Abstract
Background
The Red-headed krait (Bungarus flaviceps, Squamata: Serpentes: Elapidae) is a medically important venomous snake that inhabits South-East Asia. Although the venoms of most species of the snake genus Bungarus have been well characterized, a detailed compositional analysis of B. flaviceps is currently lacking.
Results
Here, we have sequenced 845 expressed sequence tags (ESTs) from the venom gland of a B. flaviceps. Of the transcripts, 74.8% were putative toxins; 20.6% were cellular; and 4.6% were unknown. The main venom protein families identified were three-finger toxins (3FTxs), Kunitz-type serine protease inhibitors (including chain B of β-bungarotoxin), phospholipase A2 (including chain A of β-bungarotoxin), natriuretic peptide (NP), CRISPs, and C-type lectin.
Conclusion
The 3FTxs were found to be the major component of the venom (39%). We found eight groups of unique 3FTxs and most of them were different from the well-characterized 3FTxs. We found three groups of Kunitz-type serine protease inhibitors (SPIs); one group was comparable to the classical SPIs and the other two groups to chain B of β-bungarotoxins (with or without the extra cysteine) based on sequence identity. The latter group may be functional equivalents of dendrotoxins in Bungarus venoms. The natriuretic peptide (NP) found is the first NP for any Asian elapid, and distantly related to Australian elapid NPs. Our study identifies several unique toxins in B. flaviceps venom, which may help in understanding the evolution of venom toxins and the pathophysiological symptoms induced after envenomation.
Similar content being viewed by others
Background
Snake venom is a complex mixture of biologically active proteins and peptides that exert very powerful and specific effects. This mixture is interesting from the angle of molecular evolution, as the genes encoding the venom ingredients seem to undergo some form of hypermutation resulting in accelerated evolution and a staggering diversity of isoforms [1, 2], sometimes functionally and structurally radically different. The basis for this phenomenon seems to be due to gene-duplication and diversification of existing venom genes. This results in a highly dynamic venom composition both at the interspecific and intraspecific level [3]. This allows the snake to deal with a wide array of different prey items. Snake venom is also a valuable resource for proteins and peptides that may serve as lead compounds to treat certain human disorders [4]. Examining the transcriptome of a venom gland will also reveal venom proteins that are low abundant, which is crucial to both expanding the resource of pharmaceutical compounds as well as to understand the evolution of snake venom proteins [5]. Further, cataloguing of snake venom proteins through transcriptomic analysis may help to understand the pathophysiological symptoms induced after envenomation and correlates with the venom composition [6–12].
For example, we have recently used transcriptomic analysis to show the presence of three-finger toxins (3FTxs) in viperid venom [13, 14]. By elucidating the gene structures of these toxins we could infer their relationship with the elapid 3FTxs, which helped us to understand the evolution of this toxin protein family. Hence, snake venom gland transcriptomes continue to be a valuable tool in improving our understanding of snake venom composition and evolution, management of snake bite, and the opportunity to identify and study the function of the low abundant proteins.
Kraits (Bungarus species) belong to the family Elapidae. They are one of the better studied snakes of the world. They are widely distributed across South and Southeast Asia and are highly venomous [15]. Many biologically important proteins, particularly α- bungarotoxins, κ- bungarotoxins and β- bungarotoxins, have been well characterized from the venom of Bungarus species. The first two belong to the 3FTx family, whereas the last one is a covalent heterodimer of phospholipase A2 (PLA2) and a serine protease inhibitor (SPI) -like polypeptide [16–18]. α- bungarotoxin is a highly specific toxin that binds to peripheral nicotinic acetylcholine receptors (nAChRs) and it played a key role in the isolation and characterization of mammalian nAChRs [19]. Similar to other long-chain neurotoxins, it also binds to neuronal α7 nAChRs [20]. κ- bungarotoxins specifically bind to neuronal nAChRs (α3β2, α4β2 and α3β4) [20]. On the other hand, β- bungarotoxins - the major lethal factors bind to voltage-sensitive potassium channels in the presynaptic site [21, 22].
The B. flaviceps, commonly known as the Red-headed krait, has phenotypically distinctive coloring of blue and black body, and the head, neck and tail are bright red in color. The venom of B. flaviceps is more potent than B. fasciatus but comparable in potency to B. candidus venom; the LD50 values of B. flaviceps, B. candidus and B. fasciatus venoms are 3.5 μg, 3.2 μg and 61.7 μg per kg of experimental mouse respectively [23]. Other than the isolation and characterization of β- bungarotoxin [15, 24], κ- flavitoxin [25, 26] and PLA2[15], not much information is available on the venom of B. flaviceps. Therefore, we have here examined the venom gland of B. flaviceps by using expressed sequence tags (ESTs) to explore the venom composition in detail as well as to identify novel and low abundance toxins.
Results and Discussion
Composition of cDNA Library
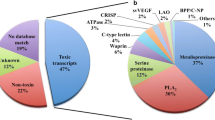
We randomly selected 845 clones from the cDNA library and isolated the plasmids. 606 clones having inserts larger than 200 bp were numbered randomly and designated with BF (Bungarus flaviceps). Sequences were categorized based on the similarity shown in the analysis results and submitted to the database (Additional file 1). Accordingly, 74.84% of the ESTs were putative toxin transcripts, 4.61% were unknown transcripts and 20.56% of the ESTs were cellular transcripts (Figure 1A). Using sequence similarities to known toxins as a guide, all the putative toxin transcripts were further classified into different toxin families. Accordingly, the B. flaviceps ESTs contained transcripts encoding for six toxin families. Most clones encoded for 3FTxs (39.29, followed by β-bungarotoxin (Chains A and B) (34.88%), and Kunitz-type SPI (21.19%). The other toxin families are less well presented; clones encoding phospholipase A2 (PLA2) (6.26% including A chains of β-bungarotoxin), and two clones each of natriuretic peptide (NP), cysteine-rich secretory protein (CRISPs), and C-type lectin (Figure 1B). 3FTx is the major transcript of this venom gland library constituting 39.29% of the toxin transcripts. Similar observation was made in Micrurus corallines venom gland library which belongs to the elapid family where the 3FTx constitutes ~52% of the toxin transcripts [12]. This indicates that 3FTx is the major toxin of elapid snakes where as in viperid venoms proteases are the major venom components (Additional file 2). By comparing the total number of sequences versus number of new toxin sequences identified we showed that the number of new toxin transcripts was reaching an asymptote. Our results can thus be considered as a representative of the overall venom composition of B. flaviceps (Figure 1C). Although this study is not exhaustive, we have identified a number of low abundance transcripts (see below). We believe when the library contains high percentage of clones (~75%) coding for toxin-like proteins, sequencing 600-1000 clones may be sufficient to identify a number of low abundant clones.
Composition of a cDNA library from B. flaviceps venom gland tissue. A) Relative abundance of genes in the cDNA library. B) Relative abundance of the toxin genes in the cDNA library. C) Graph showing the number of new toxins isolated reached a plateau when compared with the total number of sequences isolated.
Three-Finger Toxin (3FTx) Family
Three-finger toxins are a group of low-molecular weight (<10 kDa), non-enzymatic polypeptides which have 60 to 74 amino acid residues [27]. They have 8-10 cysteine residues forming four or five disulphide bridges, of which four disulphide bridges are conserved [28]. Members of this family possess similar protein structures: three β- stranded loops extending from a central core, which is made up of the four highly conserved disulphide bridges [20, 29], hence the name three-finger toxins. This 3FTxs family have been recruited in snake venom from a gene recruitment event within the SLUR/LYNX protein family [30], and their ancestral function is thought to be inhibition of the nicotinic acetylcholine receptor (nAChR) [30]. However, despite retaining the common three-finger motif, members of this family have since evolved a myriad of different functions. These include effects on: platelet function [31], different receptors associated with neurotransmission [32–38]; ion channels [39–41], viability of cardimyocytes [42] and red bloods cells [43], mitosis and apoptosis [44]; and effects on the cell membrane [45, 46]. The different 3FTx members also vary considerably in binding affinity for the different receptors [47] and are important research ligands for studying receptors [48–50].
Based on sequence similarities, 161 full length transcripts of 3FTxs were divided into 11 clusters and 4 singletons. These ESTs were segregated into eight distinct groups of toxins based on BLAST results (Figure 2). The first group has two clusters, BF601 and BF421 (46 and 2 clones) and a singleton (BF141); they all encode identical mature protein but have a single amino acid differences in the signal peptide regions and mature protein. They share 61% identity and 76% similarity to both candiduxin 1 and buntoxin (Kini RM, unpublished data) (Figure 2A). Similarly, BF9 (13 clones) encodes for a toxin that is also similar to candiduxin 1 from B. candidus (Orphan 3FTx IX subfamily; [51]) and buntoxin with 81% and 79% identity respectively (Figure 2B). Second cluster represented by clone BF9 show higher identity (81 and 79%) and similarity (85 and 84%) to the candiduxin1 and buntoxin than the above group of toxins. However the biological functions of candiduxin 1 and buntoxin are yet to be elucidated. The third group with two clusters, BF748 and BF296 (42 and 2 clones) show 51% identity to 3FTx III from Walterinnesia aegyptia[52] and 47% identity to erabutoxin a from Laticauda semifasciata (Figure 2C). In erabutoxin a Lys27, Trp29, Asp31, Arg33 and Lys47 are important in the binding to Torpedo nAChR [53, 54]. Interestingly, the toxin encoded by BF748 has 27th, 29th and 47th residues replaced with Met, Phe and Asn respectively but the 31st and 33rd residues are conserved. Lower structural similarity combined with replacement of critical residues in the functional site may confer distinct pharmacological properties or specificities to these 3FTxs. One of the clusters, BF222 (7 clones) displays sequence similarity (76% identity; 86% similarity) (Figure 2D) to bucain from B. candidus venom [55]; [56] and a neurotoxin homolog NTL4 [57]. Similar to bucain, this toxin may exhibit not-so-potent neurotoxicity.
Three-finger toxins (3FTxs) of B. flaviceps venom gland cDNA library. Similar sequences were clustered together and a representative of the each cluster is presented. B. flaviceps 3FTxs can be classified into eight major groups A through H. The deduced amino acid sequences of 3FTX isoforms of each group are shown. The amino residues which are different from the representative major isoform are shaded. The predicted signal peptide of the transcripts is shown bold. The major isoforms from each group is aligned with closely related 3FTx found in the database and the % identity and similarly are also shown. (A and B) Groups A and B are similar to candiduxin1 (Q8AY53.1) and buntoxin. Group B toxins are closer to candiduxin compared to group A toxins. C) Putative short-chain 3FTxs of B. flaviceps that are similar to 3FTx (ABX82864.1) from W.aegyptia and erabutoxin a (P60775.1). The residues of erabutoxin a that are involved in binding to nAChR receptor are shown in *. Critical functional residues in BF748 are replaced with Met, Phe and Asn (27th, 29th and 47th residues respectively). D) Group D toxins are similar to bucain (Q9YGI8.1). E) Group E non-conventional 3FTx and its alignment with candoxin (P81783.2). F) Group F non-conventional 3FTx and alignment with NTX4_BUNCA (Q6IZ95.1) and NXW1_BUNCA (Q8AY51.1). G) Group G non-conventional 3FTx and its alignment with BM14 (Q8JFX7.1) and bulongin. H) Group H 3FTx and its alignment with NXL2_BUNFL (κ-favitoxin) (P15815.1) (for details, see text).
The ESTs included 56 clones of non-conventional 3FTxs (fifth disulfide bridge in loop 1; [58]) (Figure 2E-G). Three clusters represented by BF648, BF402 and BF797 with 26, 4 and 2 clones respectively, which have few minor amino acid residue changes in the mature proteins. These transcripts encode proteins similar to candoxin with 80% identity (Figure 2E). Candoxin from B. candidus venom is known to bind reversibly to peripheral nAChRs and irreversibly to α7 nAChRs [59]. Functionally, these B. flaviceps proteins may exhibit similar, if not identical, properties. Second cluster BF11 (7 clones), BF661 (4 clones) and BF685 (single clone) shows 87% and 70% identity to two non-conventional toxins from B. candidus venom (Figure 2F). The other two isoforms are much closer (90% and 74% identity respectively). Similarly, BF533 (Singleton) showed 64% identity to both BM14 [60] and bulongin (R. M. Kini, unpublished data) from B. multicinctus and B. candidus venoms respectively (Figure 2G). BM14 binds to muscarinic M2 receptor subtypes with much lower affinities compared to typical muscarinic toxins from Dendroaspis venoms [60, 61]. In BM 14, trinitrophenylation of the Lys residues (37th and 38th) abolished its binding to muscarinic acetylcholine receptor (mAChR). However, based on the lower structural similarity and the lack of the critical Lys residues (replaced by Ile and Pro), we speculate that the protein coded by BF533 may have distinct biological properties compared to BM14 and bulongin.
Interestingly, only one singleton, BF776, shows 100% match (except for three missing amino acid residues at the N-terminal) to κ-flavitoxin (long-chain neurotoxin 2) from the venom of B. flaviceps flaviceps[25, 26] (Figure 2H). The observed truncation at the 5'end is likely to be due to its degradation of mRNA during total RNA extraction. κ- flavitoxin is a competitive neuronal acetylcholine receptor (nAChR) antagonist [25, 26]. In general, κ- neurotoxins are highly specific antagonists of α3β2 and α4β2 receptors [62]. Interestingly, we did not find a transcript encoding α- bungarotoxin.
Due to the dominating role of α-, β- and κ- bungarotoxins in Bungarus species, the functional roles of other toxins have not been systematically explored. Of the eight major groups of 3FTxs in our B. flaviceps venom gland ESTs data, only κ- neurotoxins are fairly well characterized. Therefore, it will be interesting to study these toxins and identify the molecular targets.
During our sequencing studies, we observed a number of clones with part of their sequences missing. In translated sequence of BF229 and BF249 (both singletons), loop I and part of loop II was found to be missing. When BF229 was compared to the gene of the black-and-white spitting cobra (Naja sputatrix) encoding weak neurotoxin 10 (accession number AY081762) [63], we found that exon II was missing in this clone (data not shown). Thus these clones were probably generated by an error in splicing. Premature truncation was also observed in BF846 and BF600 (both singletons). In BF846, truncation is due to a dinucleotide deletion, whereas in BF600 the insertion of adenosine nucleotide has lead to a frame shift. It is not clear whether these aberrations are artefacts of cloning and sequencing or products of independent genes, this requires further investigation. In any case, these truncated transcripts constitute about 3% of 3FTxs and may not have any influence on the overall toxicity of the venom.
Recently, by the analysis of gene structures of viperid 3FTxs, we showed that some of these toxins evolve through the phenomenon of segment switch [13]. The analysis of cDNA sequences of B. flaviceps reveals that these toxins also appear to be evolving through accelerated segment switch in exons (Additional file 1). This phenomenon, named as ASSET, may be an alternative mechanism of accelerated evolution of snake venom toxins [13, 64].
β- bungarotoxin
β-bungarotoxin is one of the major lethal components found in the venom of Bungarus species. It targets the pre-synaptic terminal, where it causes the massive release of acetylcholine resulting in subsequent exhaustion of acetylcholine and inability to conduct an impulse and finally, paralysis. It is a heterodimeric covalent protein complex [16, 17, 65] (for a review see, [64]) composed of chain A similar to PLA2 and chain B similar to Kunitz-type SPI. It is a potent presynaptic neurotoxin [16, 18]. We found three isoforms of chain A and four isoforms of chain B (for details, see below). Chain A transcripts represented 2.87% of all ESTs (13 ESTs, full length 11) and chain B transcripts 32.01% (145 ESTs, full length 96) (Figure 1B). This drastic difference in expression level is interesting, considering the fact that these two chains combine to an equimolar complex (β- bungarotoxin). This difference in expression levels requires further investigation. As the two chains belong to two different families, they will be discussed separately below.
Kunitz-type Serine Protease Inhibitor (SPI) Family
Kunitz-type SPIs are one of the major groups of snake venom proteins mainly reported from the venoms of Elapidae and Viperidae snakes and they inhibit either trypsin or chymotrypsin [66–71]. Structurally they belong to the bovine pancreatic trypsin inhibitor (BPTI) family [72, 73]. They have approximately 60 amino acid residues with six cysteine residues [74] arranged in a conserved sequence motif of C-8X-C-15X-C-4X-YGGC-12X-C-3X-C [75].
As mentioned above, the B chain of β- bungarotoxins is structurally similar to Kunitz-type SPI proteins. We found four clusters that exhibit high similarity to the B chain of β- bungarotoxins. Most common chain B isoform, BF677 (90 clones) is 100% identical to IBV_BUNFL, a chain B precursor isolated from B. flaviceps[15]. The other three isoforms (with two clones each), differ in amino acid residue from the major isoform in one or two positions (Figure 3A).
Kunitz-type SPIs of B. flaviceps venom gland cDNA library. Similar sequences were clustered together and a representative of the each cluster is presented. B. flaviceps Kunitz-type SPIs can be divided into three distinct groups (A, B and C). The deduced amino acid sequences of isoforms of each group are shown. The amino residues which are different from the representative of major isoform are shaded. The predicted signal peptide of the transcripts is shown in bold. The major isoforms from each group is aligned with closely related protein found in the database and the % identity and similarly are also shown in the figure. A) Putative clusters of β-bungarotoxin B chains of B. flaviceps and alignment with IVB_BUNFL (Q7T2Q6.1). The extra cysteine residue that is involved in the disulphide bridge with chain A is underlined. B) Putative Kunitz-type SPIs of B. flaviceps and alignment with IVBI5_BUNMU (Q1RPS9.1) chain B5 precursor of Bungarus multicinctus, IVB_BUNFL (Q7T2Q6.1) B. flaviceps, Dendrotoxin-K (P00981.2) and Dendrotoxin-I (P00979) from Dendroaspis polylepis polylepis. C) Putative Kunitz-type SPI and its truncated form. Alignment of the major isoform with Kunitz inhibitor IV (ABX82870.1) from W. aegyptia is also shown.
The second group of Kunitz-type SPI clones are similar to the B chain of β- bungarotoxins but lack the extra cysteine at the C-terminal end (Figure 3B). This extra cysteine residue is involved in interchain disulfide and is important for the complex formation of β-bungarotoxin (discussed above). The principal isoform BF548 with 48 clones shows similarity to chain B (B5) precursor of β- bungarotoxin from B. multicinctus[76]. Similarly BF130 and BF345 (both singletons) are also similar to chain B (B5) of β-bungarotoxin. Due to the absence of the extra cysteine, we believe that this polypeptide remains as monomer, similar to dendrotoxins [77] in the venom. Based on the high sequence identity and similarity to the chain B of β- bungarotoxins and dendrotoxins (Figure 3B), it is tempting to speculate that they might block voltage sensitive potassium channels [78, 79]. This needs to be further investigated.
We found two clusters, BF294 and BF539 (25 and 12 clones) as the third group of Kunitz-type SPI clones. The major isoform is structurally similar to Kunitz inhibitor IV from W. aegyptia (Figure 3C). Interestingly, the minor isoform (represented by BF539) was found to be prematurely truncated. Comparison of mRNA sequence BF539 and a Kunitz-type SPI from the King Cobra (Ophiophagus hannah) (EU246693), and to the sequence of PILP-1 from B. multicinctus[74], reveals that 87 nucleotides from exon II is lost in these clones (Additional file 4). However the stop codon and 3'UTR encoded by Exon III is conserved in this clone. This splicing error could occur due to the presence of GT (splice start site) in exon II. The truncated mature protein has 28 amino acid residues with the first pair of cysteine residues. It would be interesting to study its biological properties.
To understand the evolutionary relationships of Kunitz-type SPIs and B chains among the Bungarus species we constructed a phylogenetic tree using BPTI as an out group (Figure 4). The B. flaviceps precursors (BF677, BF664, BF842 and BF836) of B chains of β- bungarotoxins cluster separately from other two groups. The second group of transcripts (BF548, BF130 and BF345) clusters together with Kunitz-type SPI (BF294 and BF539) despite their similarities to chain B of β- bungarotoxins (Additional file 5). Thus most likely they are not the intermediates in the evolution of chain B. In the case of B. candidus and B. fasciatus, B chain cluster separately from the Kunitz-type SPIs. In B. multicinctus B chains, B1, B2, B3 and B4 cluster together and are closer to PILP-1, a SPI, whereas B chains B5 and B6 are closer to SPIs, PILP2 and PILP3.
Phospholipase A2 (PLA2) Family
PLA2 enzymes are esterolytic enzymes which hydrolyze glycerophospholipids at the sn-2 position of the glycerol backbone, resulting in the release of lysophospholipids and free fatty acids. They are 116 to 124 amino acid residues long with an approximate molecular mass of 13-14 kDa. This family of enzymes have 12-14 conserved cysteine residues which form six or seven disulphide bridges [80]. Structurally, they share a similar protein folding pattern of a α-helical core, a backbone loop and a β- wing [81]. Functionally, they have a broad array of pharmacological effects which encompass neurotoxic, myotoxic, cardiotoxic, haemolytic, convulsive, anticoagulant, antiplatelet, oedema-inducing and tissue-damaging effects (For a review see [80, 82]).
Within the ESTs, 3.30% of the transcripts belong to PLA2 family (Figure 1B). Clones encoding PLA2 were grouped into two clusters (5 and 3 clones) and two singletons (Figure 5). Prediction of signal peptide using SignalP program indicates that these sequences have the eight amino acid propeptide sequences (underlined in the Figure 6) [83]. This propeptide sequence in BF647 is represented by "SNVPPQPL" whereas in BF365 and BF161 it is "AIVPPQPL" and there are two substitutions (SN is replaced with AI). Although similar propeptides are found in other elapid PLA2 enzymes, most mature proteins do not have them. Only a small number (group IB) retain them. The PLA2 isoforms are highly similar to each other except for few amino acid substitutions in the mature protein (Figure 5A). Clone BF95 has a deletion of 32 amino acid residues (9 residues from the signal peptide and 23 residues from the N-terminal end including the propeptide) as compared to the other PLA2 isoforms. Comparison of the BF95 sequence with the sea snake (L. semfasciata) PLA2 sequence reveals that there is a deletion of 99 bp from exon II (Additional file 6). Consequence of this deletion in exon II in its activity is not known and needs further investigation. In one of the clones (data not shown) the 5' end of the transcript was missing, this might be due to degradation of the mRNA during preparation. When compared to B. flaviceps PLA2 I (BAC77655) and PLA2 II (BAC77656) sequence [15], BF647, BF365 and BF161 show 93% identity (Figure 5A).
Phospholipase A 2 of B. flaviceps venom gland cDNA library. Similar sequences were clustered together and a representative of the each cluster is presented. B. flaviceps PLA2s can be divided into two distinct groups (A and B). The deduced amino acid sequences of isoforms of each group are shown. The amino residues which are different from the representative major isoform are shaded. The predicted signal peptide of the transcripts is shown in bold and the propeptide is underlined. The major isoforms from each group is aligned with closely related protein found in the database and the % identity and similarly are also shown in the figure. A) Putative PLA2 precursors of B. flaviceps and alignment with PA22_BUNFL (Q7T2Q4.1) from B. flaviceps. B) Putative β-bungarotoxin chain A of B. flaviceps and alignment with PA2A2_BUNFL (Q7T1R1.1) from B. flaviceps. The extra cysteine residue involved in formation of disulphide bridge with chain A of β-bungarotoxin is underlined.
Natriuretic peptide in B. flaviceps venom gland cDNA library. A) The deduced amino acid sequence of the full length precursor is shown. The predicted signal peptide of the transcripts is shown in bold. The putative mature NP is boxed. B) Alignment of B. flaviceps NP with other snake venom NPs including human NP. The amino acid sequences were obtained from the database. The accession number, name of the protein and the organism are shown in the figure. The sequences were aligned manually and the gaps are filled with dashes, those amino acids which are not shown in the alignment are represented by ":". The cysteine residues involved in forming the 17 residue ring structure are highlighted in the figure.
As mentioned above, the chain A of β- bungarotoxins is structurally similar to PLA2 enzymes. We found three clusters that exhibit high identity to chain A of β- bungarotoxins (Figure 5B). BF1 (6 clones) which has few amino acid residue changes both in the mature protein as well as in the signal peptide as compared to BF284 (3 clones) and BF555 (2 clones). But BF284 and BF555 have two residue differences; one amino acid substitution in the signal peptide and two others in the mature protein. Cluster BF1 is 100% identical to Chain A2 of β-bungarotoxin from B. flaviceps reported earlier whereas BF284 and BF555 show 97% identity to Chain A1 in the mature protein [15]. Multiple isoforms of Chain A of β- bungarotoxin has been reported from Bungarus species. Five isoforms (A1-A5) have been reported from a single species of B. multicinctus snake [84], but we have observed only three isoforms in B. flaviceps transcriptome.
Natriuretic Peptide Family
Natriuretic peptides (NPs) are endogenous hormones initially found in mammals. Three mammalian NPs have been identified and characterized so far, which includes atrial NP (ANP), B-type NP (BNP) and C-type NP (CNP) [85–90]. In NPs a 17-residue ring structure, formed by an intra-molecular disulphide bond, is a highly conserved feature. Functionally, NPs are involved in various physiological processes such as regulation of water and electrolyte balance, cardiovascular system and cell growth [91–93]. Physiologically, in mammals NPs exhibit potent hypotensive and vasorelaxant properties and contributing to sodium and water retention [90, 94–96]. The first NP of snake venom was isolated from Dendroaspis angusticeps venom and is known as Dendroaspis natriuretic peptide (DNP) [97]. It lowers the blood pressure through vasodilation [98]. Interestingly two clones (0.44%) encoding natriuretic peptides were identified in B. flaviceps library; one of them was partial while the other (BF131) was a full length transcript (Figure 6A). This is the first full length mRNA sequence of an elapid NP precursor. When compared to CNP precursors of crotalid snakes, B. flaviceps NP precursor lacks the BPP domain and part of the linker sequence (data not shown) similar to colubrid NP precursor reported from the Duvernoy's (venom) gland transcriptome of Philodryas olfersii[99]. However, P. olfersii encodes for CNP type molecule, whereas B. flaviceps NP belongs to the ANP/BNP family due to the C-terminal extension (Figure 6B). This is the first report of NPs in any Asian elapid. We constructed a phylogenetic tree to understand the evolutionary relationships of ANP/BNP found in different snakes. B. flavipceps ANP/BNP was found to be closely related to African mamba (Dendroaspis) and coral snake (Micrurus) NPs, but distantly related to the Australian elapid counterparts (Figure 7). However the presence of these transcripts in the protein level need to confirmed through proteomic approaches.
Other families
CRISPs (2 clones), and C-type lectins (2 clones) transcripts were found, in a level of 0.44% of the total ESTs (Additional file 7). The CRISPs transcripts showed high similarity to latisemin reported from Laticauda semifasciata, oharin precursor from Ophiophagus hannah, pseudechetoxin-like protein precursor from Oxyuranus scutellatus and to kaouthin-2 precursor from Naja kaouthia[100–102]. Latisemin, ophanin and tigrin block potassium-stimulated smooth muscle contraction [102]. In CRISPs N-terminal pathogenesis-related protein-1 (PR-1) domain and the C-terminal cysteine-rich domain are conserved. PR-1 is known to be important for recognition of the target molecule and this domain. C-type lectins are non-enzymatic proteins which interact with carbohydrate moieties in the presence of Ca2+ ions and usually possess the highly conserved carbohydrate recognition domain (CRD) [103, 104]. One full length and other partial C-type lectins were obtained in this library. The full length transcripts have seven cysteine residues hence they are likely to exist as covalent homodimers in the venom [105]. Analysis of the translated sequence reveals a Ca2+ binding site [105, 106], suggesting that it is a Ca2+-dependent C-type lectin.
Cellular transcripts
The cellular transcripts (125 EST) constitute about 20.56% of the total transcripts. Some of the important house-keeping ESTs include: ribosomal protein (24 clones), protein Sec61 beta subunit (4 clones) involved in protein transport, eukaryotic translation initiation factor (3 clones) and eukaryotic translation elongation factor 1 beta 2 (3 clones), which are involved in protein transcription and translation. The proteins encoded by these transcripts are involved in protein synthesis and secretion. Other house-keeping transcripts involved in cellular functions were: NADH dehydrogenase (3 clones); ubiquitin C (2 clones); ADP-ribosylation factor-like (2 clones); Cytoplasmic actin type 5; ubiquinol-cytochrome c reductase; sodium-dependent dicarboxylate transporter; Succinate-CoA ligase; Type 1 glutamine amidotransferase; L-lactate dehydrogenase; and a DNA bindng protein (zinc finger) (1 clone each).
Conclusion
Analysis of 845 ESTs of the venom gland of the Red-headed krait (B. flaviceps) shows that 3FTxs and B chains of β-bungarotoxins are the main components of the venom. Most of the 3FTxs transcripts described here are structurally different from the previously characterized 3FTxs and hence they are likely to show distinct biological activities. We found a group of Kunitz-type SPIs highly similar to B chain of the β-bungarotoxin, but without the extra cysteine involved in interchain disulfide. Identification of ANP/BNP underscores the importance of transcriptome analysis in identifying low abundant proteins. Thus our study provides a platform to initiate the characterization of several novel proteins found in B. flaviceps venom gland.
Methods
Collection of venom glands and liver
A juvenile specimen of Bungarus flaviceps (Malaysia) was collected in Malaysia and imported to Europe. Venom was first obtained manually by sliding pipette tips over each fang, a technique standard for milking smaller elapid snakes. Venom was lyophilized and stored at -80°C. Four days later, when mRNA production is assumed to be maximal [107], the snake was anesthetized with Zoletil (Zolazepam and Tiletamine) and sacrificed by decapitation. The venom glands and liver were carefully but quickly dissected, cut into small pieces of 2-3 mm, and stored in RNAlater solution (Ambion, City, Country) in a ten-fold ratio to the volume of the glands, and stored at -80°C up until use.
Construction of cDNA library and DNA sequencing
Total RNA was extracted from both the venom glands using the RNeasy® Mini kit from Qiagen (Valencia, CA, USA). The quality of the RNA was tested using gel electrophoresis. Using Creator™ SMART™ cDNA library construction kit from Clontech Laboratories (Palo Alto, CA, USA), the following was achieved: (i) synthesis of first strand cDNAs from the total RNAs; (ii) synthesis of double-stranded cDNAs from the first strand cDNAs; and (iii) purification of double-stranded cDNAs using the CHROMA SPIN-400 column. Purified double-stranded cDNAs were cloned into the pCR®2.1-TOPO® vector from Invitrogen (Carlsbad, CA, USA). Recombinant TOPO vector was transformed into DH5α competent cells. After plating onto ampicillin/IPTG/X-gal Luria Broth (LB) agar plates, the transformed DH5α cells were subjected to blue/white screening. Individual white colonies were randomly selected and grown in LB added with ampicillin. Plasmids were then purified using the GeneAll® Exprep™ Plasmid Quick kit from GeneAll Biotechnology Co., Ltd (Songpa-gu, Seoul, Korea). The presence and size of inserts in the plasmids were confirmed using EcoRI digestion. Plasmids containing inserts larger than 200 bp were selected for DNA sequencing. The plasmids were sequenced using the chain termination method [108] using the ABI PRISM® BigDye Terminator v3.1 Cycle Sequencing kit and ABI PRISM® 3100 automated DNA sequencer from Applied Biosystem (Foster City, CA, USA). Representative full length cDNA sequences from each cluster or singleton were submitted to NCBI database. The accession numbers for 3FTx families were from GU190789 to GU190804, Kunitz-type SPI GU190805 to GU190810 β-bungarotoxin B Chain GU1908011 to GU190814, PLA2 GU190815 to GU190817, β-bungarotoxin B Chain GU190818 to GU190820, Natriuretic peptide GU190821 and C-type lectin GU190822.
Bioinformatics analyses
Vector and adaptor sequences were removed from the DNA sequences before translating them in all three frames for identification of the open reading frame (ORF). The trimmed DNA sequences and their correct ORF protein sequences were queried against nucleotide and protein databases using NCBI BLASTn and BLASTp http://blast.ncbi.nlm.nih.gov/Blast.cgi respectively to predict their putative functions. SignalP 3.0 server http://www.cbs.dtu.dk/services/SignalP/ was used for the prediction of signal peptide of the translated sequences. The similarity of transcripts to known toxin sequences available in the database was used as the first criterion for cataloguing transcripts of putative toxins. If the transcripts are novel with an ORF and do not show any similarity with any sequences of the database then we have used two criteria to identify potential novel toxins. The protein product should have a signal peptide (as venom proteins are secretory proteins) and rich in Cys residues (most venom toxins are rich in Cys residues). Multiple sequence alignments (MSA) were carried out using either Clustalx or DNAMAN version 4.15 (Lynnon Corporation, Pointe-Claire, Quebec, Canada; http://www.ebi.ac.uk/Tools/clustalw2/) or manually. Protein sequences were obtained from the NCBI database and sequence alignments were done using Clustalx. Phylogenetic trees were generated using Clustalx which employs Bootstrap N-J tree method and the tree was viewed using tree viewer.
References
Nakashima K, Nobuhisa I, Deshimaru M, Nakai M, Ogawa T, Shimohigashi Y, et al: Accelerated evolution in the protein-coding regions is universal in crotalinae snake venom gland phospholipase A2 isozyme genes. Proc Natl Acad Sci USA. 1995, 92: 5605-5609. 10.1073/pnas.92.12.5605
Kini RM, Chan YM: Accelerated evolution and molecular surface of venom phospholipase A2 enzymes. J Mol Evol. 1999, 48: 125-132. 10.1007/PL00006450
Daltry JC, Wuster W, Thorpe RS: Diet and snake venom evolution. Nature. 1996, 379: 537-540. 10.1038/379537a0
Koh DC, Armugam A, Jeyaseelan K: Snake venom components and their applications in biomedicine. Cell Mol Life Sci. 2006, 63: 3030-3041. 10.1007/s00018-006-6315-0
Junqueira-de-Azevedo IL, Ching AT, Carvalho E, Faria F, Nishiyama MY, Ho PL, et al: Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: implications for snake toxin repertoire evolution. Genetics. 2006, 173: 877-889. 10.1534/genetics.106.056515
Junqueira-de-Azevedo IL, Ho PL: A survey of gene expression and diversity in the venom glands of the pitviper snake Bothrops insularis through the generation of expressed sequence tags (ESTs). Gene. 2002, 299: 279-291. 10.1016/S0378-1119(02)01080-6
Qinghua L, Xiaowei Z, Wei Y, Chenji L, Yijun H, Pengxin Q, et al: A catalog for transcripts in the venom gland of the Agkistrodon acutus: identification of the toxins potentially involved in coagulopathy. Biochem Biophys Res Commun. 2006, 341: 522-531. 10.1016/j.bbrc.2006.01.006
Cidade DA, Simao TA, Davila AM, Wagner G, Junqueira-de-Azevedo IL, Ho PL, et al: Bothrops jararaca venom gland transcriptome: analysis of the gene expression pattern. Toxicon. 2006, 48: 437-461. 10.1016/j.toxicon.2006.07.008
Neiva M, Arraes FB, de Souza JV, Radis-Baptista G, Prieto da Silva AR, Walter ME, et al: Transcriptome analysis of the Amazonian viper Bothrops atrox venom gland using expressed sequence tags (ESTs). Toxicon. 2009, 53: 427-436. 10.1016/j.toxicon.2009.01.006
Zhang B, Liu Q, Yin W, Zhang X, Huang Y, Luo Y, et al: Transcriptome analysis of Deinagkistrodon acutus venomous gland focusing on cellular structure and functional aspects using expressed sequence tags. BMC Genomics. 2006, 7: 152- 10.1186/1471-2164-7-152
Francischetti IM, My-Pham V, Harrison J, Garfield MK, Ribeiro JM: Bitis gabonica (Gaboon viper) snake venom gland: toward a catalog for the full-length transcripts (cDNA) and proteins. Gene. 2004, 337: 55-69. 10.1016/j.gene.2004.03.024
Leao LI, Ho PL, Junqueira-de-Azevedo IL: Transcriptomic basis for an antiserum against Micrurus corallinus (coral snake) venom. BMC Genomics. 2009, 10: 112- 10.1186/1471-2164-10-112
Doley R, Pahari S, Mackessy SP, Kini RM: Accelerated exchange of exon segments in Viperid three-finger toxin genes (Sistrurus catenatus edwardsii; Desert Massasauga). BMC Evol Biol. 2008, 8: 196- 10.1186/1471-2148-8-196
Pahari S, Mackessy SP, Kini MR: The venom gland transcriptome of the Desert Massasauga Rattlesnake (Sistrurus catenatus edwardsii): towards an understanding of venom composition among advanced snakes (Superfamily Colubroidea). BMC Mol Biol. 2007, 8: 115- 10.1186/1471-2199-8-115
Yanoshita R, Ogawa Y, Murayama N, Omori-Satoh T, Saguchi K, Higuchi S, et al: Molecular cloning of the major lethal toxins from two kraits (Bungarus flaviceps and Bungarus candidus). Toxicon. 2006, 47: 416-424. 10.1016/j.toxicon.2005.12.004
Chang CC, Lee CY: ISOLATION OF NEUROTOXINS FROM THE VENOM OF BUNGARUS MULTICINCTUSAND THEIR MODES OF NEUROMUSCULAR BLOCKING ACTION. Arch Int Pharmacodyn Ther. 1963, 144: 241-257.
Chu CC, Li SH, Chen YH: Resolution of isotoxins in the beta-bungarotoxin family. J Chromatogr A. 1995, 694: 492-497. 10.1016/0021-9673(94)01173-C
Su MJ, Chang CC: Presynaptic effects of snake venom toxins which have phospholipase A2 activity (beta-bungarotoxin, taipoxin, crotoxin). Toxicon. 1984, 22: 631-640. 10.1016/0041-0101(84)90003-5
Chiappinelli VA: Kappa-bungarotoxin: a probe for the neuronal nicotinic receptor in the avian ciliary ganglion. Brain Res. 1983, 277: 9-22. 10.1016/0006-8993(83)90902-2
Tsetlin V: Snake venom alpha-neurotoxins and other 'three-finger' proteins. Eur J Biochem. 1999, 264: 281-286. 10.1046/j.1432-1327.1999.00623.x
Benishin CG: Potassium channel blockade by the B subunit of beta-bungarotoxin. Mol Pharmacol. 1990, 38: 164-169.
Harvey AL, Karlsson E: Protease inhibitor homologues from mamba venoms: facilitation of acetylcholine release and interactions with prejunctional blocking toxins. Br J Pharmacol. 1982, 77: 153-161.
Chanhome L, Wongtongkam N, Khow O, Pakmanee N, Omori-Satoh T, Sitprija V: Genus specific neutralization of Bungarus snake venoms by Thai Red Cross banded krait antivenom. J Nat Toxins. 1999, 8: 135-140.
Khow O, Chanhome L, Omori-Satoh T, Sitprija V: Isolation of the major lethal toxin in the venom of Bungarus flaviceps. Toxicon. 2002, 40: 463-469. 10.1016/S0041-0101(01)00235-5
Grant GA, Frazier MW, Chiappinelli VA: Amino acid sequence of kappa-flavitoxin: establishment of a new family of snake venom neurotoxins. Biochemistry. 1988, 27: 3794-3798. 10.1021/bi00410a041
Chiappinelli VA, Wolf KM, DeBin JA, Holt IL: Kappa-flavitoxin: isolation of a new neuronal nicotinic receptor antagonist that is structurally related to kappa-bungarotoxin. Brain Res. 1987, 402: 21-29. 10.1016/0006-8993(87)91043-2
Dufton MJ, Hider RC: Structure and pharmacology of elapid cytotoxins. Pharmacol Ther. 1988, 36: 1-40. 10.1016/0163-7258(88)90111-8
Endo T, Tamiya N: Structure-function relationship of postsynaptic neurotoxins from snake venoms. Snake Toxins. 1991, 165-222.
Menez A: Functional architectures of animal toxins: a clue to drug design?. Toxicon. 1998, 36: 1557-1572. 10.1016/S0041-0101(98)00148-2
Fry BG: From genome to "venome": molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005, 15: 403-420. 10.1101/gr.3228405
McDowell RS, Dennis MS, Louie A, Shuster M, Mulkerrin MG, Lazarus RA: Mambin, a potent glycoprotein IIb-IIIa antagonist and platelet aggregation inhibitor structurally related to the short neurotoxins. Biochemistry. 1992, 31: 4766-4772. 10.1021/bi00135a004
Marchot P, Prowse CN, Kanter J, Camp S, Ackermann EJ, Radic Z, et al: Expression and activity of mutants of fasciculin, a peptidic acetylcholinesterase inhibitor from mamba venom. J Biol Chem. 1997, 272: 3502-3510. 10.1074/jbc.272.6.3502
Marchot P, Bourne Y, Prowse CN, Bougis PE, Taylor P: Inhibition of mouse acetylcholinesterase by fasciculin: crystal structure of the complex and mutagenesis of fasciculin. Toxicon. 1998, 36: 1613-1622. 10.1016/S0041-0101(98)00154-8
jas-Bailador F, Costa G, Dajas F, Emmett S: Effects of alpha-erabutoxin, alpha-bungarotoxin, alpha-cobratoxin and fasciculin on the nicotine-evoked release of dopamine in the rat striatum in vivo. Neurochem Int. 1998, 33: 307-312. 10.1016/S0197-0186(98)00033-3
Carsi JM, Valentine HH, Potter LT: m2-toxin: A selective ligand for M2 muscarinic receptors. Mol Pharmacol. 1999, 56: 933-937.
Carsi JM, Potter LT: m1-toxin isotoxins from the green mamba (Dendroaspis angusticeps) that selectively block m1 muscarinic receptors. Toxicon. 2000, 38: 187-198. 10.1016/S0041-0101(99)00141-5
Gong N, Armugam A, Jeyaseelan K: Postsynaptic short-chain neurotoxins from Pseudonaja textilis. cDNA cloning, expression and protein characterization. Eur J Biochem. 1999, 265: 982-989. 10.1046/j.1432-1327.1999.00800.x
Gong N, Armugam A, Jeyaseelan K: Molecular cloning, characterization and evolution of the gene encoding a new group of short-chain alpha-neurotoxins in an Australian elapid, Pseudonaja textilis. FEBS Lett. 2000, 473: 303-310. 10.1016/S0014-5793(00)01549-0
Chaki S, Muramatsu M, Ushiyama Y, Otomo S: Purification and partial characterization of K+ channel blockers from the venom of Dendroaspis angusticeps. Neurochem Int. 1992, 20: 553-558. 10.1016/0197-0186(92)90034-O
Albrand JP, Blackledge MJ, Pascaud F, Hollecker M, Marion D: NMR and restrained molecular dynamics study of the three-dimensional solution structure of toxin FS2, a specific blocker of the L-type calcium channel, isolated from black mamba venom. Biochemistry. 1995, 34: 5923-5937. 10.1021/bi00017a022
Kini RM, Caldwell RA, Wu QY, Baumgarten CM, Feher JJ, Evans HJ: Flanking proline residues identify the L-type Ca2+ channel binding site of calciseptine and FS2. Biochemistry. 1998, 37: 9058-9063. 10.1021/bi9802723
Jang JY, Krishnaswamy T, Kumar S, Jayaraman G, Yang PW, Yu C: Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid. Biochemistry. 1997, 36: 14635-14641. 10.1021/bi971107a
Fryklund L, Eaker D: Complete amino acid sequence of a nonneurotoxic hemolytic protein from the venom of Haemachatus haemachates (African ringhals cobra). Biochemistry. 1973, 12: 661-667. 10.1021/bi00728a015
Choudhury SR, Gomes A, Gomes A, Dattagupta JK, Sen U: Purification, crystallization and preliminary X-ray structural studies of a 7.2 kDa cytotoxin isolated from the venom of Daboia russelli russelli of the Viperidae family. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006, 62: 292-294. 10.1107/S1744309106005963
Dubovskii PV, Lesovoy DM, Dubinnyi MA, Konshina AG, Utkin YN, Efremov RG, et al: Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins. Biochem J. 2005, 387: 807-815. 10.1042/BJ20041814
Ploug M, Ellis V: Structure-function relationships in the receptor for urokinase-type plasminogen activator. Comparison to other members of the Ly-6 family and snake venom alpha-neurotoxins. FEBS Lett. 1994, 349: 163-168. 10.1016/0014-5793(94)00674-1
Kornisiuk E, Jerusalinsky D, Cervenansky C, Harvey AL: Binding of muscarinic toxins MTx1 and MTx2 from the venom of the green mamba Dendroaspis angusticeps to cloned human muscarinic cholinoceptors. Toxicon. 1995, 33: 11-18. 10.1016/0041-0101(94)00161-Z
Jerusalinsky D, Harvey A, Karlsson E, Potter L: Workshop: the use of muscarinic toxins in the study of muscarinic receptors. Life Sci. 1997, 60: 1161-1162. 10.1016/S0024-3205(97)00061-1
Kanda T, Yoshihara T, Miyazaki M, Kaneko T: Neuromuscular junctions of the posterior cricoarytenoid muscle in the cat. Acta Otolaryngol Suppl. 1983, 393: 25-32. 10.3109/00016488309129574
Unwin N: Nicotinic acetylcholine receptor at 9 A resolution. J Mol Biol. 1993, 229: 1101-1124. 10.1006/jmbi.1993.1107
Fry BG, Wuster W, Kini RM, Brusic V, Khan A, Venkataraman D, et al: Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003, 57: 110-129. 10.1007/s00239-003-2461-2
Tsai HY, Wang YM, Tsai IH: Cloning, characterization and phylogenetic analyses of members of three major venom families from a single specimen of Walterinnesia aegyptia. Toxicon. 2008, 51: 1245-1254. 10.1016/j.toxicon.2008.02.012
Pillet L, Tremeau O, Ducancel F, Drevet P, Zinn-Justin S, Pinkasfeld S, et al: Genetic engineering of snake toxins. Role of invariant residues in the structural and functional properties of a curaremimetic toxin, as probed by site-directed mutagenesis. J Biol Chem. 1993, 268: 909-916.
Tremeau O, Lemaire C, Drevet P, Pinkasfeld S, Ducancel F, Boulain JC, et al: Genetic engineering of snake toxins. The functional site of Erabutoxin a, as delineated by site-directed mutagenesis, includes variant residues. J Biol Chem. 1995, 270: 9362-9369. 10.1074/jbc.270.16.9362
Watanabe L, Nirthanan S, Rajaseger G, Polikarpov I, Kini RM, Arni RK: Crystallization and preliminary X-ray analysis of bucain, a novel toxin from the Malayan krait Bungarus candidus. Acta Crystallogr D Biol Crystallogr. 2002, 58: 1879-1881. 10.1107/S0907444902011022
Murakami MT, Kini RM, Arni RK: Crystal structure of bucain, a three-fingered toxin from the venom of the Malayan krait (Bungarus candidus). Protein Pept Lett. 2009, 16: 1473-1477. 10.2174/092986609789839304
Qian YC, Fan CY, Gong Y, Yang SL: cDNA cloning and sequence analysis of six neurotoxin-like proteins from Chinese continental banded krait. Biochem Mol Biol Int. 1998, 46: 821-828.
Nirthanan S, Gopalakrishnakone P, Gwee MC, Khoo HE, Kini RM: Non-conventional toxins from Elapid venoms. Toxicon. 2003, 41: 397-407. 10.1016/S0041-0101(02)00388-4
Nirthanan S, Charpantier E, Gopalakrishnakone P, Gwee MC, Khoo HE, Cheah LS, et al: Candoxin, a novel toxin from Bungarus candidus, is a reversible antagonist of muscle (alphabetagammadelta) but a poorly reversible antagonist of neuronal alpha 7 nicotinic acetylcholine receptors. J Biol Chem. 2002, 277: 17811-17820. 10.1074/jbc.M111152200
Chung C, Wu BN, Yang CC, Chang LS: Muscarinic toxin-like proteins from Taiwan banded krait (Bungarus multicinctus) venom: purification, characterization and gene organization. Biol Chem. 2002, 383: 1397-1406. 10.1515/BC.2002.158
Harvey AL, Kornisiuk E, Bradley KN, Cervenansky C, Duran R, Adrover M, et al: Effects of muscarinic toxins MT1 and MT2 from green mamba on different muscarinic cholinoceptors. Neurochem Res. 2002, 27: 1543-1554. 10.1023/A:1021660708187
Chiappinelli VA, Weaver WR, McLane KE, Conti-Fine BM, Fiordalisi JJ, Grant GA: Binding of native kappa-neurotoxins and site-directed mutants to nicotinic acetylcholine receptors. Toxicon. 1996, 34: 1243-1256. 10.1016/S0041-0101(96)00110-9
Jeyaseelan K, Poh SL, Nair R, Armugam A: Structurally conserved alpha-neurotoxin genes encode functionally diverse proteins in the venom of Naja sputatrix. FEBS Lett. 2003, 553: 333-341. 10.1016/S0014-5793(03)01039-1
Doley R, Kini RM: Protein complexes in snake venom. Cell Mol Life Sci. 2009, 66: 2851-2871. 10.1007/s00018-009-0050-2
Strong PN, Goerke J, Oberg SG, Kelly RB: beta-Bungarotoxin, a pre-synaptic toxin with enzymatic activity. Proc Natl Acad Sci USA. 1976, 73: 178-182. 10.1073/pnas.73.1.178
Ritonja A, Meloun B, Gubensek F: The primary structure of Vipera ammodytes venom trypsin inhibitor I. Biochim Biophys Acta. 1983, 748: 429-435.
Shafqat J, Beg OU, Yin SJ, Zaidi ZH, Jornvall H: Primary structure and functional properties of cobra (Naja naja naja) venom Kunitz-type trypsin inhibitor. Eur J Biochem. 1990, 194: 337-341. 10.1111/j.1432-1033.1990.tb15622.x
Shafqat J, Zaidi ZH, Jornvall H: Purification and characterization of a chymotrypsin Kunitz inhibitor type of polypeptide from the venom of cobra (Naja naja naja). FEBS Lett. 1990, 275: 6-8. 10.1016/0014-5793(90)81426-O
Chang L, Chung C, Huang HB, Lin S: Purification and characterization of a chymotrypsin inhibitor from the venom of Ophiophagus hannah (King Cobra). Biochem Biophys Res Commun. 2001, 283: 862-867. 10.1006/bbrc.2001.4878
Cheng YC, Yan FJ, Chang LS: Taiwan cobra chymotrypsin inhibitor: cloning, functional expression and gene organization. Biochim Biophys Acta. 2005, 1747: 213-220.
Hokama Y, Iwanaga S, Tatsuki T, Suzuki T: Snake venom proteinase inhibitors. III. Isolation of five polypeptide inhibitors from the venoms of Hemachatus haemachatus (Ringhal's corbra) and Naja nivea (Cape cobra) and the complete amino acid sequences of two of them. J Biochem. 1976, 79: 559-578.
Laskowski M: Protein inhibitors of serine proteinases--mechanism and classification. Adv Exp Med Biol. 1986, 199: 1-17.
Laskowski M, Kato I: Protein inhibitors of proteinases. Annu Rev Biochem. 1980, 49: 593-626. 10.1146/annurev.bi.49.070180.003113
Chang LS, Wang JJ, Cheng YC, Chou WM: Genetic organization of Bungarus multicinctus protease inhibitor-like proteins. Toxicon. 2008, 51: 1490-1495. 10.1016/j.toxicon.2008.03.025
Gojobori T, Ikeo K: Molecular evolution of serine protease and its inhibitor with special reference to domain evolution. Philos Trans R Soc Lond B Biol Sci. 1994, 344: 411-415. 10.1098/rstb.1994.0080
Cheng YC, Chen KC, Lin SK, Chang LS: Divergence of genes encoding B chains of beta-bungarotoxins. Toxicon. 2006, 47: 322-329. 10.1016/j.toxicon.2005.11.009
Harvey AL, Karlsson E: Dendrotoxin from the venom of the green mamba, Dendroaspis angusticeps. A neurotoxin that enhances acetylcholine release at neuromuscular junction. Naunyn Schmiedebergs Arch Pharmacol. 1980, 312: 1-6. 10.1007/BF00502565
Danse JM, Rowan EG, Gasparini S, Ducancel F, Vatanpour H, Young LC, et al: On the site by which alpha-dendrotoxin binds to voltage-dependent potassium channels: site-directed mutagenesis reveals that the lysine triplet 28-30 is not essential for binding. FEBS Lett. 1994, 356: 153-158. 10.1016/0014-5793(94)01235-0
Smith LA, Reid PF, Wang FC, Parcej DN, Schmidt JJ, Olson MA, et al: Site-directed mutagenesis of dendrotoxin K reveals amino acids critical for its interaction with neuronal K+ channels. Biochemistry. 1997, 36: 7690-7696. 10.1021/bi963105g
Kini RM: Anticoagulant proteins from snake venoms: structure, function and mechanism. Biochem J. 2006, 397: 377-387. 10.1042/BJ20060302
Arni RK, Ward RJ: Phospholipase A2--a structural review. Toxicon. 1996, 34: 827-841. 10.1016/0041-0101(96)00036-0
Kini RM: Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon. 2005, 45: 1147-1161. 10.1016/j.toxicon.2005.02.018
Pearson JA, Tyler MI, Retson KV, Howden ME: Studies on the subunit structure of textilotoxin, a potent presynaptic neurotoxin from the venom of the Australian common brown snake (Pseudonaja textilis). 2. The amino acid sequence and toxicity studies of subunit D. Biochim Biophys Acta. 1991, 1077: 147-150.
Chang LS, Wu PF, Chang CC: cDNA sequence analysis and mutagenesis studies on the A chain of beta-bungarotoxin from Taiwan banded krait. J Protein Chem. 1996, 15: 755-761. 10.1007/BF01887150
Currie MG, Geller DM, Cole BR, Siegel NR, Fok KF, Adams SP, et al: Purification and sequence analysis of bioactive atrial peptides (atriopeptins). Science. 1984, 223: 67-69. 10.1126/science.6419347
de Bold AJ, Borenstein HB, Veress AT, Sonnenberg H: A rapid and potent natriuretic response to intravenous injection of atrial myocardial extract in rats. Reprinted from Life Sci. 28:89-94, 1981. J Am Soc Nephrol. 2001, 12: 403-409.
Flynn TG, de Bold ML, de Bold AJ: The amino acid sequence of an atrial peptide with potent diuretic and natriuretic properties. Biochem Biophys Res Commun. 1983, 117: 859-865. 10.1016/0006-291X(83)91675-3
Misono KS, Grammer RT, Fukumi H, Inagami T: Rat atrial natriuretic factor: isolation, structure and biological activities of four major peptides. Biochem Biophys Res Commun. 1984, 123: 444-451. 10.1016/0006-291X(84)90250-X
Sudoh T, Kangawa K, Minamino N, Matsuo H: A new natriuretic peptide in porcine brain. Nature. 1988, 332: 78-81. 10.1038/332078a0
Sudoh T, Minamino N, Kangawa K, Matsuo H: C-type natriuretic peptide (CNP): a new member of natriuretic peptide family identified in porcine brain. Biochem Biophys Res Commun. 1990, 168: 863-870. 10.1016/0006-291X(90)92401-K
Brenner BM, Ballermann BJ, Gunning ME, Zeidel ML: Diverse biological actions of atrial natriuretic peptide. Physiol Rev. 1990, 70: 665-699.
Espiner EA, Richards AM, Yandle TG, Nicholls MG: Natriuretic hormones. Endocrinol Metab Clin North Am. 1995, 24: 481-509.
Tsukada T, Rankin JC, Takei Y: Involvement of drinking and intestinal sodium absorption in hyponatremic effect of atrial natriuretic peptide in seawater eels. Zoolog Sci. 2005, 22: 77-85. 10.2108/zsj.22.77
Hama N, Itoh H, Shirakami G, Suga S, Komatsu Y, Yoshimasa T, et al: Detection of C-type natriuretic peptide in human circulation and marked increase of plasma CNP level in septic shock patients. Biochem Biophys Res Commun. 1994, 198: 1177-1182. 10.1006/bbrc.1994.1166
Ogawa Y, Nakao K, Itoh H, Suga S, Imura H: [Molecular biology of the natriuretic peptide system]. Nippon Rinsho. 1992, 50: 2885-2892.
Collins SP, Ronan-Bentle S, Storrow AB: Diagnostic and prognostic usefulness of natriuretic peptides in emergency department patients with dyspnea. Ann Emerg Med. 2003, 41: 532-545. 10.1067/mem.2003.113
Schweitz H, Vigne P, Moinier D, Frelin C, Lazdunski M: A new member of the natriuretic peptide family is present in the venom of the green mamba (Dendroaspis angusticeps). J Biol Chem. 1992, 267: 13928-13932.
Johns DG, Ao Z, Heidrich BJ, Hunsberger GE, Graham T, Payne L, et al: Dendroaspis natriuretic peptide binds to the natriuretic peptide clearance receptor. Biochem Biophys Res Commun. 2007, 358: 145-149. 10.1016/j.bbrc.2007.04.079
Ching AT, Rocha MM, Paes Leme AF, Pimenta DC, de Fatima DF, Serrano SM, et al: Some aspects of the venom proteome of the Colubridae snake Philodryas olfersii revealed from a Duvernoy's (venom) gland transcriptome. FEBS Lett. 2006, 580: 4417-4422. 10.1016/j.febslet.2006.07.010
Yamazaki Y, Hyodo F, Morita T: Wide distribution of cysteine-rich secretory proteins in snake venoms: isolation and cloning of novel snake venom cysteine-rich secretory proteins. Arch Biochem Biophys. 2003, 412: 133-141. 10.1016/S0003-9861(03)00028-6
Matsunaga Y, Yamazaki Y, Hyodo F, Sugiyama Y, Nozaki M, Morita T: Structural divergence of cysteine-rich secretory proteins in snake venoms. J Biochem. 2009, 145: 365-375. 10.1093/jb/mvn174
Yamazaki Y, Brown RL, Morita T: Purification and cloning of toxins from elapid venoms that target cyclic nucleotide-gated ion channels. Biochemistry. 2002, 41: 11331-11337. 10.1021/bi026132h
Lu Q, Navdaev A, Clemetson JM, Clemetson KJ: Snake venom C-type lectins interacting with platelet receptors. Structure-function relationships and effects on haemostasis. Toxicon. 2005, 45: 1089-1098. 10.1016/j.toxicon.2005.02.022
Xu Q, Wu XF, Xia QC, Wang KY: Cloning of a galactose-binding lectin from the venom of Trimeresurus stejnegeri. Biochem J. 1999, 341 (Pt 3): 733-737. 10.1042/0264-6021:3410733
Morita T: Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon. 2005, 45: 1099-1114. 10.1016/j.toxicon.2005.02.021
Drickamer K: Recognition of complex carbohydrates by Ca(2+)-dependent animal lectins. Biochem Soc Trans. 1993, 21: 456-459.
Rotenberg D, Bamberger ES, Kochva E: Studies on ribonucleic acid synthesis in the venom glands of Vipera palaestinae (Ophidia, Reptilia). Biochem J. 1971, 121: 609-612.
Sanger F, Nicklen S, Coulson AR: DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977, 74: 5463-5467. 10.1073/pnas.74.12.5463
Acknowledgements
This work was supported by a grant to RMK from the Biomedical Research Council, Agency for Science, Technology and Research, Singapore.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
ASS and RD worked on the project and drafted the manuscript. RD analyzed the resulted and interpreted. FJV supplied the tissue samples and interpreted the results. RMK is the principal investigator who designed the experiment, analyzed the data and critically reviewed the manuscript. All the authors have approved the final version of the manuscript.
Electronic supplementary material
12867_2009_505_MOESM1_ESM.PDF
Additional file 1:Transcripts submitted to database showing similarity to snake venom protein family.Numbers of transcripts, accession number, search programme and E values are shownin the table. (PDF 17 KB)
12867_2009_505_MOESM2_ESM.PDF
Additional file 2:% of venom proteins families observed in venom gland transcriptome. Comparison of different toxin families observed in transcriptome of elapid and viperidvenom gland. (PDF 33 KB)
12867_2009_505_MOESM3_ESM.PDF
Additional file 3:ASSET in 3FTxs of B. flaviceps. Alignment of 3FTx of B. flaviceps showing Accelerated Segment Switch in Exon toalter Targeting (ASSET). The segments which are similar are shown in same color where as segments which are dissimilar are shown in different color. The clone name and the number of clones are also shown in the figure. (PDF 9 KB)
12867_2009_505_MOESM4_ESM.PDF
Additional file 4:Premature truncated kunitz type SPI from B. flaviceps. Comparison of protein and nucleotide of truncated kunitz type SPI from B. flaviceps with EU246693 from Ophiophagus hannah. Exons are highlighted with different colors, Exon I is highlighted with red color, Exon II with Blue and Exon III is in grey color. 87 Nucleotides are deleted from the exon II of BF539 as shown with dashes in the figure. Comparison of the mRNA sequence of BF539 with BF294 reveals that a dinucleotide "GT" (underlined and highlighted in red letter) is present at the end of the exon II of BF539. The splicing error could be due to change in this base substitution. However the exon III is intact as stop codon and one of the amino acid residue is encoded by the exon III. (PDF 13 KB)
12867_2009_505_MOESM5_ESM.PDF
Additional file 5:Phylogenetic relationship of B chain and Kunitz SPI of different Bungarus species. Kunitz type SPI and B chain of β-bungarotoxin of Bungarus sp was obtained from the database and phylogenetic tree was constructed to understand the relationship between kunitz SPI and B chain of β-bungarotoxin. (PDF 15 KB)
12867_2009_505_MOESM6_ESM.PDF
Additional file 6:Comparison of BF95 from B. flaviceps with Laticauda semifasciata PLA2(AB062439). Exons are highlighted with different colors, Exon I is highlighted in green color; Exon II in magenta; Exon III in dark blue and Exon IV in grey. In BF95, part of the exon II (99 bp) is missing as shown in the figure with dashes. (PDF 14 KB)
12867_2009_505_MOESM7_ESM.PDF
Additional file 7:CRISPs and C-type lectins. A) Transcripts encoding CRISPs (BF53), found in this venom gland cDNA library. B) C-type lectins (BF53) found in this venom gland cDNA library. One of the C-type lectin found in this venom gland cDNA library was truncated in the 5' end (BF764). The amino acid residues of C-type lectin involved in binding to the Ca2+ are highlighted with green color and the cysteine residues are highlighted. (PDF 11 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Siang, A.S., Doley, R., Vonk, F.J. et al. Transcriptomic analysis of the venom gland of the red-headed krait (Bungarus flaviceps) using expressed sequence tags. BMC Molecular Biol 11, 24 (2010). https://doi.org/10.1186/1471-2199-11-24
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2199-11-24