied by atomic force microscopy [22–24]. These studies have identified two distinct classes of cruciforms. One class of cruciforms, denoted as unfolded, have a square planar conformation characterized by a 4-fold symmetry in which adjacent arms are nearly perpendicular to one another. The second class comprises a folded (or stacked) conformation where the adjacent arms form an acute angle with the main DNA strands (Figure 2). Two of the three structural motifs inherent to cruciforms, the branch point and stem, are also found in Holliday junctions. Holliday junctions are formed during recombination, double-strand break repair, and fork reversal during replication. Resolving Holliday junctions is a critical process for maintaining genomic stability [25, 26]. These junctions are resolved by a class of structure-specific nucleases: the junction-resolving enzymes.
Changes associated with transition from the linear to cruciform state in the p53 target sequence from the p21 promoter. The promoter sequence contains a 20 bp p53 target sequence with 7 bp long inverted repeat (red), (A) as linear DNA and (B) as an inverted repeat as a cruciform structure. In the cruciform structure, the p53 target sequence is presented as stems and loops.
Conformations of a cruciform structure. Conformations of a cruciform can vary from (A) "unfolded" with 4-fold symmetry to (B) bent, and to (C) "stacked" with 4 chains of DNA in close vicinity. D) Topology of a Holliday junction stabilized by a psoralen cross-linking agent (PDBID 467D). Here, the junction takes the form of an anti-parallel stacked x-structure.
Cruciforms are not thermodynamically stable in naked linear DNA due to branch migration [27]. Cruciform structure formation in vivo has been shown in both prokaryotes and eukaryotes using several methodological approaches. The presence of the cruciform structure was first described in circular plasmid DNA where the negative superhelix density can stabilize cruciform formation. Plasmids with native superhelical density usually contain cruciform structures in vitro and in vivo[28]. For example, higher order structure in the pT181 plasmid was shown to exist in vivo using bromoacetaldehyde treatment [29]. Deletion of the sequence which forms this structure at the ori site leads either to a reduction or failure in replication [30]. Similarly, deletion of the cruciform binding domain in 14-3-3 proteins results in reduced origin binding which affects the initiation of DNA replication in budding yeast [31]. Monoclonal antibodies against cruciform structures have also been used successfully to isolate cruciform-containing segments of genomic DNA. Furthermore, these sequences were able to replicate autonomously when transfected into HeLa cells [32]. Stabilization of the cruciform structures by monoclonal antibodies 2D3 and 4B4, with anti-cruciform DNA specificity, resulted in a 2- to 6-fold enhancement of replication in vivo[33]. 14-3-3 sigma was found to associate in vivo with the monkey origins of DNA replication ors8 and ors12 in a cell cycle-dependent manner, as assayed by a chromatin immunoprecipitation (ChIP) assay that involved formaldehyde cross-linking, followed by immunoprecipitation with anti-14-3-3 sigma antibody and quantitative PCR [34]. Similarly, the 14-3-3 protein homologs from Saccharomyces cerevisiae, Bmh1p and Bmh2p, have cruciform DNA-binding activity and associate in vivo with ARS307 [35]. Several studies show that transcription is regulated directly by the presence of cruciform structure in vivo. Another example includes the ability of the d(AT)n-d(AT)n insert to spontaneously adopt a cruciform state in E. coli, resulting in a block of protein synthesis [36]. Using site-directed mutational analysis and P1 nuclease mapping, it was demonstrated that the formation of a cruciform structure is required for the repression of enhancer function in transient transfection assays and that Alu elements may contribute to regulation of the CD8 alpha gene enhancer through the formation of secondary structure that disrupts enhancer function [37]. Transcriptionally driven negative supercoiling also mediates cruciform formation in vivo and enhanced cruciform formation correlates with an elevation in promoter activity [38]. It was also shown that the secondary DNA structures of the ATF/CREB element play a vital role in protein-DNA interactions and its cognate transcription factors play a predominant role in the promoter activity of the RNMTL1 gene [39]. Hypo-methylation of inverted repeats by the Dam methylase show that these sequences are consistent with an unusual secondary structure, such as DNA cruciform or hairpin in vivo[40]. The in vivo effects of cruciform formation during transcription have been studied in detail by Krasilnikov et al. [4]. Interestingly hairpin-capped linear DNA (in which the replication of hairpin-capped DNA and cruciform formation and resolution play central roles) was stably maintained for months in a human cancer cell line as numerous extra-chromosomal episomes [41]. Long palindromes can also induce DNA breaks after assuming a cruciform structure. Palindromes in S. cerevisiae are resolved, in vivo, by structure-specific enzymes. In vivo resolution requires either the Mus81 endonuclease or, as a substitute, the bacterial HJ resolvase RusA. These findings provide confirmation of cruciform extrusion and resolution in the context of eukaryotic chromatin [42]. Taken together, these studies show that cruciforms have been detected in vivo using a variety of independent techniques and that they are an intriguing and integral phenomenon of DNA biology and biochemistry.
Proteins involved in interactions with cruciform structures
Junction-resolving enzymes
There are a large number of proteins that recognize cruciforms (summarized in Table 1) and, of these, the junction-resolving enzymes have been studied extensively. These proteins have been identified in many organisms from bacteria (and their phages) to yeast, archea and mammals [43]. The majority of the junction-resolving enzymes can be divided into one of two superfamilies [44]. Those in the first class target specific DNA sequences for enzymatic activity, although they will bind equally well to junctions of any sequence. This superfamily includes E. coli RuvC, the yeast integrases, Cce1, Ydc2, and RnaseH. The second group includes the phage T7, endonuclease RecU, the Hjc and Hje resolving enzymes, the MutH protein family and related restriction enzymes. The x-ray structures of the junction-resolving enzymes in complex with 4-way junctions highlight the flexibility inherent to DNA (Figure 3) [25] in that these enzymes recognize and distort the junction. This enables them to carry out such key roles as the cleavage of allogene DNAs and maintenance of genomic stability to name but a few. The recognition of non-B-DNA structure by junction-resolving enzymes has been the subject of several reviews [25, 43, 45, 46].
Crystal structure of the E. coli RuvA tetramer in complex with a Holliday junction (PDBID 1C7Y). A) The Holliday junction is depressed at the center where it makes close contacts with RuvA. Each of the arms outside of the junction center takes on a standard beta-DNA conformation B) Rotation of A) by 90°.
Proteins involved in transcription and DNA repair
The maintenance of a cell's genomic stability is achieved through several independent mechanisms. Arguably, the most important of these mechanisms is DNA repair. Protein binding to damaged DNA and to the local alternative DNA structures is therefore a key function of these processes. The promoter regions of genes are often characterized by presence of inverted repeats that are capable of forming cruciforms in vivo. A number of DNA-binding proteins, such as those of the HMGB-box family [47], Rad54 [48], BRCA1 protein [49, 50], as well as PARP-1 (poly(ADP-ribose) polymerase-1) [51], display only a weak sequence preference but bind preferentially to cruciform structures. Moreover, some proteins can induce the formation of cruciform structures upon DNA binding [51, 52]. Among the DNA repair proteins which bind to cruciforms are the junction-resolving enzymes Ruv and RuvB [53, 54], DNA helicases [55], XPG protein [56], and multifunctional proteins like HMG-box proteins [57] BRCA1, 14-3-3 protein family including homolog's Bmh1 and Bmh2 from S. cerevisiae, and GF14 from plants. Footprinting analysis of the gonadotropin-releasing hormone gene promoter region indicated the human estrogen receptor (ER) to be another potential cruciform binding protein. In this case, extrusion of the cruciform structure allowed the estrogen response elements motifs to be accessed by the ER protein [58].
PARP-1
PARP-1 is an abundant, nuclear, zinc-finger protein present in ~ 1 enzyme per 50 nucleosomes. It has a high affinity for damaged DNA and becomes catalytically active upon binding to DNA breaks [59]. In the absence of DNA damage, the presence of PARP-1 leads to the perturbation of histone-DNA contacts allowing DNA to be accessible to regulatory factors [60]. PARP-1 activity is also linked to the coordination of chromatin structure and gene expression in Drosophila [61]. It was reported that PARP can bind to the DNA hairpins in heteroduplex DNA and that the auto-modification of PARP in the presence of NAD+ inhibited its hairpin binding activity. Atomic force microscopy studies revealed that, in vitro, PARP protein has a preference for the promoter region of the PARP gene in superhelical DNA where the dyad symmetry elements form hairpins (Figure 4) [62]. PARP-1 recognizes distortions in the DNA backbone allowing it to bind to three- and four-way junctions [63]. Kinetic analysis has revealed that the structural features of non-B form DNA are important for PARP-1 catalysis activated by undamaged DNA. The order of PARP-1's substrate preference has been shown to be: cruciforms > loops > linear DNA. These results suggest a link between PARP-1 binding to cruciforms structures in the genome and its function in the modulation of chromatin structure in cellular processes. Moreover, it was shown that the binding of PARP-1 to DNA can induce changes in DNA topology as was demonstrated using plasmid DNA targets [51].
AFM and SFM images of proteins binding to a cruciform structure. A) AFM images of PARP-1 binding to supercoiled pUC8F14 plasmid DNA containing a 106 bp inverted repeat. PARP-1 binds to the end of the hairpin arm (white arrow). Images show 300 × 300 nm2 surface areas (reprinted with permission from [51]. B) The interaction between p53CD and supercoiled DNA gives rise to cruciform structures. Shown is an SFM image of complex formed between p53CD and sc pXG(AT)34 plasmid DNA at a molar ratio of 2.5; the complexes were mounted in the presence of 10 mM MgAc2. The scale bars represent 200 nm (reprinted with permission from [132].
P53
P53 is arguably one of the most intensively studied tumor suppressor genes. More than 50% of all human tumors contain p53 mutations and the inactivation of this gene plays a critical role in the induction of malignant transformation [64]. Sequence-specific DNA binding is crucial for p53 function. P53 target sequences, which consist of two copies of the sequence 5'-RRRC(A/T)(T/A)GYYY-3, often form inverted repeats [65]. It was reported that p53 binding is temperature sensitive and dependent on DNA fragment length [66, 67]. Moreover, it was demonstrated, in vivo, that p53 binding to its target sequence is highly dependent on the presence of an inverted repeat at the target site. Preferential binding of p53 to superhelical DNA has also been described [68, 69]. Non-canonical DNA structures such as mismatched duplexes, cruciform structures [70], bent DNA [71], structurally flexible chromatin DNA [13], hemicatenated DNA [72], DNA bulges, three- and four-way junctions [73], or telomeric t-loops [74] can all be bound selectively by p53. There is a strong correlation between the cruciform-forming targets and an enhancement of p53 DNA binding [75]. Target sequences capable of forming cruciform structures in topologically constrained DNA bound p53 with a remarkably higher affinity than did the internally asymmetrical target site [76]. These results implicate DNA topology as having an important role in the complex, with possible implications in modulation of the p53 regulon.
Chromatin-associated proteins
The chromatin-associated proteins cover a broad spectrum of the proteins localized in the cell nucleus. They are partly involved in modulating chromatin structure, but are also implicated in a range of processes associated with DNA function. They fine-tune transcriptional events (DEK, BRCA1) and are involved in both DNA repair and replication (HMG proteins, Rad51, Rad51ap, topoisomerases). Another family of enzymes deemed important in these processes is that of topoisomerases. These enzymes occur in all known organisms and play crucial roles in the remodeling of DNA topology. Topoisomerase I binds to Holliday junctions [77], and topoisomerase II recognizes and cleaves cruciform structures [78] and interacts with the HMGB1 protein [57]. These processes are particularly important for maintaining genomic stability due to their ability to diffuse the stresses that are levied upon a DNA molecule during transcription, replication and the resolving of long cruciforms that would otherwise hinder DNA chain separation. The Rad54 protein plays an important role during homologous recombination in eukaryotes [79]. Yeast and human Rad54 bind specifically to Holliday junctions and promote branch migration [80]. The binding preference for the open conformation of the X-junction appears to be common for many proteins that bind to Holliday junctions. Human Rad54 binds preferentially to the open conformation of branched DNA as opposed to the stacked conformation [48]. Similarly, RAD51AP1, the RAD51 accessory protein, specifically stimulates joint molecule formation through the combination of structure-specific DNA binding and by interacting with RAD51. RAD51AP1 has a particular affinity for branched-DNA structures that are obligatory intermediates during joint molecule formation [81]. The recognition of branched structures during homologous recombination is a critical step in this process.
DEK
The human DEK protein is an abundant nuclear protein of 375 amino acids that occurs in numbers greater than 1 million copies per nucleus [82]. Its interactions with transcriptional activators and repressors suggest that DEK may have a role in the formation of transcription complexes at promoter and enhancer sites [reviewed in [83]]. The binding of DEK to DNA is not sequence specific and DEK has a clear preference for supercoiled and four-way junctions [84]. Work with isolated and recombinant DEK has shown that it has intrinsic DNA-binding activity with a preference for four-way junction and superhelical DNA over linear DNA and introduces positive supercoils into relaxed circular DNA [83, 85]. DEK has two DNA-binding domains. The first domain is centrally located and harbors a conserved sequence element, the SAF (scaffold attachment factor). The second DNA-binding domain is located at the C-terminus of DEK which is also post-translationally modified by phosphorylation. In fact, the DNA-binding properties of DEK are clearly influenced by phosphorylation as phosphorylated DEK binds with a weaker affinity to DNA than does unmodified DEK and induces the formation of DEK multimers [86, 87]. DEK's monomeric SAF box (residues 137-187) does not appear to interact with DNA in solution. However, when many SAF boxes are brought into close proximity, it cooperativity drives DNA binding. A DEK construct spanning amino acids 87-187 binds to DNA much like the intact DEK preferring four-way DNA junctions over linear DNA. This fragment forms large aggregates in the presence of DNA and is also able to introduce supercoils into relaxed circular DNA. Interestingly, the 87-187 amino acid peptide induces negative DNA supercoils [88].
BRCA1
BRCA1 is a multifunctional tumor suppressor protein having roles in cell cycle progression, transcription, DNA repair and chromatin remodeling. Mutations to the BRCA1 gene are associated with a significant increase in the risk of breast cancer. The function of BRCA1 likely involves interactions with both DNA and an array of proteins. BRCA1 associates directly with RAD51 and both proteins co-localize to discrete subnuclear foci that redistribute to sites of DNA damage under genotoxic stress [89]. BRCA1 also co-localizes with phosphorylated H2AX (γH2AX) in response to double strand breaks [90].
The central region of human BRCA1 binds strongly to negatively supercoiled plasmid DNA with native superhelical density [50] and binds with high affinity to cruciform DNA [91]. The BRCA1 cruciform DNA complex must dissociate to allow the nuclease complex to work in DNA recombinational repair of double stranded breaks. BRCA1 also acts as a scaffold for assembly of the Rad51 ATPase which is responsible for homologous recombination in somatic cells. The full-length BRCA1 protein binds strongly to supercoiled plasmid DNA and to junction DNA. The difference in affinity was on the order of 6- to 7-fold between linear and junction DNA in reactions containing physiological levels of magnesium [92]. BRCA1 230-534 binds with a higher affinity to four-way junction DNA as compared to duplex and single-stranded DNA [91]. Residues 340-554 of BRCA1 have been identified as the minimal DNA-binding region [93]. The highest affinity among the different DNA targets which mimic damaged DNA (four-way junction DNA, DNA mismatches, DNA bulges and linear DNA) was for DNA four-way junctions. To this end, a 20-fold excess of linear DNA was unable to compete off any of the BRCA1 230-534 bound to DNA molecules mimicking damaged DNA [49]. Furthermore, the loss of the BRCA1 gene prevents cell survival after exposure to DNA cross-linkers such as mitomycin C [94]. These results speak to the importance of BRCA1's ability to recognize cruciform structures.
HMGB family
The high mobility-group (HMG) proteins are a family of abundant and ubiquitous non-histone proteins that are known to bind to eukaryotic chromatin. The three HMG protein families comprise the (a) HMGA proteins (formerly HMGI/Y) containing A/T-hook DNA-binding motifs, (b) HMGB proteins (formerly HMG1/2) containing HMG-box domain(s), and (c) HMGN proteins (formerly HMG14/17) containing a nucleosome-binding domain [95].
HMGB proteins bind DNA in a sequence independent manner and are known to bind to certain DNA structures (four-way junctions, DNA minicircles, cis-platinated DNA, etc.) with high affinity as compared to linear DNA [96, 97]. The chromatin architectural protein HMGB1 can bind with extremely high affinity to DNA structures that form DNA loops [72], while other studies have shown that the HMG box of different proteins can induce DNA bending [98–100]. The HMG box is an 80 amino acid domain found in a variety of eukaryotic chromosomal proteins and transcription factors. HMG box binding to DNA is associated with distortions in DNA structure. Members of the HMG protein family are involved in transcription [101–103] and DNA repair [57, 104, 105]. The HMG protein T160 was found to be co-localized with DNA replication foci [106]. The fact that all HMG box domains bind to four-way DNA junctions suggests that a common feature in the binding targets of this protein family must exist. Single HMG box domains interact exclusively with the open square form of the junction, and conditions that stabilize the stacked × structure conformation significantly weaken the HMG box DNA interaction [107]. Binding of the isolated A domain of HMGB1 protein to four-way junction DNA substrates is abolished by mutation of both Lys2 and Lys11 together to alanine, indicating that these residues play an important role in DNA binding [108].
Proteins involved in replication
Transient transitions from B-DNA to cruciform structures are correlated with DNA replication and transcription [109]. It has been shown that cruciforms serve as recognition signals at or near eukaryotic origins of DNA replication [110–112]. There are a large number of proteins involved in replication which bind to cruciform structures (see Table 1). We focus here primarily on the 14-3-3 protein family and MLL and WRN proteins. We will comment briefly on other systems of interest.
S16 is a structure-specific DNA-binding protein displaying preferential binding for cruciform DNA structures [113]. The AF10 protein binds cruciform DNA via a specific interaction with an AT-hook motif and is localized to the nucleus by a defined bipartite nuclear localization signal in the N-terminal region [114]. The structural maintenance of chromosomes (SMC) protein family, with members from lower and higher eukaryotes, may be divided into four subfamilies (SMC1 to SMC4) and two SMC-like protein subfamilies (SMC5 and SMC6) [115–117]. Members of this family are implicated in a large range of activities that modulate chromosome structure and organization. Smc1 and smc2 proteins have a high affinity for cruciform DNA molecules and for AT-rich DNA fragments including fragments from the scaffold-associated regions [118]. The baculovirus very late expression factor 1 (VLF-1), a member of the integrase protein family, does not bind to single and double strand structures, but it does bind (listed with increasing affinity) to Y-forks, three-way junctions and cruciform structures. This protein is involved in the processing of branched DNA molecules at the late stages of viral genome replication [119].
14-3-3
The 14-3-3 protein family consists of a highly conserved and widely distributed group of dimeric proteins which occur as multiple isoforms in eukaryotes [120]. There are at least seven distinct 14-3-3 genes in vertebrates, giving rise to nine isoforms (α, β, γ, δ, ε, ζ, η, σ and τ) and at least another 20 have been identified in yeast, plants, amphibians and invertebrates [110]. A striking feature of the 14-3-3 proteins is their ability to bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors. This plethora of proteins allows 14-3-3s to modulate a wide variety of vital regulatory processes, including mitogenic signal transduction, apoptosis and cell cycle regulation [121]. The 14-3-3 proteins are found mainly within the nucleus and are involved in eukaryotic DNA replication via binding to the cruciform DNA that forms transiently at replication origins at the onset of the S phase [122].
14-3-3 cruciform binding activity was first observed in proteins purified from sheep's brain. More recently, immunofluorescence analyses showed that 14-3-3 isoforms with cruciform-binding activity are present in HeLa cells [123]. The direct interaction with cruciform DNA was confirmed with 14-3-3 isoforms β, γ, σ, ε, and ζ [34]. 14-3-3 analogs with cruciform-specific binding are also found in yeast (Bmh1 and Bmh2) and plants (GF14) [35].
The prevalence of the 14-3-3 family proteins in all eukaryotes combined with a high degree of sequence conservation between species is indicative of their importance. Genetic studies have shown that knocking out the yeasts homologs of the 14-3-3 proteins is lethal [124]. Moreover, 14-3-3 proteins are involved in interactions with numerous transcription factors and it has been reported that several of the 14-3-3 proteins functions are associated with its cruciform binding properties.
Mixed lineage leukemia (MLL) protein
The MLL gene encodes a putative transcription factor with regions of homology to several other proteins including the zinc fingers and the so-called "AT-hook" DNA-binding motif of high mobility group proteins [125]. The 11q23 chromosomal translocation, found in both acute lymphoid and myeloid leukemias, results in disruption of the MLL gene. Leukemogenesis is often correlated with alternations in chromatin structure brought about by either a gain or loss in function of the regulatory factors due to their being disrupted by chromosomal translocations. The MLL gene, a target of such translocation events, forms a chimeric fusion product with a variety of partner genes [126].
The MLL AT-hook domain binds cruciform DNA, recognizing the structure rather than the sequence of the target DNA. This interaction can be antagonized both by Hoechst 33258 dye and distamycin. In a nitrocellulose protein-DNA binding assay, the MLL AT-hook domain was shown to bind to AT-rich SARs, but not to non-SAR DNA fragments [125]. MLL appears to be involved in chromatin-mediated gene regulation. In translocations involving MLL, the loss of the activation domain combined with the retention of a repression domain alters the expression of downstream target genes, thus suggesting a potential mechanism of action for MLL in leukemia [126]. AF10 translocations to the vicinity of genes other than MLL also result in myeloid leukemia. A biochemical analysis of the MLL partner gene AF10 showed that its AT-hook motif is able to bind to cruciform DNA, but not to double-stranded DNA, and that it forms a homo-tetramer in vitro[114].
WRN
The Werner syndrome protein belongs to the RecQ family of evolutionary conserved 3' → 5' DNA helicases [127]. WRN encodes a single polypeptide of 162 kDa that contains 1432 amino acids. Prokaryotes and lower eukaryotes generally have one RecQ member while higher eukaryotes possess multiple members and five homologs have been identified in human cells. All RecQ members share a conserved helicase core with one or two additional C-terminal domains, the RQC (RecQ C-terminal) and HRDC (helicase and RNaseD C-terminal) domains. These domains bind both to proteins and DNA. Eukaryotic RecQ helicases have N- and C-terminal extensions that are involved in protein-protein interactions and have been postulated to lend unique functional characteristics to these proteins [55, 128]. WRN has been shown to bind at replication fork junctions and to Holliday junction structures. Binding to junction DNA is highly specific because little or no WRN binding is visualized at other sites along these substrates [129]. Upon binding to DNA, WRN assembles into a large complex composed of four monomers.
Cruciform binding proteins and disease
The recognition of DNA junctions and cruciform structures is critical for genomic stability and for the regulation of basic cellular processes. The resolution of Holliday junctions and long cruciforms is necessary for genomic stability where the dysregulation of these proteins can lead to DNA translocations, deletions, loss of genomics stability and carcinogenesis. The large numbers of proteins which bind to these DNA structures work together to keep the genome intact. We believe that the formation of cruciform structures serves as a marker for the proper timing and initiation of some very basic biological processes. The mutations and epigenetic modifications that alter the propensity for cruciform formation can have drastic consequences for cellular processes. Thus, it is unsurprising that the dysregulation of cruciform binding proteins is often associated with the pathology of disease.
As stated above, the cruciform binding proteins including p53, BRCA1, WRN and the proto-oncogenes DEK, MLL and HMG are also associated with cancer development and/or progression. Some of these proteins play such important roles that their mutation and/or inactivation result in severe genomic instability and sometimes lethality. For example, Brca1 -/- mouse embryonic stem cells show spontaneous chromosome breakage, profound genomic instability and hypersensitivity to a variety of damaging agents (e.g. γ radiation) all of which suggests a defect in DNA repair. The connection between the BRCA1 mutation and breast cancer is well known. P53's transcriptional regulation is fine-tuned by its timely binding to promoter elements. The formation of a cruciform structure in p53 recognition elements may be an important determinant of p53 transcription activity.
The dHMGI(Y) family of "high mobility group" non-histone proteins comprises architectural transcription factors whose over expression is highly correlated with carcinogenesis, increased malignancy and metastatic potential of tumors in vivo[95]. 14-3-3 proteins are related to several diseases, including cancer, Alzeheimer's disease, the neurological Miller Dieker and Spinocerebellar ataxia type 1 diseases, and spongiform encephalopathy. The deletion of 14-3-3σ in human colorectal cancer cells leads to the loss of the DNA damage checkpoint control [130]. The human DEK protein was discovered as a fusion with a nuclear pore protein in a subset of patients with acute myeloid leukemia. It was also identified as an autoantigen in a relatively high percentage of patients with autoimmune diseases. In addition, DEK mRNA levels are higher in transcriptionally active and proliferating cells than in resting cells, and elevated mRNA levels are found in several transformed and cancer cells [6, 7]. Werner syndrome is an autosomal recessive disorder characterized by features of premature aging and a high incidence of uncommon cancers [127]. The Werner syndrome protein (WRN) plays central roles in maintaining the genomic stability of organisms [131]. Individuals harboring mutations in WRN have a rare, autosomal recessive genetic disorder manifested by early onset of symptoms characteristic of aged individuals.
Conclusions
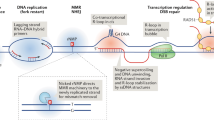
Cruciform structures are fundamentally important for a wide range of biological processes, including DNA transcription, replication, recombination, control of gene expression and genome organization. The putative mechanistic roles of cruciform binding proteins in transcription, DNA replication, and DNA repair are shown in Figure 5. Alternative DNA structures, including cruciforms, are often formed at sites of negatively supercoiled DNA by perfect or imperfect inverted repeats of 6 or more nucleotides. Longer DNA palindromes present a threat to genomic stability as they are recognized by junction-resolving enzymes. Shorter palindromic sequences are essential for basic processes like DNA replication and transcription. The presence of cruciform structures may also play an important role in epigenetics, such that cruciform structures are protected from DNA methylation. For example, the Dam methylase is not able to modify its GATC target site when it occurs in a cruciform or hairpin conformation. The center of a long perfect palindrome located in bacteriophage lambda has also been shown to be methylation-resistant in vivo[40]. Moreover, the centers of long palindromes are hypo-methylated as compared to identical sequences in non-palindromic conformations [40]. To this end, transient cruciforms can directly influence DNA methylation and therefore provide another layer for regulation of the DNA code. Proteins that bind to cruciforms can be divided into several categories. In addition to a well defined group of junction-resolving enzymes, we have classified cruciform binding proteins into groups involved in transcription and DNA repair (PARP, BRCA1, p53, 14-3-3), chromatin-associated proteins (DEK, BRCA1, HMG protein family, topoisomerases), and proteins involved in replication (MLL, WRN, 14-3-3, helicases) (see Table 1). Within these groups are proteins indispensable for cell viability, as well as tumor suppressors, proto-oncogenes and DNA remodeling proteins. Similarly, triplet repeat expansion, a phenomenon important in several genetic diseases, including Friedreich's ataxia, cardiomyopathy, myotonic dystrophy type I and other neurological disorders, can change the spectrum of cruciform binding proteins. Lastly, single nucleotide polymorphisms and/or insertion/deletion mutations at inverted repeats located in promoter sites can also influence cruciform formation, which might be manifested through altered gene regulation. A deeper understanding of the processes related to the formation and function of alternative DNA structures will be an important component to consider in the post-genomic era.
Scheme of the putative mechanistic roles of cruciform binding proteins in transcription, DNA replication, and DNA repair. A) A model for the structure-specific binding of transcription factors to a cognate palindrome-type cruciform implicated in transcription. The equilibrium between classic B-DNA and the higher order cruciform favors duplex DNA, but, when cruciform binding proteins are present, they either preferentially bind to and stabilize the cruciform or bind to the classic form and convert it to the cruciform. This interaction results in both an initial melting of the DNA region covered by transcription factor and an extension of the melt region in both directions. The melting region continues to extend in response to the needs of the active transcription machinery. B) A model for the initiation of replication enhanced by extrusion to a cruciform structure. Dimeric cruciform binding proteins interact with and stabilize the cruciform structure. The replisome is assembled concomitantly and is assumed to include polymerases, single-strand binding proteins and helicases. C) Model for the influence of cruciform binding proteins on DNA structure in DNA damage regulation. Naked cruciforms are sensitive to DNA damage and are covered by proteins in order to protect these sequences from being cleaved. In these cases, a deficiency in cruciform binding proteins can lead to DNA breaks. Here, cruciform-DNA complexes can also serve as scaffolds to recruit the DNA damage machinery.
Abbreviations
- scDNA:
-
supercoiled DNA.
References
Smith GR: Meeting DNA palindromes head-to-head. Genes Dev. 2008, 22 (19): 2612-2620. 10.1101/gad.1724708
Palecek E: Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991, 26: 151-226. 10.3109/10409239109081126
van Holde K, Zlatanova J: Unusual DNA structures, chromatin and transcription. Bioessays. 1994, 16 (1): 59-68. 10.1002/bies.950160110
Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM: Large-scale effects of transcriptional DNA supercoiling in vivo. J Mol Biol. 1999, 292 (5): 1149-1160. 10.1006/jmbi.1999.3117
Mikheikin AL, Lushnikov AY, Lyubchenko YL: Effect of DNA supercoiling on the geometry of holliday junctions. Biochemistry. 2006, 45 (43): 12998-13006. 10.1021/bi061002k
Limanskaia O, Limanskii AP: Distribution of potentially hairpin-loop structures in the genome of bovine retroviruses. Vopr Virusol. 2009, 54 (4): 27-32.
Werbowy K, Cieslinski H, Kur J: Characterization of a cryptic plasmid pSFKW33 from Shewanella sp. 33B. Plasmid. 2009, 62 (1): 44-49. 10.1016/j.plasmid.2009.03.003
Pearson CE, Zorbas H, Price GB, Zannis-Hadjopoulos M: Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. J Cell Biochem. 1996, 63 (1): 1-22. 10.1002/(SICI)1097-4644(199610)63:1<1::AID-JCB1>3.0.CO;2-3
Aranda A, Perez-Ortin JE, Benham CJ, Del Olmo ML: Analysis of the structure of a natural alternating d(TA)n sequence in yeast chromatin. Yeast. 1997, 13 (4): 313-326. 10.1002/(SICI)1097-0061(19970330)13:4<313::AID-YEA93>3.0.CO;2-8
Bates AD, Maxwell A: DNA Topology. 2005, Oxford: Oxford University Press, second
Mani P, Yadav VK, Das SK, Chowdhury S: Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009, 4 (2): e4399- 10.1371/journal.pone.0004399
Lin CT, Lyu YL, Liu LF: A cruciform-dumbbell model for inverted dimer formation mediated by inverted repeats. Nucleic Acids Res. 1997, 25 (15): 3009-3016. 10.1093/nar/25.15.3009
Kim E, Deppert W: The complex interactions of p53 with target DNA: we learn as we go. Biochem Cell Biol. 2003, 81 (3): 141-150. 10.1139/o03-046
Drolet M: Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006, 59 (3): 723-730. 10.1111/j.1365-2958.2005.05006.x
Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR: Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004, 5 (11): R87- 10.1186/gb-2004-5-11-r87
Mazur SJ, Sakaguchi K, Appella E, Wang XW, Harris CC, Bohr VA: Preferential binding of tumor suppressor p53 to positively or negatively supercoiled DNA involves the C-terminal domain. J Mol Biol. 1999, 292 (2): 241-249. 10.1006/jmbi.1999.3064
Brazdova M, Palecek J, Cherny DI, Billova S, Fojta M, Pecinka P, Vojtesek B, Jovin TM, Palecek E: Role of tumor suppressor p53 domains in selective binding to supercoiled DNA. Nucleic Acids Res. 2002, 30 (22): 4966-4974. 10.1093/nar/gkf616
Campos J, Gonzalez-Quintela A, Quinteiro C, Gude F, Perez LF, Torre JA, Vidal C: The -159C/T polymorphism in the promoter region of the CD14 gene is associated with advanced liver disease and higher serum levels of acute-phase proteins in heavy drinkers. Alcohol Clin Exp Res. 2005, 29 (7): 1206-1213. 10.1097/01.ALC.0000171977.25531.7A
Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR: The structure of supercoiled intermediates in DNA replication. Cell. 1998, 94 (6): 819-827. 10.1016/S0092-8674(00)81740-7
Vologodskii AV, Cozzarelli NR: Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994, 23: 609-643. 10.1146/annurev.bb.23.060194.003141
Vologodskii A, Cozzarelli NR: Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996, 70 (6): 2548-2556. 10.1016/S0006-3495(96)79826-0
Lyubchenko YL: DNA structure and dynamics: an atomic force microscopy study. Cell Biochem Biophys. 2004, 41 (1): 75-98. 10.1385/CBB:41:1:075
Kurahashi H, Inagaki H, Yamada K, Ohye T, Taniguchi M, Emanuel BS, Toda T: Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J Biol Chem. 2004, 279 (34): 35377-35383. 10.1074/jbc.M400354200
Shlyakhtenko LS, Potaman VN, Sinden RR, Lyubchenko YL: Structure and dynamics of supercoil-stabilized DNA cruciforms. J Mol Biol. 1998, 280 (1): 61-72. 10.1006/jmbi.1998.1855
Declais AC, Lilley DM: New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr Opin Struct Biol. 2008, 18 (1): 86-95. 10.1016/j.sbi.2007.11.001
Tolmasky ME, Colloms S, Blakely G, Sherratt DJ: Stability by multimer resolution of pJHCMW1 is due to the Tn1331 resolvase and not to the Escherichia coli Xer system. Microbiology. 2000, 146 (Pt 3): 581-589.
Shlyakhtenko LS, Hsieh P, Grigoriev M, Potaman VN, Sinden RR, Lyubchenko YL: A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J Mol Biol. 2000, 296 (5): 1169-1173. 10.1006/jmbi.2000.3542
Panayotatos N, Fontaine A: A native cruciform DNA structure probed in bacteria by recombinant T7 endonuclease. J Biol Chem. 1987, 262 (23): 11364-11368.
Noirot P, Bargonetti J, Novick RP: Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci USA. 1990, 87 (21): 8560-8564. 10.1073/pnas.87.21.8560
Yamaguchi K, Yamaguchi M: The replication origin of pSC101: the nucleotide sequence and replication functions of the ori region. Gene. 1984, 29 (1-2): 211-219. 10.1016/0378-1119(84)90181-1
Yahyaoui W, Callejo M, Price GB, Zannis-Hadjopoulos M: Deletion of the cruciform binding domain in CBP/14-3-3 displays reduced origin binding and initiation of DNA replication in budding yeast. BMC Mol Biol. 2007, 8: 27- 10.1186/1471-2199-8-27
Bell D, Sabloff M, Zannis-Hadjopoulos M, Price G: Anti-cruciform DNA affinity purification of active mammalian origins of replication. Biochim Biophys Acta. 1991, 1089 (3): 299-308.
Zannis-Hadjopoulos M, Frappier L, Khoury M, Price GB: Effect of anti-cruciform DNA monoclonal antibodies on DNA replication. Embo J. 1988, 7 (6): 1837-1844.
Alvarez D, Novac O, Callejo M, Ruiz MT, Price GB, Zannis-Hadjopoulos M: 14-3-3sigma is a cruciform DNA binding protein and associates in vivo with origins of DNA replication. J Cell Biochem. 2002, 87 (2): 194-207. 10.1002/jcb.10294
Callejo M, Alvarez D, Price GB, Zannis-Hadjopoulos M: The 14-3-3 protein homologues from Saccharomyces cerevisiae, Bmh1p and Bmh2p, have cruciform DNA-binding activity and associate in vivo with ARS307. J Biol Chem. 2002, 277 (41): 38416-38423. 10.1074/jbc.M202050200
Haniford DB, Pulleyblank DE: Transition of a cloned d(AT)n-d(AT)n tract to a cruciform in vivo. Nucleic Acids Res. 1985, 13 (12): 4343-4363. 10.1093/nar/13.12.4343
Hanke JH, Hambor JE, Kavathas P: Repetitive Alu elements form a cruciform structure that regulates the function of the human CD8 alpha T cell-specific enhancer. J Mol Biol. 1995, 246 (1): 63-73. 10.1006/jmbi.1994.0066
Dayn A, Malkhosyan S, Mirkin SM: Transcriptionally driven cruciform formation in vivo. Nucleic Acids Res. 1992, 20 (22): 5991-5997. 10.1093/nar/20.22.5991
Xu J, De Zhu J, Ni M, Wan F, Gu JR: The ATF/CREB site is the key element for transcription of the human RNA methyltransferase like 1(RNMTL1) gene, a newly discovered 17p13.3 gene. Cell Res. 2002, 12 (3-4): 177-197. 10.1038/sj.cr.7290124
Allers T, Leach DR: DNA palindromes adopt a methylation-resistant conformation that is consistent with DNA cruciform or hairpin formation in vivo. J Mol Biol. 1995, 252 (1): 70-85. 10.1006/jmbi.1994.0476
Harada S, Uchida M, Shimizu N: Episomal high copy number maintenance of hairpin-capped DNA bearing a replication initiation region in human cells. J Biol Chem. 2009, 284 (36): 24320-24327. 10.1074/jbc.M109.008128
Cote AG, Lewis SM: Mus81-dependent double-strand DNA breaks at in vivo-generated cruciform structures in S. cerevisiae. Mol Cell. 2008, 31 (6): 800-812. 10.1016/j.molcel.2008.08.025
Lilley DM, White MF: The junction-resolving enzymes. Nat Rev Mol Cell Biol. 2001, 2 (6): 433-443. 10.1038/35073057
Aravind L, Makarova KS, Koonin EV: SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000, 28 (18): 3417-3432. 10.1093/nar/28.18.3417
Khuu PA, Voth AR, Hays FA, Ho PS: The stacked-X DNA Holliday junction and protein recognition. J Mol Recognit. 2006, 19 (3): 234-242. 10.1002/jmr.765
Lilley DM: Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000, 33 (2): 109-159. 10.1017/S0033583500003590
Stefanovsky VY, Moss T: The cruciform DNA mobility shift assay: a tool to study proteins that recognize bent DNA. Methods Mol Biol. 2009, 543: 537-546. 10.1007/978-1-60327-015-1_31
Mazina OM, Rossi MJ, Thomaa NH, Mazin AV: Interactions of human rad54 protein with branched DNA molecules. J Biol Chem. 2007, 282 (29): 21068-21080. 10.1074/jbc.M701992200
Naseem R, Webb M: Analysis of the DNA binding activity of BRCA1 and its modulation by the tumour suppressor p53. PLoS ONE. 2008, 3 (6): e2336- 10.1371/journal.pone.0002336
Brazda V, Jagelska EB, Liao JC, Arrowsmith CH: The central region of BRCA1 binds preferentially to supercoiled DNA. J Biomol Struct Dyn. 2009, 27 (1): 97-104.
Chasovskikh S, Dimtchev A, Smulson M, Dritschilo A: DNA transitions induced by binding of PARP-1 to cruciform structures in supercoiled plasmids. Cytometry A. 2005, 68 (1): 21-27.
Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T: TRF2 promotes, remodels and protects telomeric Holliday junctions. Embo J. 2009, 28 (6): 641-651. 10.1038/emboj.2009.11
Shiba T, Iwasaki H, Nakata A, Shinagawa H: SOS-inducible DNA repair proteins, RuvA and RuvB, of Escherichia coli: functional interactions between RuvA and RuvB for ATP hydrolysis and renaturation of the cruciform structure in supercoiled DNA. Proc Natl Acad Sci USA. 1991, 88 (19): 8445-8449. 10.1073/pnas.88.19.8445
Iwasaki H, Takahagi M, Nakata A, Shinagawa H: Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992, 6 (11): 2214-2220. 10.1101/gad.6.11.2214
van Brabant AJ, Stan R, Ellis NA: DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet. 2000, 1: 409-459. 10.1146/annurev.genom.1.1.409
Wakasugi M, Reardon JT, Sancar A: The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997, 272 (25): 16030-16034. 10.1074/jbc.272.25.16030
Stros M, Bacikova A, Polanska E, Stokrova J, Strauss F: HMGB1 interacts with human topoisomerase IIalpha and stimulates its catalytic activity. Nucleic Acids Res. 2007, 35 (15): 5001-5013. 10.1093/nar/gkm525
Klungland H, Andersen O, Kisen G, Alestrom P, Tora L: Estrogen receptor binds to the salmon GnRH gene in a region with long palindromic sequences. Mol Cell Endocrinol. 1993, 95 (1-2): 147-154. 10.1016/0303-7207(93)90040-Q
Benjamin RC, Gill DM: Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA. A comparison of DNA molecules containing different types of strand breaks. J Biol Chem. 1980, 255 (21): 10502-10508.
Rouleau M, Aubin RA, Poirier GG: Poly(ADP-ribosyl)ated chromatin domains: access granted. J Cell Sci. 2004, 117 (Pt 6): 815-825.
Tulin A, Chinenov Y, Spradling A: Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003, 56: 55-83.
Soldatenkov VA, Chasovskikh S, Potaman VN, Trofimova I, Smulson ME, Dritschilo A: Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem. 2002, 277 (1): 665-670.
Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA: Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005, 280 (17): 17076-17083. 10.1074/jbc.M413483200
Dey A, Verma CS, Lane DP: Updates on p53: modulation of p53 degradation as a therapeutic approach. Br J Cancer. 2008, 98 (1): 4-8. 10.1038/sj.bjc.6604098
Kim E, Rohaly G, Heinrichs S, Gimnopoulos D, Meissner H, Deppert W: Influence of promoter DNA topology on sequence-specific DNA binding and transactivation by tumor suppressor p53. Oncogene. 1999, 18 (51): 7310-7318. 10.1038/sj.onc.1203139
Brazda V, Jagelska EB, Fojta M, Palecek E: Searching for target sequences by p53 protein is influenced by DNA length. Biochem Biophys Res Commun. 2006, 341 (2): 470-477. 10.1016/j.bbrc.2005.12.202
Brazda V, Muller P, Brozkova K, Vojtesek B: Restoring wild-type conformation and DNA-binding activity of mutant p53 is insufficient for restoration of transcriptional activity. Biochem Biophys Res Commun. 2006, 351 (2): 499-506. 10.1016/j.bbrc.2006.10.065
Palecek E, Vlk D, Stankova V, Brazda V, Vojtesek B, Hupp TR, Schaper A, Jovin TM: Tumor suppressor protein p53 binds preferentially to supercoiled DNA. Oncogene. 1997, 15 (18): 2201-2209. 10.1038/sj.onc.1201398
Brazda V, Palecek J, Pospisilova S, Vojtesek B, Palecek E: Specific modulation of p53 binding to consensus sequence within supercoiled DNA by monoclonal antibodies. Biochem Biophys Res Commun. 2000, 267 (3): 934-939. 10.1006/bbrc.1999.2056
Degtyareva N, Subramanian D, Griffith JD: Analysis of the binding of p53 to DNAs containing mismatched and bulged bases. J Biol Chem. 2001, 276 (12): 8778-8784. 10.1074/jbc.M006795200
Nagaich AK, Appella E, Harrington RE: DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J Biol Chem. 1997, 272 (23): 14842-14849. 10.1074/jbc.272.23.14842
Stros M, Muselikova-Polanska E, Pospisilova S, Strauss F: High-affinity binding of tumor-suppressor protein p53 and HMGB1 to hemicatenated DNA loops. Biochemistry. 2004, 43 (22): 7215-7225. 10.1021/bi049928k
Subramanian D, Griffith JD: Modulation of p53 binding to Holliday junctions and 3-cytosine bulges by phosphorylation events. Biochemistry. 2005, 44 (7): 2536-2544. 10.1021/bi048700u
Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T: Mammalian telomeres end in a large duplex loop. Cell. 1999, 97 (4): 503-514. 10.1016/S0092-8674(00)80760-6
Jagelska EB, Brazda V, Pecinka P, Palecek E, Fojta M: DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites. Biochem J. 2008, 412 (1): 57-63. 10.1042/BJ20071648
Jagelska EB, Pivonkova H, Fojta M, Brazda V: The potential of the cruciform structure formation as an important factor influencing p53 sequence-specific binding to natural DNA targets. Biochem Biophys Res Commun. 2010, 391 (3): 1409-1414. 10.1016/j.bbrc.2009.12.076
Hede MS, Petersen RL, Frohlich RF, Kruger D, Andersen FF, Andersen AH, Knudsen BR: Resolution of Holliday junction substrates by human topoisomerase I. J Mol Biol. 2007, 365 (4): 1076-1092. 10.1016/j.jmb.2006.10.050
Lee GE, Kim JH, Chung IK: Topoisomerase II-mediated DNA cleavage on the cruciform structure formed within the 5'upstream region of the human beta-globin gene. Mol Cells. 1998, 8 (4): 424-430.
Heyer WD, Li X, Rolfsmeier M, Zhang XP: Rad54: the Swiss Army knife of homologous recombination?. Nucleic Acids Res. 2006, 34 (15): 4115-4125. 10.1093/nar/gkl481
Bugreev DV, Mazina OM, Mazin AV: Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006, 442 (7102): 590-593. 10.1038/nature04889
Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R: RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell. 2007, 28 (3): 468-481. 10.1016/j.molcel.2007.08.025
Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C: Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001, 276 (28): 26317-26323. 10.1074/jbc.M100162200
Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R: The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004, 343 (1): 1-9. 10.1016/j.gene.2004.08.029
Waldmann T, Baack M, Richter N, Gruss C: Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003, 31 (23): 7003-7010. 10.1093/nar/gkg864
Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C: The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000, 14 (11): 1308-1312.
Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C: Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004, 24 (13): 6011-6020. 10.1128/MCB.24.13.6011-6020.2004
Kappes F, Scholten I, Richter N, Gruss C, Waldmann T: Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004, 24 (13): 6000-6010. 10.1128/MCB.24.13.6000-6010.2004
Bohm F, Kappes F, Scholten I, Richter N, Matsuo H, Knippers R, Waldmann T: The SAF-box domain of chromatin protein DEK. Nucleic Acids Res. 2005, 33 (3): 1101-1110. 10.1093/nar/gki258
Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM: Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997, 90 (3): 425-435. 10.1016/S0092-8674(00)80503-6
Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM: A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000, 10 (15): 886-895. 10.1016/S0960-9822(00)00610-2
Sturdy A, Naseem R, Webb M: Purification and characterisation of a soluble N-terminal fragment of the breast cancer susceptibility protein BRCA1. J Mol Biol. 2004, 340 (3): 469-475. 10.1016/j.jmb.2004.05.005
Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M: Direct DNA binding by Brca1. Proc Natl Acad Sci USA. 2001, 98 (11): 6086-6091. 10.1073/pnas.111125998
Naseem R, Sturdy A, Finch D, Jowitt T, Webb M: Mapping and conformational characterization of the DNA-binding region of the breast cancer susceptibility protein BRCA1. Biochem J. 2006, 395 (3): 529-535. 10.1042/BJ20051646
De la Torre C, Pincheira J, Lopez-Saez JF: Human syndromes with genomic instability and multiprotein machines that repair DNA double-strand breaks. Histol Histopathol. 2003, 18 (1): 225-243.
Banks GC, Li Y, Reeves R: Differential in vivo modifications of the HMGI(Y) nonhistone chromatin proteins modulate nucleosome and DNA interactions. Biochemistry. 2000, 39 (28): 8333-8346. 10.1021/bi000378+
Grasser KD, Teo SH, Lee KB, Broadhurst RW, Rees C, Hardman CH, Thomas JO: DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1). Eur J Biochem. 1998, 253 (3): 787-795. 10.1046/j.1432-1327.1998.2530787.x
Agresti A, Bianchi ME: HMGB proteins and gene expression. Curr Opin Genet Dev. 2003, 13 (2): 170-178. 10.1016/S0959-437X(03)00023-6
Deckert J, Khalaf RA, Hwang SM, Zitomer RS: Characterization of the DNA binding and bending HMG domain of the yeast hypoxic repressor Rox1. Nucleic Acids Res. 1999, 27 (17): 3518-3526. 10.1093/nar/27.17.3518
Phillips NB, Nikolskaya T, Jancso-Radek A, Ittah V, Jiang F, Singh R, Haas E, Weiss MA: Sry-directed sex reversal in transgenic mice is robust with respect to enhanced DNA bending: comparison of human and murine HMG boxes. Biochemistry. 2004, 43 (22): 7066-7081. 10.1021/bi049920a
Dragan AI, Read CM, Makeyeva EN, Milgotina EI, Churchill ME, Crane-Robinson C, Privalov PL: DNA binding and bending by HMG boxes: energetic determinants of specificity. J Mol Biol. 2004, 343 (2): 371-393. 10.1016/j.jmb.2004.08.035
Stros M, Polanska E, Struncova S, Pospisilova S: HMGB1 and HMGB2 proteins up-regulate cellular expression of human topoisomerase IIalpha. Nucleic Acids Res. 2009, 37 (7): 2070-2086. 10.1093/nar/gkp067
Stefanovsky VY, Langlois F, Bazett-Jones D, Pelletier G, Moss T: ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF. Biochemistry. 2006, 45 (11): 3626-3634. 10.1021/bi051782h
Harrer M, Luhrs H, Bustin M, Scheer U, Hock R: Dynamic interaction of HMGA1a proteins with chromatin. J Cell Sci. 2004, 117 (Pt 16): 3459-3471.
Boulikas T: Evolutionary consequences of nonrandom damage and repair of chromatin domains. J Mol Evol. 1992, 35 (2): 156-180.
Kamashev D, Balandina A, Rouviere-Yaniv J: The binding motif recognized by HU on both nicked and cruciform DNA. Embo J. 1999, 18 (19): 5434-5444. 10.1093/emboj/18.19.5434
Hertel L, De Andrea M, Bellomo G, Santoro P, Landolfo S, Gariglio M: The HMG protein T160 colocalizes with DNA replication foci and is down-regulated during cell differentiation. Exp Cell Res. 1999, 250 (2): 313-328. 10.1006/excr.1999.4495
JR P, Norman DG, Bramham J, Bianchi ME, Lilley DM: HMG box proteins bind to four-way DNA junctions in their open conformation. Embo J. 1998, 17 (3): 817-826. 10.1093/emboj/17.3.817
Assenberg R, Webb M, Connolly E, Stott K, Watson M, Hobbs J, Thomas JO: A critical role in structure-specific DNA binding for the acetylatable lysine residues in HMGB1. Biochem J. 2008, 411 (3): 553-561. 10.1042/BJ20071613
Pearson CE, Zorbas H, Price GB, Zannis-Hadjopoulos M: Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. J Cell Biochem. 1996, 63 (1): 1-22. 10.1002/(SICI)1097-4644(199610)63:1<1::AID-JCB1>3.0.CO;2-3
Zannis-Hadjopoulos M, Yahyaoui W, Callejo M: 14-3-3 cruciform-binding proteins as regulators of eukaryotic DNA replication. Trends Biochem Sci. 2008, 33 (1): 44-50. 10.1016/j.tibs.2007.09.012
Kim E, Lane CE, Curtis BA, Kozera C, Bowman S, Archibald JM: Complete sequence and analysis of the mitochondrial genome of Hemiselmis andersenii CCMP644 (Cryptophyceae). BMC Genomics. 2008, 9: 215- 10.1186/1471-2164-9-215
Omberg L, Meyerson JR, Kobayashi K, Drury LS, Diffley JF, Alter O: Global effects of DNA replication and DNA replication origin activity on eukaryotic gene expression. Mol Syst Biol. 2009, 5: 312-
Bonnefoy E: The ribosomal S16 protein of Escherichia coli displaying a DNA-nicking activity binds to cruciform DNA. Eur J Biochem. 1997, 247 (3): 852-859. 10.1111/j.1432-1033.1997.t01-1-00852.x
Linder B, Newman R, Jones LK, Debernardi S, Young BD, Freemont P, Verrijzer CP, Saha V: Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J Mol Biol. 2000, 299 (2): 369-378. 10.1006/jmbi.2000.3766
Peterson CL: The SMC family: novel motor proteins for chromosome condensation?. Cell. 1994, 79 (3): 389-392. 10.1016/0092-8674(94)90247-X
Palecek J, Vidot S, Feng M, Doherty AJ, Lehmann AR: The Smc5-Smc6 DNA repair complex. bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits. J Biol Chem. 2006, 281 (48): 36952-36959. 10.1074/jbc.M608004200
Hirano T: SMC proteins and chromosome mechanics: from bacteria to humans. Philos Trans R Soc Lond B Biol Sci. 2005, 360 (1455): 507-514. 10.1098/rstb.2004.1606
Akhmedov AT, Frei C, Tsai-Pflugfelder M, Kemper B, Gasser SM, Jessberger R: Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998, 273 (37): 24088-24094. 10.1074/jbc.273.37.24088
Mikhailov VS, Rohrmann GF: Binding of the baculovirus very late expression factor 1 (VLF-1) to different DNA structures. BMC Mol Biol. 2002, 3: 14- 10.1186/1471-2199-3-14
Aitken A: 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006, 16 (3): 162-172. 10.1016/j.semcancer.2006.03.005
Fu H, Subramanian RR, Masters SC: 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000, 40: 617-647. 10.1146/annurev.pharmtox.40.1.617
Zannis-Hadjopoulos M, Sibani S, Price GB: Eucaryotic replication origin binding proteins. Front Biosci. 2004, 9: 2133-2143. 10.2741/1369
Todd A, Cossons N, Aitken A, Price GB, Zannis-Hadjopoulos M: Human cruciform binding protein belongs to the 14-3-3 family. Biochemistry. 1998, 37 (40): 14317-14325. 10.1021/bi980768k
van Heusden GP, van der Zanden AL, Ferl RJ, Steensma HY: Four Arabidopsis thaliana 14-3-3 protein isoforms can complement the lethal yeast bmh1 bmh2 double disruption. FEBS Lett. 1996, 391 (3): 252-256. 10.1016/0014-5793(96)00746-6
Broeker PL, Harden A, Rowley JD, Zeleznik-Le N: The mixed lineage leukemia (MLL) protein involved in 11q23 translocations contains a domain that binds cruciform DNA and scaffold attachment region (SAR) DNA. Curr Top Microbiol Immunol. 1996, 211: 259-268.
Zeleznik-Le NJ, Harden AM, Rowley JD: 11q23 translocations split the "AT-hook" cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994, 91 (22): 10610-10614. 10.1073/pnas.91.22.10610
Ozgenc A, Loeb LA: Current advances in unraveling the function of the Werner syndrome protein. Mutat Res. 2005, 577 (1-2): 237-251.
Hanada K, Hickson ID: Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007, 64 (17): 2306-2322. 10.1007/s00018-007-7121-z
Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD: The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J Biol Chem. 2008, 283 (36): 24478-24483. 10.1074/jbc.M803370200
Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B: 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999, 401 (6753): 616-620. 10.1038/44188
Chu WK, Hickson ID: RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009, 9 (9): 644-654. 10.1038/nrc2682
Jett SD, Cherny DI, Subramaniam V, Jovin TM: Scanning force microscopy of the complexes of p53 core domain with supercoiled DNA. J Mol Biol. 2000, 299 (3): 585-592. 10.1006/jmbi.2000.3759
Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H: Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. Embo J. 1991, 10 (13): 4381-4389.
Biertumpfel C, Yang W, Suck D: Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. 2007, 449 (7162): 616-620. 10.1038/nature06152
Pan PS, Curtis FA, Carroll CL, Medina I, Liotta LA, Sharples GJ, McAlpine SR: Novel antibiotics: C-2 symmetrical macrocycles inhibiting Holliday junction DNA binding by E. coli RuvC. Bioorg Med Chem. 2006, 14 (14): 4731-4739. 10.1016/j.bmc.2006.03.028
Fogg JM, Schofield MJ, Declais AC, Lilley DM: Yeast resolving enzyme CCE1 makes sequential cleavages in DNA junctions within the lifetime of the complex. Biochemistry. 2000, 39 (14): 4082-4089. 10.1021/bi992785v
Garcia AD, Otero J, Lebowitz J, Schuck P, Moss B: Quaternary structure and cleavage specificity of a poxvirus holliday junction resolvase. J Biol Chem. 2006, 281 (17): 11618-11626. 10.1074/jbc.M600182200
Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T: A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005, 435 (7045): 1059-1066. 10.1038/nature03657
Declais AC, Liu J, Freeman AD, Lilley DM: Structural recognition between a four-way DNA junction and a resolving enzyme. J Mol Biol. 2006, 359 (5): 1261-1276. 10.1016/j.jmb.2006.04.037
Guan C, Kumar S: A single catalytic domain of the junction-resolving enzyme T7 endonuclease I is a non-specific nicking endonuclease. Nucleic Acids Res. 2005, 33 (19): 6225-6234. 10.1093/nar/gki921
Hadden JM, Declais AC, Carr SB, Lilley DM, Phillips SE: The structural basis of Holliday junction resolution by T7 endonuclease I. Nature. 2007, 449 (7162): 621-624. 10.1038/nature06158
Spiro C, McMurray CT: Switching of DNA secondary structure in proenkephalin transcriptional regulation. J Biol Chem. 1997, 272 (52): 33145-33152. 10.1074/jbc.272.52.33145
Middleton CL, Parker JL, Richard DJ, White MF, Bond CS: Substrate recognition and catalysis by the Holliday junction resolving enzyme Hje. Nucleic Acids Res. 2004, 32 (18): 5442-5451. 10.1093/nar/gkh869
Lyu YL, Lin CT, Liu LF: Inversion/dimerization of plasmids mediated by inverted repeats. J Mol Biol. 1999, 285 (4): 1485-1501. 10.1006/jmbi.1998.2419
Giraud-Panis MJ, Lilley DM: Near-simultaneous DNA cleavage by the subunits of the junction-resolving enzyme T4 endonuclease VII. Embo J. 1997, 16 (9): 2528-2534. 10.1093/emboj/16.9.2528
Macmaster R, Sedelnikova S, Baker PJ, Bolt EL, Lloyd RG, Rafferty JB: RusA Holliday junction resolvase: DNA complex structure--insights into selectivity and specificity. Nucleic Acids Res. 2006, 34 (19): 5577-5584. 10.1093/nar/gkl447
Owen BA, W HL, McMurray CT: The nucleotide binding dynamics of human MSH2-MSH3 are lesion dependent. Nat Struct Mol Biol. 2009, 16 (5): 550-557. 10.1038/nsmb.1596
Surtees JA, Alani E: Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J Mol Biol. 2006, 360 (3): 523-536. 10.1016/j.jmb.2006.05.032
Chang JH, Kim JJ, Choi JM, Lee JH, Cho Y: Crystal structure of the Mus81-Eme1 complex. Genes Dev. 2008, 22 (8): 1093-1106. 10.1101/gad.1618708
Ehmsen KT, Heyer WD: Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008, 36 (7): 2182-2195. 10.1093/nar/gkm1152
Taylor ER, McGowan CH: Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci USA. 2008, 105 (10): 3757-3762. 10.1073/pnas.0710291105
Fouche N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD: The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem. 2006, 281 (49): 37486-37495. 10.1074/jbc.M608778200
Lee JH, Park CJ, Arunkumar AI, Chazin WJ, Choi BS: NMR study on the interaction between RPA and DNA decamer containing cis-syn cyclobutane pyrimidine dimer in the presence of XPA: implication for damage verification and strand-specific dual incision in nucleotide excision repair. Nucleic Acids Res. 2003, 31 (16): 4747-4754. 10.1093/nar/gkg683
Sekelsky JJ, Hollis KJ, Eimerl AI, Burtis KC, Hawley RS: Nucleotide excision repair endonuclease genes in Drosophila melanogaster. Mutat Res. 2000, 459 (3): 219-228.
Lee S, Cavallo L, Griffith J: Human p53 binds Holliday junctions strongly and facilitates their cleavage. J Biol Chem. 1997, 272 (11): 7532-7539. 10.1074/jbc.272.11.7532
Ma B, Levine AJ: Probing potential binding modes of the p53 tetramer to DNA based on the symmetries encoded in p53 response elements. Nucleic Acids Res. 2007, 35 (22): 7733-7747. 10.1093/nar/gkm890
Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ: Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005, 25 (11): 4476-4487. 10.1128/MCB.25.11.4476-4487.2005
Rass U, Kemper B: Crp1p, a new cruciform DNA-binding protein in the yeast Saccharomyces cerevisiae. J Mol Biol. 2002, 323 (4): 685-700. 10.1016/S0022-2836(02)00993-2
van Houte LP, Chuprina VP, van der Wetering M, Boelens R, Kaptein R, Clevers H: Solution structure of the sequence-specific HMG box of the lymphocyte transcriptional activator Sox-4. J Biol Chem. 1995, 270 (51): 30516-30524. 10.1074/jbc.270.51.30516
Pearson CE, Ruiz MT, Price GB, Zannis-Hadjopoulos M: Cruciform DNA binding protein in HeLa cell extracts. Biochemistry. 1994, 33 (47): 14185-14196. 10.1021/bi00251a030
Nakamura Y, Yoshioka K, Shirakawa H, Yoshida M: HMG box A in HMG2 protein functions as a mediator of DNA structural alteration together with box B. J Biochem. 2001, 129 (4): 643-651.
Culard F, Gervais A, de Vuyst G, Spotheim-Maurizot M, Charlier M: Response of a DNA-binding protein to radiation-induced oxidative stress. J Mol Biol. 2003, 328 (5): 1185-1195. 10.1016/S0022-2836(03)00361-9
Tripathi P, Pal D, Muniyappa K: Saccharomyces cerevisiae Hop1 protein zinc finger motif binds to the Holliday junction and distorts the DNA structure: implications for holliday junction migration. Biochemistry. 2007, 46 (44): 12530-12542. 10.1021/bi701078v
Tripathi P, Anuradha S, Ghosal G, Muniyappa K: Selective binding of meiosis-specific yeast Hop1 protein to the holliday junctions distorts the DNA structure and its implications for junction migration and resolution. J Mol Biol. 2006, 364 (4): 599-611. 10.1016/j.jmb.2006.08.096
Rene B, Fermandjian S, Mauffret O: Does topoisomerase II specifically recognize and cleave hairpins, cruciforms and crossovers of DNA?. Biochimie. 2007, 89 (4): 508-515. 10.1016/j.biochi.2007.02.011
Dip R, Naegeli H: More than just strand breaks: the recognition of structural DNA discontinuities by DNA-dependent protein kinase catalytic subunit. Faseb J. 2005, 19 (7): 704-715. 10.1096/fj.04-3041rev
Bonnefoy E, Takahashi M, Yaniv JR: DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J Mol Biol. 1994, 242 (2): 116-129. 10.1006/jmbi.1994.1563
Pinson V, Takahashi M, Rouviere-Yaniv J: Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J Mol Biol. 1999, 287 (3): 485-497. 10.1006/jmbi.1999.2631
Acknowledgements
This work was supported by the GACR (301/10/1211), by the MEYS CR (LC 06035) and by the Academy of Science of the Czech Republic (grants M200040904, AV0Z50040507 and AV0Z50040702).
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Brázda, V., Laister, R.C., Jagelská, E.B. et al. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Molecular Biol 12, 33 (2011). https://doi.org/10.1186/1471-2199-12-33
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2199-12-33