Abstract
Background
Campylobacter is a major cause of human disease worldwide and poultry are identified as a significant source of this pathogen. Most disease in humans is associated with the consumption of contaminated poultry or cross-contamination with other foods. The primary drugs of choice for treatment of human campylobacteriosis include erythromycin and ciprofloxacin. In this study, we investigated the prevalence of resistance to erythromycin and ciprofloxacin in Campylobacter isolates recovered from turkey carcasses at two processing plants in the Upper Midwest US. Further analysis of a subset of isolates was carried out to assess resistance and genotype profiles.
Results
Campylobacter isolates from plant A (n = 439; including 196 C. coli and 217 C. jejuni) and plant B (n = 362, including 281 C. coli and 62 C. jejuni) were tested for susceptibility to ciprofloxacin and erythromycin using agar dilution. C. coli were more frequently resistant than C. jejuni in both plants, including resistance to ciprofloxacin (28% of C. jejuni and 63% of C. coli, plant B; and 11% of C. coli, plant A). Erythromycin resistance was low among C. jejuni (0% plant A and 0.3% plant B) compared to C. coli (41%, plant A and 17%, plant B). One hundred resistant and susceptible isolates were selected for additional antimicrobial susceptibility testing, restriction fragment length polymorphism analysis of the flaA gene (fla typing), and pulsed-field gel electrophoresis (PFGE). Fla-PFGE types obtained (n = 37) were associated with a specific plant with the exception of one type that was isolated from both plants. C. coli isolates (n = 65) were grouped into 20 types, while C. jejuni isolates (n = 35) were grouped into 17 types. Most isolates with identical fla-PFGE patterns shared identical or very similar antimicrobial resistance profiles. PFGE alone and composite analysis using fla-PFGE with resistance profiles separated C. jejuni and C. coli into distinct groups.
Conclusion
Ciprofloxacin and erythromycin resistance in Campylobacter recovered from processed turkey occurred more frequently among C. coli than C. jejuni. Fla-PFGE types were associated with a particular species, antimicrobial resistance profiles, and a specific plant. Molecular subtyping in this study provided more information about the relationships among antimicrobial-resistant Campylobacter at the processing level.
Similar content being viewed by others
Background
Campylobacter spp. are one of the major causes of human gastroenteritis worldwide and are estimated to cause over two million cases of illness annually in the U.S. [1]. Greater than 95% of human infections are due to C. jejuni or C. coli [2]. Human disease is characterized by diarrhea, fever, and abdominal cramping [3]. Campylobacteriosis is most often associated with the handling and consumption of raw or undercooked poultry [2–4].
In poultry, Campylobacter is considered a commensal organism [4]. When colonized poultry enter the processing plant, contamination of the carcass and processed product can result [4]. Turkey is an important reservoir of Campylobacter; studies have reported prevalence rates of 65-95% in U.S. turkeys at production [5–7]. In a study from our lab, the prevalence of Campylobacter was 34.9% from two turkey processing plants [8], while at the retail level, the organism has been detected in 1.0-15% of samples tested [9, 10].
Human campylobacteriosis is generally self-limiting, although in severe cases it requires antimicrobial therapy. Erythromycin and ciprofloxacin are often the drugs of choice [11]. Fluoroquinolones such as ciprofloxacin have been used for first-line treatment of bacterial gastroenteritis in the absence of a microbiological diagnosis [3]. However, an increase in fluoroquinolone-resistant Campylobacter infections in humans has been documented worldwide [12–14], and may be associated with fluoroquinolone use in food animals [12, 15, 16]. Although the approval of enrofloxacin (a fluoroquinolone) for use in poultry was withdrawn by the U.S. Food and Drug Administration in 2005, it is possible that fluoroquinolone-resistant Campylobacter will persist in poultry flocks [17]. Macrolides such as erythromycin have been the preferred treatment for Campylobacter infections [3, 13]; however, increasing resistance to erythromycin among Campylobacter has been documented, particularly in C. coli [12, 18–20]. The duration of illness, risk of invasive illness, or poorer treatment response has been shown to be greater for patients infected with quinolone- or macrolide-resistant Campylobacter [16, 21–23]; although Wassenaar et al. [24] did not find these effects associated with fluoroquinolone-resistant Campylobacter infections. In Campylobacter, resistance to quinolones and macrolides is primarily associated with mutations in the gyrA and 23S rRNA genes, respectively [20, 25]. The involvement of the CmeABC multidrug efflux pump in resistance to both classes of antimicrobials has also been recognized [26, 27].
Information about antimicrobial resistance of Campylobacter at different levels of production is important for the development of control strategies for this pathogen. In addition, differentiation of antimicrobial-resistant strains is necessary to investigate the epidemiology of resistance. Restriction fragment length polymorphism (RFLP) analysis of the flaA gene (fla typing) and pulsed-field gel electrophoresis (PFGE) are two genotyping methods used to successfully differentiate Campylobacter strains [28, 29]. This study was conducted to assess the ciprofloxacin and erythromycin resistance in Campylobacter isolated from turkey at the processing level. Fla typing, PFGE, and antimicrobial susceptibility testing were used to characterize a subset of ciprofloxacin- and/or erythromycin-resistant and susceptible Campylobacter isolates obtained from pre and post chill turkey carcasses and chill water.
Results
Antimicrobial susceptibility testing
Figure 1A and 1B shows the MICs of 801 Campylobacter isolates to ciprofloxacin and erythromycin. Few isolates were co-resistant to both antimicrobials (2 from plant A [0.45% of plant A isolates] and 9 from plant B [2.5% of plant B isolates]). Resistant isolates were recovered from carcasses at pre chill and post chill at both plants. No significant difference (P > 0.01) was observed between the number of ciprofloxacin-resistant or erythromycin-resistant isolates obtained from either process stage at plant A (Table 1).
Antimicrobial susceptibility profiles of Campylobacter isolates (n = 801). Isolates from plant A (n = 439; open bars) and plant B (n = 362; black bars) were tested for antimicrobial susceptibility using agar dilution. A. The frequency of MICs obtained for ciprofloxacin. The arrow denotes the breakpoint of ≥ 4 μg/ml. B. The frequency of MICs obtained for erythromycin. The arrow denotes the breakpoint of ≥ 32 μg/ml.
Differences were observed in the frequency of resistance among C. coli compared to C. jejuni (Table 2). C. coli were more likely to be erythromycin-resistant compared to C. jejuni (41% plant A and 17% plant B compared to 0.0%, plant A and 0.3%, plant B) (P < 0.01). C. coli were also more likely to be ciprofloxacin-resistant compared to C. jejuni in both plant A (C. coli, 11%; C. jejuni, 0.0%) and plant B (C. coli, 63%; C. jejuni, 28%) (P < 0.01).
Additional antimicrobial susceptibility testing conducted on a subset of isolates selected for subtyping (n = 100) found that isolates from plant A (n = 51; C. jejuni, 8; C. coli, 43) were resistant to tetracycline (100%), nalidixic acid (49%; C. jejuni, 2; C. coli, 23), kanamycin (41%; C. jejuni, 0; C. coli, 21), and streptomycin (41%; C. jejuni, 0; C. coli, 21), while those from plant B (n = 49; C. jejuni, 27; C. coli, 22) were resistant to nalidixic acid (94%; C. jejuni, 24; C. coli, 22), tetracycline (86%; C. jejuni, 26; C. coli, 16), kanamycin (20%; C. jejuni, 9; C. coli, 1) and streptomycin (18%; C. jejuni, 0; C. coli, 9). Sixteen different drug resistance profiles were identified, with most isolates displaying resistance to more than one agent (Figure 2). None of the isolates were resistant to all six agents tested. The two most prevalent multiple resistance profiles observed were 1) ciprofloxacin, nalidixic acid and tetracycline for 25 isolates (most common profile among C. jejuni) and 2) ciprofloxacin, nalidixic acid, kanamycin and tetracycline for 25 isolates (most common profile among C. coli)
Antimicrobial resistance profiles and frequency among selected Campylobacter isolates (n = 100). C. jejuni (n = 35; open bars) and C. coli (n = 65; black bars) isolates were tested for antimicrobial resistance using agar dilution. Presence of a letter indicates resistance, with C = ciprofloxacin, N = nalidixic acid, E = erythromycin, S = streptomycin, K = kanamycin, and T = tetracycline.
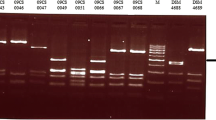
Flatyping and pulsed-field gel electrophoresis
All of the isolates examined (n = 100) tested positive for the flaA gene and 24 different fla types were observed. Twenty-six PFGE types were observed. Fla typing separated the isolates into three major groups at 50% similarity (data not shown), while PFGE separated them into two major groups at 30% similarity (Figure 3). Similar fla types were found in isolates originating from different plants (types A, B, K, M and X). Two PFGE types were detected in isolates from both plants (types 10 and 28). Thirty-seven combined fla-PFGE types were obtained, 22 of which contained only single isolates (Figure 4). Plant A isolates were grouped into 16 fla-PFGE types and plant B isolates were grouped into 22 fla-PFGE types. Fla-PFGE types were unique to a particular plant with the exception of M10, which was isolated from both plants on different days in the same month. M10 was also isolated once from plant A in the previous month. In both plants, some isolates obtained from different sampling stages (pre or post chill) had identical fla-PFGE types.
Six fla types were observed for C. jejuni isolates, while fourteen fla types were observed for C. coli isolates. Four fla types within two of the three major clusters included isolates of C. jejuni and C. coli (data not shown). Using PFGE, C. jejuni isolates were divided into 13 PFGE types, while C. coli were also divided into 13 PFGE types. The two major clusters obtained with PFGE generally separated the two species (Figure 3). Combined fla-PFGE types were unique to a particular species. C. coli isolates (n = 65) were grouped into 20 fla-PFGE types; three of these fla-PFGE types (B4, L18, and P2) contained 62% of the total C. coli isolates. C. jejuni isolates (n = 35) were grouped into 17 fla-PFGE types; one fla-PFGE type (I3) contained 29% of the C. jejuni isolates, while the other fla-PFGE types included no more than 3 C. jejuni isolates each.
Antimicrobial resistance profiles and combined fla-PFGE types are shown in Figure 4. Thirty-seven isolates with the same fla-PFGE type had identical resistance profiles, including fla-PFGE types J28, D28, I30, I3, P2, V2, R9, and T6. Forty-one isolates with the same fla-PFGE type had either identical resistance profiles or very similar resistance profiles, including fla-PFGE types B4, U9, F22, L18, M10, X11, and O20. Within some fla-PFGE types, the MICs for the antimicrobials varied, generally between one to four dilutions (data not shown). On occasion, different fla-PFGE types from the same plant had identical antimicrobial resistance profiles (Figure 4). For example, types A14 and J28 from plant B were both resistant to ciprofloxacin, nalidixic acid, and tetracycline.
Composite analysis (Figure 4) using fla typing, PFGE, and antimicrobial resistance profiles separated the isolates into 30 distinct types. At 43% similarity, three major clusters (I, II, and III) were evident. One isolate was not clustered into any of these three groups. The majority of isolates in group II were C. coli, while all of the isolates in groups I and III were C. coli and C. jejuni, respectively.
The numerical index of discrimination (D) was used to evaluate the results of fla typing, PFGE, and antimicrobial resistance profiling. The discrimination index was highest for fla-PFGE analysis (D = 0.9321) followed by PFGE (D = 0.9147), composite data (all three methods, D = 0.9137), fla typing (D = 0.9119), and antimicrobial resistance profiling (D = 0.8430).
Discussion
Campylobacter isolates from two turkey processing plants in the upper Midwest were examined for susceptibility to ciprofloxacin and erythromycin, antimicrobial agents used for the treatment of human campylobacteriosis. Although co-resistance to both antimicrobials was low, resistance was detected and differences were observed in the frequency of resistance in relation to species. C. coli from plant A (41%) and plant B (17%) were more likely to be erythromycin-resistantcompared to C. jejuni (plant A, 0.0%; plant B, 0.3%) (P < 0.01). Similarly, other studies on Campylobacter isolated from poultry have reported that erythromycin resistance occurs more frequently in C. coli than C. jejuni [6, 9, 18, 30–32]. The occurrence of erythromycin resistance among C. coli isolated from the processing environment in this study (41%, plant A and 17%, plant B) was greater in comparison to 11.8% and 12.5% for C. coli from retail turkey in the U.S. [9] and Germany [33], respectively. Erythromycin resistance among C. jejuni in this study was very low, similar to the aforementioned reports on retail turkey where resistance was 0% for C. jejuni in both countries [9, 33]. In contrast, 6.4% of C. jejuni obtained from turkeys at a Belgian slaughterhouse were resistant [32].
In this study, the frequency of ciprofloxacin resistance was also found to be higher in C. coli (plant A, 11%; plant B, 63%) compared to C. jejuni (plant A, 0.0%; plant B, 28%) (P < 0.01). Others have reported a higher occurrence of fluoroquinolone resistance in C. coli compared to C. jejuni as well [6, 19, 30, 34]. In comparison to previous studies conducted at different parts of the production system, ciprofloxacin resistance at plant B (28% in C. jejuni and 63% in C. coli) was similar to U.S. turkeys at the farm level [6, 35], Belgian turkey at slaughter [32] and retail turkey in Germany [33].
Resistance to multiple antimicrobial agents was observed in most of the Campylobacter isolates selected for molecular profiling (Figures 2 and 4). Most isolates were resistant to 3 or 4 agents. The most frequent resistance profile observed among C. jejuni isolates was to ciprofloxacin, nalidixic acid, and tetracycline. This profile was also reported as the most common multidrug resistance pattern for human Campylobacter isolates received through NARMS from 1997-2001 [13]. In this study, the most common multiple resistance pattern among C. coli isolated from turkey was resistance to ciprofloxacin, nalidixic acid, kanamycin, and tetracycline. These findings differ from reports by Lee et al. [36] and Luangtongkum et al. [6], where resistance profiles of ciprofloxacin, nalidixic acid, erythromycin, streptomycin, kanamycin, and tetracycline resistance predominated in C. coli from turkeys.
In addition to expanded antimicrobial resistance testing, fla typing and PFGE were used to further characterize antimicrobial-resistant C. jejuni and C. coli from processed turkey. It was observed that most of the Campylobacter isolates with identical fla-PFGE types had the same antimicrobial resistance profiles, a finding also noted by Ge et al. using PFGE [30]; however, analysis of additional antimicrobial-sensitive strains would be indicated. For subtyping C. jejuni and C. coli in this study, the greatest discrimination index was obtained using fla-PFGE together. Similarly, Nayak et al. [35] found a combination of subtyping methods for Campylobacter isolated from turkey farms had a greater discriminatory value than a single method. In the current study, fla typing failed to distinguish completely between the two Campylobacter species, a finding also noted by others [37–39]. In contrast, PFGE showed greater discrimination in separating the two species, which can be attributed to its ability to detect whole genome restriction site polymorphisms [29]. In addition to discriminatory value, other characteristics of these molecular typing methods should be acknowledged, which have been reviewed elsewhere [28, 29, 37, 40, 41]. Fla typing is a useful tool for subtyping campylobacters [39, 42], and has the advantages of being simple, quick, and low cost [28, 29, 42]. Nayak et al. reported that fla typing was more suitable than PFGE for typing C. coli isolated from turkey farms [35]. However, the potential for recombination within the fla genes is a drawback of using fla typing alone or for long-term studies [29, 43]. For this reason, and because fla typing is generally less discriminatory than PFGE, it is recommended to use fla typing in conjunction with other typing methods [29, 41]. PFGE is highly discriminatory and well-accepted for typing campylobacters, although it is laborious and can be expensive [29, 37]. PFGE profiles may also be affected by genetic instability in Campylobacter [28, 29].
In this study, the genetic diversity of antimicrobial-resistant strains varied between C. coli and C. jejuni. One fla-PFGE type (I3) contained 29% of the C. jejuni isolates while the remaining 16 fla-PFGE types contained one to three isolates each. In contrast, most of the C. coli isolates (62%) were grouped into only three fla-PFGE types, suggesting less diversity among C. coli. Bae et al. [44] demonstrated that PFGE types of antimicrobial-resistant C. coli from cattle were less diverse than those of C. jejuni, and Nayak et al. [35] reported a similar effect among antimicrobial-resistant C. coli and C. jejuni from turkey farms. Wesley et al. [7] described the opposite case, that C. coli from turkeys were more diverse than C. jejuni based on PFGE, although antimicrobial resistance was not determined.
The Campylobacter isolates examined in this study originated from turkey carcasses at either the pre or post chill stages of processing. The prevalence of ciprofloxacin or erythromycin resistance was similar from either stage in plant A. In contrast, Berrang et al. found that the numbers of erythromycin-resistant C. jejuni on broiler carcasses were reduced after chilling, and suggested further study to determine whether this resistance influences the ability of Campylobacter to endure immersion chilling [45]. In the current study, several of the same fla-PFGE types were recovered from both stages, indicating that some ciprofloxacin- and/or erythromycin-resistant strains were present beyond chilling. Information about antimicrobial-resistant Campylobacter on post-chill turkey product is limited and further study is needed.
Most of the fla-PFGE types (36 of 37) in the current study were unique to a particular plant. Similarly, Rasschaert et al. [46] demonstrated that most fla-PFGE types obtained from broilers at three processing plants were unique within a particular plant. The two plants participating in the current study were located approximately 150 miles apart in different states and were not likely to receive turkeys from the same farms. Isolation of the same fla-PFGE type (M10) from both plants may suggest a common source of this type, and warrants further investigation. However, it must be noted that the isolates subtyped for this study comprised a small portion of the entire Campylobacter collection (n = 801) tested, which may influence the frequency of fla-PFGE types obtained and is a limitation of our study.
Clustering using PFGE alone or fla-PFGE in conjunction with resistance profiles separated C. jejuni and C. coli into different groups. The diversity of genetic profiles, in conjunction with differences in resistance profiles by species, further supports the importance of considering C. jejuni and C. coli separately in epidemiological investigations [7, 30, 47, 48]. Although C. jejuni is implicated in most campylobacteriosis cases, human illness attributed to C. coli is also recognized [13, 47, 49, 50]. C. coli is often associated with pigs; but was prevalent in turkeys in our previous study [8] and those of others [7, 51]. In Denmark, poultry, but not pigs, were associated with human C. coli infections [48].
Conclusion
This study found that ciprofloxacin and erythromycin resistance was present in Campylobacter recovered from processed turkey in the Upper Midwest, and the prevalence differed significantly between C. jejuni and C. coli. Resistance observed in these strains has the potential to complicate the effectiveness of treatment for poultry-acquired Campylobacter infections in humans should they remain on the processed product. Molecular subtyping using fla typing and PFGE provided additional information on antimicrobial-resistant Campylobacter from processed turkey. Fla-PFGE types were relatively diverse and associated with a specific plant and species. Some ciprofloxacin and/or erythromycin resistant isolates with the same fla-PFGE types were recovered from processing both before and after chilling. Factors contributing to the occurrence of antimicrobial-resistant Campylobacter in processed turkey warrant further investigation.
Methods
Campylobacter isolates
Campylobacter isolates in this study (n = 801, Table 2) were obtained from two unrelated Midwestern processing plants (A and B) prior to the FDA ban of enrofloxacin use in poultry [8]. Plant A received turkeys from independent producers belonging to a farmers' cooperative, while plant B received turkeys from producers under contract with a large turkey processing company. Isolates were recovered and identified by Logue et al. as previously described [8]. Briefly, isolates were recovered from whole carcass swabs collected from randomly selected carcasses at two points on the processing line: pre chill and post chill, from plants visited monthly over a period of 12 months [8]. Samples of the chill water were also collected. Birds sampled on a single day were usually from one supplier or farm. Throughout all parts of the study, isolates were removed from -80°C storage in Brucella broth (Becton Dickinson, Cockeysville, Md.) with 20% glycerol and cultured onto sheep blood agar (BBL Prepared Media Trypticase Soy Agar II, 5% Sheep Blood; Becton Dickinson, Sparks, Md.). All cultures were incubated in a microaerobic environment of approximately 14% CO2 and 6% O2 generated by Pack-Micro Aero (Mitsubishi Gas Chemical, New York, N.Y.).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing on all isolates (n = 801) was conducted using the agar dilution method [52, 53] with testing ranges of 0.008-4 μg/ml for ciprofloxacin (Serologicals Proteins, Kankakee, Ill.) and 0.06-32 μg/ml for erythromycin (Sigma Chemical, St. Louis, Mo.). C. jejuni ATCC #33560 was used as a quality control strain [11, 53]. Resistance breakpoints were ≥ 4 μg/ml for ciprofloxacin and ≥ 32 μg/ml for erythromycin [54]. Isolates (n = 241) with an MIC of > 4 μg/ml for ciprofloxacin and/or an MIC of > 32 μg/ml for erythromycin were re-tested with extended antimicrobial concentrations of 0.5-32 μg/ml for ciprofloxacin and 2.0-128 μg/ml for erythromycin.
One hundred isolates (n = 51, plant A and n = 49, plant B) were selected for further characterization. A similar number of isolates from each plant and processing stage were included in the subset, comprised of 58 ciprofloxacin-resistant isolates, 22 erythromycin-resistant isolates, 10 co-resistant isolates, and 10 isolates sensitive to these two antimicrobials. C. jejuni and C. coli species identification was confirmed using multiplex PCR as described previously [55]. Testing for susceptibility against tetracycline, streptomycin, kanamycin and nalidixic acid was conducted using the agar dilution method [52, 53]. The test ranges used were 0.06-32 μg/ml for tetracycline (Sigma), 0.125-64 μg/ml for streptomycin (Sigma) and kanamycin (Amresco, Solon, Ohio), and 0.25-128 μg/ml for nalidixic acid (Sigma). The quality control strain used was C. jejuni ATCC #33560 [11, 53]. For streptomycin and kanamycin testing, Escherichia coli ATCC #25922 and C. jejuni ATCC #33560 were included. Campylobacter isolates were defined as resistant or sensitive based on breakpoints of ≥ 16 μg/ml for tetracycline, ≥ 64 μg/ml for nalidixic acid, and ≥ 64 μg/ml for streptomycin and kanamycin [54, 56].
Flatyping
Fla typing (n = 100) was carried out using the method of Nachamkin et al. [57] with minor modifications. Whole cell lysate [58] was used as the template. PCR amplification was performed in a Mastercycler gradient 5331 thermocycler (Eppendorf, Hamburg, Germany). C. jejuni ATCC #700819 was used as the positive control, and sterile water was substituted for the DNA template as the negative control. To confirm the presence of the 1.7 kb flaA amplicon, 10 μl of the PCR product was subjected to gel electrophoresis followed by ethidium bromide staining and UV transillumination. DdeI (Promega, Madison, Wis.) was used to digest 5 μl of the flaA PCR product according to the manufacturer's instructions at 37°C for 12-16 h overnight. Digested samples were electrophoresed on a 2% agarose gel, followed by staining in 0.5 μg/ml ethidium bromide solution and UV transillumination. A 100 bp ladder (Promega) was used as a molecular size standard.
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) was performed using the PulseNet method [59] with slight modifications. Salmonella enterica serotype Braenderup H9812 (ATCC #BAA-664) was used as the molecular weight size standard. Restriction digestion of each sample plug slice was carried out in a 100 μl mixture containing 85 μl sterile water, 10 μl 10× J buffer, 4 μl of 10 U/μl SmaI (Promega), and 1 μl BSA at 25°C for 3 h. Electrophoresis was performed using the Chef Mapper system (Bio-Rad, Hercules, Calif.) and the following conditions: auto algorithm function (50 kb low molecular weight and 400 kb high molecular weight), run time 18 h, initial switch time 6.76 s and final switch time 38.35 s. Gels were stained with 1 μg/ml ethidium bromide solution for 30 min, destained in 500 ml reagent grade water for 60-90 min with water changes every 20 min, and viewed under UV transillumination.
Documentation and analysis of flatyping and PFGE patterns
Gels were photographed and recorded as digital TIFF images using an Alpha-Innotech imager (Alpha Innotech, San Leandro, Calif.). Images were analyzed with Fingerprinting II Informatix software (Version 3.0, Bio-Rad). Band matching and cluster analysis was performed using an unweighted pair group method with arithmetic averages (UPGMA) and the Dice coefficient with 1% optimization and tolerance levels. Based on the dendrogram obtained from the cluster analysis, letters were assigned to designate fla types and numbers were assigned to designate PFGE types. Isolates with > 90% similarity were assigned to the same fla type or PFGE type. Composite cluster analysis including fla typing, PFGE, and antimicrobial resistance testing data was performed using the Fingerprinting II Informatix software. The composite dendrogram was determined by UPGMA using the average from the experiment as a coefficient for similarity and correction for internal weights.
Statistical analysis
The χ2 test was used to analyze the significance of the difference between ciprofloxacin and erythromycin resistance rates, including C. jejuni compared to C. coli in each plant, and pre chill compared to post chill in plant A. An α of 0.01 was used for statistical significance.
The discriminatory ability of fla typing, PFGE, antimicrobial resistance profiling, and composite analysis was calculated using the numerical index of discrimination (D) according to the method of Hunter and Gaston [60]. The discriminatory index represents the probability that two unrelated strains sampled from the test population will be placed into different typing groups [60].
Abbreviations
- C:
-
ciprofloxacin
- D :
-
diversity index
- E:
-
erythromycin
- FDA:
-
Food and Drug Administration
- Fla:
-
flagellin
- K:
-
kanamycin
- MIC:
-
minimum inhibitory concentration
- N:
-
nalidixic acid
- NARMS:
-
National Antimicrobial Resistance Monitoring System
- PFGE:
-
pulsed-field gel electrophoresis
- S:
-
streptomycin
- T:
-
tetracycline.
References
Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV: Food-related illness and death in the United States. Emerg Infect Dis. 1999, 5: 607-625. 10.3201/eid0505.990502.
Butzler JP: Campylobacter, from obscurity to celebrity. Clin Microbiol Infect. 2004, 10: 868-876. 10.1111/j.1469-0691.2004.00983.x.
Allos BM: Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001, 32: 1201-1206. 10.1086/319760.
Jacobs-Reitsma W: Campylobacter in the food supply. Campylobacter American Society for Microbiology, Washington, D.C. Edited by: Nachamkin I, Blaser MJ. 2000, 467-481. 2
Cox NA, Stern NJ, Craven SE, Berrang ME, Musgrove MT: Prevalence of Campylobacter and Salmonella in the cecal droppings of turkeys during production. J Appl Poult Res. 2000, 9: 542-545.
Luangtongkum T, Morishita TY, Ison AJ, Huang S, McDermott PF, Zhang Q: Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl Environ Microbiol. 2006, 72: 3600-3607. 10.1128/AEM.72.5.3600-3607.2006.
Wesley IV, Muraoka WT, Trampel DW, Hurd HS: Effect of preslaughter events on prevalence of Campylobacter jejuni and Campylobacter coli in market-weight turkeys. Appl Environ Microbiol. 2005, 71: 2824-2831. 10.1128/AEM.71.6.2824-2831.2005.
Logue CM, Sherwood JS, Elijah LM, Olah PA, Dockter MR: The incidence of Campylobacter spp. on processed turkey from processing plants in the midwestern United States. J Appl Microbiol. 2003, 95: 234-241. 10.1046/j.1365-2672.2003.01969.x.
U.S. Food and Drug Administration/Center for Veterinary Medicine: National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Retail meat annual report, 2005. 2007, U.S. Food and Drug Administration, Rockville, MD
Zhao C, Ge B, De Villena J, Sudler R, Yeh E, Zhao S, White DG, Wagner D, Meng J: Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the greater Washington, D.C. area. Appl Environ Microbiol. 2001, 67: 5431-5436. 10.1128/AEM.67.12.5431-5436.2001.
McDermott PF, Bodeis SM, Aarestrup FM, Brown S, Traczewski M, Fedorka-Cray P, Wallace M, Critchley IA, Thornsberry C, Graff S, Flamm R, Beyer J, Shortridge D, Piddock LJ, Ricci V, Johnson MM, Jones RN, Reller B, Mirrett S, Aldrobi J, Rennie R, Brosnikoff C, Turnbull L, Stein G, Schooley S, Hanson RA, Walker RD: Development of a standardized susceptibility test for Campylobacter with quality-control ranges for ciprofloxacin, doxycycline, erythromycin, gentamicin, and meropenem. Microb Drug Resist. 2004, 10: 124-131. 10.1089/1076629041310064.
Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I: Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: Resistance mechanisms and trends in human isolates. Emerg Infect Dis. 2001, 7: 24-34. 10.3201/eid0701.010104.
Gupta A, Nelson JM, Barrett TJ, Tauxe RV, Rossiter SP, Friedman CR, Joyce KW, Smith KE, Jones TF, Hawkins MA, Shiferaw B, Beebe JL, Vugia DJ, Rabatsky-Ehr T, Benson JA, Root TP, Angulo FJ: Antimicrobial resistance among Campylobacter strains, United States, 1997-2001. Emerg Infect Dis. 2004, 10: 1102-1109.
Hein I, Schneck C, Knogler M, Feierl G, Pless P, Kofer J, Achmann R, Wagner M: Campylobacter jejuni isolated from poultry and humans in Styria, Austria: epidemiology and ciprofloxacin resistance. Epidemiol Infect. 2003, 130: 377-386.
Smith KE, Bender JB, Osterholm MT: Antimicrobial resistance in animals and relevance to human infections. Campylobacter, American Society for Microbiology, Washington, D.C. Edited by: Nachamkin I, Blaser MJ. 2000, 483-495. 2
Smith KE, Besser JM, Hedberg CW, Leano FT, Bender JB, Wicklund JH, Johnson BP, Moore KA, Osterholm MT: Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N Engl J Med. 1999, 340: 1525-1532. 10.1056/NEJM199905203402001.
Nelson JM, Chiller TM, Powers JH, Angulo FJ: Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin Infect Dis. 2007, 44: 977-980. 10.1086/512369.
Avrain L, Humbert F, L'Hospitalier R, Sanders P, Vernozy-Rozand C, Kempf I: Antimicrobial resistance in Campylobacter from broilers: association with production type and antimicrobial use. Vet Microbiol. 2003, 96: 267-276. 10.1016/j.vetmic.2003.07.001.
Bae W, Kaya KN, Hancock DD, Call DR, Park YH, Besser TE: Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington State. Appl Environ Microbiol. 2005, 71: 169-174. 10.1128/AEM.71.1.169-174.2005.
Gibreel A, Taylor DE: Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 2006, 58: 243-255. 10.1093/jac/dkl210.
Engberg J, Neimann J, Nielsen EM, Aarestrup FM, Fussing V: Quinolone-resistant Campylobacter infections in Denmark: risk factors and clinical consequences. Emerg Infect Dis. 2004, 10: 1056-1063.
Helms M, Simonsen J, Olsen KEP, Mølbak K: Adverse health events associated with antimicrobial drug resistance in Campylobacter species: a registry-based cohort study. J Infect Dis. 2005, 191: 1050-1055. 10.1086/428453.
Nelson JM, Smith KE, Vugia DJ, Rabatsky-Ehr T, Segler SD, Kassenborg HD, Zansky SM, Joyce K, Marano N, Hoekstra RM, Angulo FJ: Prolonged diarrhea due to ciprofloxacin-resistant Campylobacter infection. J Infect Dis. 2004, 190: 1150-1157. 10.1086/423282.
Wassenaar TM, Kist M, de Jong A: Re-analysis of the risks attributed to ciprofloxacin-resistant Campylobacter jejuni infections. Int J Antimicrob Agents. 2007, 30: 195-201. 10.1016/j.ijantimicag.2007.01.019.
Ge B, McDermott PF, White DG, Meng J: Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2005, 49: 3347-3354. 10.1128/AAC.49.8.3347-3354.2005.
Lin J, Yan M, Sahin O, Pereira S, Chang Y-J, Zhang Q: Effect of macrolide usage on emergence of erythromycin-resistant Campylobacter isolates in chickens. Antimicrob Agents Chemother. 2007, 51 (5): 1678-1686. 10.1128/AAC.01411-06.
Luo N, Sahin O, Lin J, Michel LO, Zhang Q: In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother. 2003, 47: 390-394. 10.1128/AAC.47.1.390-394.2003.
Fitzgerald C, Stanley K, Andrew S, Jones K: Use of pulsed-field gel electrophoresis and flagellin gene typing in identifying clonal groups of Campylobacter jejuni and Campylobacter coli in farm and clinical environments. Appl Environ Microbiol. 2001, 67: 1429-1436. 10.1128/AEM.67.4.1429-1436.2001.
Newell DG, Frost JA, Duim B, Wagenaar JA, Madden RH, Plas van der J, On SLW: New developments in the subtyping of Campylobacter species. Campylobacter, American Society for Microbiology, Washington, D.C. Edited by: Nachamkin I, Blaser MJ. 2000, 27-44. 2
Ge B, White DG, McDermott PF, Girard W, Zhao S, Hubert S, Meng J: Antimicrobial-resistant Campylobacter species from retail raw meats. Appl Environ Microbiol. 2003, 69: 3005-3007. 10.1128/AEM.69.5.3005-3007.2003.
Larkin C, van Donkersgoed C, Mahdi A, Johnson P, McNab B, Odumeru J: Antibiotic resistance of Campylobacter jejuni and Campylobacter coli isolated from hog, beef, and chicken carcass samples from provincially inspected abattoirs in Ontario. J Food Prot. 2006, 69: 22-26.
Van Looveren M, Daube G, De Zutter L, Dumont J-M, Lammens C, Wijdooghe M, Vandamme P, Jouret M, Cornelis M, Goossens H: Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J Antimicrob Chemother. 2001, 48: 235-240. 10.1093/jac/48.2.235.
Luber P, Wagner J, Hahn H, Bartelt E: Antimicrobial resistance in Campylobacter jejuni and Campylobacter coli strains isolated in 1991 and 2001-2002 from poultry and humans in Berlin, Germany. Antimicrob Agents Chemother. 2003, 47: 3825-3830. 10.1128/AAC.47.12.3825-3830.2003.
Bywater R, Deluyker H, Deroover E, de Jong A, Marion H, McConville M, Rowan T, Shryock T, Shuster D, Thomas V, Vallé M, Walters J: A European survey of antimicrobial susceptibility among zoonotic and commensal bacteria isolated from food-producing animals. J Antimicrob Chemother. 2004, 54: 744-754. 10.1093/jac/dkh422.
Nayak R, Stewart T, Nawaz M, Cerniglia C: In vitro antimicrobial susceptibility, genetic diversity and prevalence of UDP-glucose 4-epimerase (galE) gene in Campylobacter coli and Campylobacter jejuni from turkey production facilities. Food Microbiol. 2006, 23: 379-392. 10.1016/j.fm.2005.04.007.
Lee BC, Reimers N, Barnes HJ, D'lima C, Carver D, Kathariou S: Strain persistence and fluctuation of multiple-antibiotic resistant Campylobacter coli colonizing turkeys over successive production cycles. Foodborne Pathog Dis. 2005, 2: 103-110. 10.1089/fpd.2005.2.103.
de Boer P, Duim B, Rigter A, Plas van der J, Jacobs-Reitsma WF, Wagenaar JA: Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 2000, 38: 1940-1946.
Ertaş HB, Çetinkaya B, Muz A, Öngör H: Genotyping of broiler-originated Campylobacter jejuni and Campylobacter coli isolates using fla typing and random amplified polymorphic DNA methods. Int J Food Microbiol. 2004, 94: 203-209. 10.1016/S0168-1605(03)00312-X.
Harrington CS, Moran L, Ridley AM, Newell DG, Madden RH: Inter-laboratory evaluation of three flagellin PCR/RFLP methods for typing Campylobacter jejuni and C. coli: The CampyNet experience. J Appl Microbiol. 2003, 95: 1321-1333. 10.1046/j.1365-2672.2003.02101.x.
Nielsen EM, Engberg J, Fussing V, Petersen L, Brogren C, On SLW: Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J Clin Microbiol. 2000, 38: 3800-3810.
Wassenaar TM, Newell DG: Genotyping of Campylobacter spp. Appl Environ Microbiol. 2000, 66: 1-9. 10.1128/AEM.66.1.1-9.2000.
VanWorth C, McCrea BA, Tonooka KH, Boggs CL, Schrader JS: Diversity of flaA genotypes among Campylobacter jejuni isolated from six niche-market poultry species at farm and processing. J Food Prot. 2006, 69: 299-307.
Harrington CS, Thomson-Carter FM, Carter PE: Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997, 35: 2386-2392.
Bae W, Hancock DD, Call DR, Park YH, Berge ACB, Finger RM, Sischo WM, Besser TE: Dissemination of antimicrobial resistant strains of Campylobacter coli and Campylobacter jejuni among cattle in Washington State and California. Vet Microbiol. 2007, 122: 306-315. 10.1016/j.vetmic.2007.01.020.
Berrang ME, Ladely SR, Meinersmann RJ, Fedorka-Cray PJ: Subtherapeutic tylosin phosphate in broiler feed affects Campylobacter on carcasses during processing. Poult Sci. 2007, 86: 1229-1233.
Rasschaert G, Houf K, van Hende J, de Zutter L: Campylobacter contamination during poultry slaughter in Belgium. J Food Prot. 2006, 69: 27-33.
Gillespie IA, O'Brien SJ, Frost JA, Adak GK, Horby P, Swan AV, Painter MJ, Neal KR: A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg Infect Dis. 2002, 8: 937-942.
Siemer BL, Nielsen EM, On SLW: Identification and molecular epidemiology of Campylobacter coli isolates from human gastroenteritis, food, and animal sources by amplified fragment length polymorphism analysis and Penner serotyping. Appl Environ Microbiol. 2005, 71: 1953-1958. 10.1128/AEM.71.4.1953-1958.2005.
Dasti JI, Groß U, Pohl S, Lugert R, Weig M, Schmidt-Ott R: Role of the plasmid-encoded tet (O) gene in tetracycline-resistant clinical isolates of Campylobacter jejuni and Campylobacter coli. J Med Microbiol. 2007, 56: 833-837. 10.1099/jmm.0.47103-0.
Tam CC, O'Brien SJ, Adak GK, Meakins SM, Frost JA: Campylobacter coli-- an important foodborne pathogen. J Infect. 2003, 47: 28-32. 10.1016/S0163-4453(03)00042-2.
Smith K, Reimers N, Barnes HJ, Lee BC, Siletzky R, Kathariou S: Campylobacter colonization of sibling turkey flocks reared under different management conditions. J Food Prot. 2004, 67: 1463-1468.
Ge B, Bodeis S, Walker RD, White DG, Zhao S, McDermott PF, Meng J: Comparison of the Etest and agar dilution for in vitro antimicrobial susceptibility testing of Campylobacter. J Antimicrob Chemother. 2002, 50: 487-494. 10.1093/jac/dkf162.
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A2. 2002, Clinical and Laboratory Standards Institute, Wayne, PA
Clinical and Laboratory Standards Institute. 2006: Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. Approved guideline M45-A. 2006, Clinical and Laboratory Standards Institute, Wayne, PA
Cloak OM, Fratamico PM: A multiplex polymerase chain reaction of the differentiation of Campylobacter jejuni and Campylobacter coli from a swine processing facility and characterization of isolates by pulsed-field gel electrophoresis and antibiotic resistance profiles. J Food Prot. 2002, 65: 266-273.
U.S. Food and Drug Administration/Center for Veterinary Medicine: National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Retail meat annual report, 2004. 2006, U.S. Food and Drug Administration, Rockville, MD.
Nachamkin I, Ung H, Patton CM: Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J Clin Microbiol. 1996, 34: 277-281.
Marmur J: A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961, 3: 208-218.
Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ: Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J Clin Microbiol. 2001, 39: 1889-1894. 10.1128/JCM.39.5.1889-1894.2001.
Hunter PR, Gaston MA: Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988, 26: 2465-2466.
Acknowledgements
The authors gratefully acknowledge the U.S. Food and Drug Administration for financial and technical assistance. We also thank Curt Doetkott, North Dakota State University (NDSU), for statistical consultation and Dr. Mohamed Fakhr, University of Tulsa, for assistance with data analysis and manuscript review.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
CML and JSS isolated and characterized the campylobacters recovered from poultry; EML carried out the antimicrobial resistance assays and molecular analysis; JMM carried out molecular and software analysis. All authors read and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lutgen, E.M., McEvoy, J.M., Sherwood, J.S. et al. Antimicrobial resistance profiling and molecular subtyping of Campylobacterspp. from processed turkey. BMC Microbiol 9, 203 (2009). https://doi.org/10.1186/1471-2180-9-203
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-9-203