Abstract
Background
Group A streptococcus (GAS) causes a wide variety of life threatening diseases in humans and the incidence of such infections is high in developing countries like India. Although distribution of emm types of GAS in India has been described, there is a lack of data describing either the comparative distribution of emm types in throat versus skin isolates, or the distribution of certain virulence factors amongst these isolates. Therefore in the present study we have monitored the emm type pattern of Group A streptococcus throat and skin isolates from India. Additionally, the association of these isolates with closely related sic (crs), a multifunctional compliment binding virulence factor, was also explored.
Results
Of the 94 (46 throat and 48 skin) isolates analyzed, 37 emm types were identified. The most frequently observed emm types were emm49 (8.5%) and emm112 (7.5%) followed by 6.5% each of emm1-2, emm75, emm77, and emm81. Out of 37 emm types, 27 have been previously reported and rest were isolated for the first time in the Indian Community. The predominant emm types of throat (emm49 and emm75) samples were different from those of skin (emm44, emm81 and emm112) samples. After screening all the 94 isolates, the crs gene was found in six emm1-2 (crs1-2) isolates, which was confirmed by DNA sequencing and expression analysis. Despite the polymorphic nature of crs, no intravariation was observed within crs1-2. However, insertions and deletions of highly variable sizes were noticed in comparison to CRS isolated from other emm types (emm1.0, emm57). CRS1-2 showed maximum homology with CRS57, but the genomic location of crs1-2 was found to be the same as that of sic1.0. Further, among crs positive isolates, speA was only present in skin samples thus suggesting possible role of speA in tissue tropism.
Conclusion
Despite the diversity in emm type pattern of throat and skin isolates, no significant association between emm type and source of isolation was observed. The finding that the crs gene is highly conserved even in two different variants of emm1-2 GAS (speA +ve and -ve) suggests a single allele of crs may be prevalent in the highly diverse throat and skin isolates of GAS in India.
Similar content being viewed by others
Background
Group A streptococcus (GAS, Streptococcus pyogenes) causes various diseases ranging from mild impetigo, pharyngitis and scarlet fever to more severe and serious sequelae such as rheumatic fever (RF), rheumatic heart disease (RHD) and acute glomerulonephritis [1]. The incidence of severe GAS diseases is high in children aged between 5–15 years and is more common in developing countries [2]. The prevalence of RF/RHD is known to vary from 0.3 to 5.4 children per 1000 in India [3].
Diversity in GAS strain is reflected not only among types of strains circulating in a particular community, but also the virulence factors associated with them [1]. GAS express a variety of virulence factors such as M protein, Streptolysin O and S, C5a peptidase, streptococcal pyrogenic exotoxins (Spe), streptococcal protective antigen (Spa) and streptococcal inhibitor of complement (SIC). Some of these virulence factors like Spa [4] or SIC [5] are found to be restricted in their distribution to specific emm types.
The complement binding protein, SIC, was first described by Akesson et al, 1996 [6] in M1 and later its variants were reported in M12, M55 and M57 [5, 7]. Originally, SIC was characterized as an inhibitor of complement function that interferes with the function of the membrane attack complex by binding to one or more protein components associated with the complex. Subsequently SIC has also been shown to inhibit antimicrobial activity of lysozyme, secretory leukocyte proteinase inhibitor (SLPI), α and β-defensins, and Cathelicidin LL-37, which are components of the innate immune system [6, 8, 9].
The gene encoding SIC (sic) is highly polymorphic, both between different emm types and within strains of the same emm type [10]. Two forms of this gene have been identified, the closely related sic gene (crs) present in emm1 and emm57, and distantly related sic gene (drs) isolated from emm12 and emm55 [5]. Despite previous reports suggesting crs is associated with only emm1 and emm57 GAS isolates, Ma et al (2002) reported the association of the crs gene with other emm types including emm2, emm4, emm12, emm28, emm75, emm89, emm94 and emm112, leading to question about the distribution of crs [11].
Information regarding the circulating emm type is available from community screening [12] as well as from hospital data [13, 14] in India. However, there is a lack of information regarding the distribution of emm types among strains isolated from different sites (throat and skin) and their virulence factors. Earlier, we have reported the presence of toxins [15] in GAS strains from our country and very recently, unraveled the conserved nature of other form of sic i.e. the drs gene [16]. Except for the presence of variable numbers of repeats, drs was found to be conserved not only within Indian isolates but also within the isolates from other countries. Here, we have explored for the first time the presence of the crs gene in emm1-2 isolates of Group A streptococcus from throat and skin infections. Although this study showed the conserved nature of the crs1-2 gene among Indian isolates, unlike drs, crs1-2 was found to be highly polymorphic when compared to isolates from other countries.
Results and discussion
Characterization of GAS strains by emmtyping
For the first time, the emm type distribution of both throat and skin isolates of Group A streptococcus from India was studied and compared. 94 isolates associated with either throat or skin were categorized into 37 different emm types (Table 1). The majority of throat isolates were of emm49 and emm75, whereas emm44, emm81 and emm112 were mostly associated with skin infection. Only eleven emm types were found to be common in both throat and skin isolates. The distribution of emm types among throat and skin samples was different to the study of McGregor et al (2004) which involved samples belonging to geographic regions far away from Indian subcontinent [17]. However our data is quite similar to reports from other Asian countries, such as Japan [18] and Nepal[19].
The most frequently observed emm types (Table 1) among all isolates in this study were emm49 (8.5%), emm112 (7.4%) followed by 6.3% of emm1-2, emm75, emm77 and emm81. These most frequent emm types were not only different from previously reported most prevalent emm types in India [12–14] but also from epidemiological studies of isolates from other countries like Japan [18], Taiwan [20], Germany [21], Australia [22] and United States [23]. The difference in the most prevalent emm types of this study in comparison to earlier Indian reports may be due to the fact that the most prevalent serotypes within a population changes over time, which can be predicted by continuing surveillance [24]. Additionally, in this study both throat and skin isolates were studied, whereas earlier studies, involved throat samples only. Out of 37 emm types, 27 have been described in earlier reports from India, where as 16 were identical to emm types reported from Hong Kong [25] and interestingly, 29 to emm types of Ethiopia [26]. Identification for the first time of new emm types associated with skin infections in the Indian community further justifies the inclusion of skin isolates in this study.
On the basis of differences in amino acid sequence of the test strain from the parent strain in the type specific region of the emm gene, 37 types were subdivided in to 38 subtypes [27]. This observation is in contrast to the report available from Mexico where 31 types were differentiated into 66 subtypes [28]. In this study, out of 37 types only six isolates of emm81 could be divided into two subtypes i.e 81.1(3) and 81.2(3) (Table 1).
This study involved a small sampling of isolates from one area of a highly diverse country. The diversity in emm types reflect the extent of heterogeneity which exists among the strains prevalent in India. Only 11 emm types of this study correspond to emm types used in multivalent vaccine that is under trial in the USA [23]. This supports the development of a multivalent vaccine specific for this particular region covering all emm types prevalent in the Indian community. However development of such strategies needs further investigation with more samples belonging to each part of India.
Screening of isolates for the crs (closely related sic) gene
To elude the host defense and establish infection, GAS produces a number of virulence factors, including streptococcal inhibitor of complement (SIC). The polymorphic extra cellular complement binding protein SIC has not only pathological but also epidemiological significance. We have studied the sic gene distribution by screening ninety four GAS isolates. The six emm1-2 isolates (Table 1) were positive for the presence of the crs gene (crs1-2) specific ~830-bp fragment (Fig 1A), while in Japan the sic gene was isolated from 10 different emm types [11]. Therefore, the isolation rate (6.5%) of the crs gene in this study was found to be less compared to a study (77.3%) made in Japan [11]. Our data reports for the first time the presence of the crs gene from emm type 1–2, which is a distinct type, not a subtype of emm1.0.
Characterization of CRS. (A) Screening of crs gene. Lanes: M, 100-bp Ladder (NEB, USA); 1, M1 reference strain used as positive control; 2–7, representative clinical emm1-2 isolates; (-), negative control (without template). B. Multiple sequence analysis of CRS1-2 from representative isolate, SIC1.0 (AP1 strain from Sweden) and CRS57 (reported from Australia). C. Western Blot analysis of CRS. Lanes: 1, M1 reference strain used as positive control; 2, negative control; 3, representative clinical emm1-2 isolate.
crsgene sequence and phylogenetic analysis
The crs gene product was further confirmed by sequence analysis using modified internal primers [29], which yielded a 912 bp full-length crs gene sequence (Accession number EF543156 – EF543161). The six crs positive isolates did not show intravariation either at the DNA or at the amino acid level. However, like sic1.0, crs1-2 also possessed a short repeat region (SRR), central long repeat region (LRR) and C proximal Proline rich region (PRR) as reported earlier [6]. A number of mutations such as insertions and deletions were observed in CRS1-2 throughout the sequence in comparison to SIC1.0 [6] and CRS57 [7]. Similar to SIC1.0, a deletion of five amino acids (GWSGD) was observed in CRS1-2 in comparison to CRS57 at position 40. However, an insertion of 27 amino acids (EWPEDDWSEDDWSNDYWSKYSWSSDKE) at position 82, similar to CRS57 in comparison to SIC1.0 has been noticed in CRS1-2. On the other hand a deletion of 29 amino acids (GALGTGYEKRDDWGGPGTVATDPYTPPYG) at position 165 makes CRS1-2 unique from both CRS1.0 and CRS57 (Fig 1B). Insertion and deletion sequences monitored in CRS1-2 in comparison to SIC1.0 and CRS57 is different from the 29 amino acid insertion (PPYGGALGTGYEKRDDWGGPGTVATDPYT) and the 31 amino acid deletion (GLSKYDRSGVGLSQYGWSQYGWSSDKEEWPE) sequence, most commonly observed in different alleles of sic1.0 [29]. It is likely, because this gene is under strong natural selection pressure [10], and as such harbors significant sequence variation and is highly divergent. The high number of allelic variations in sic is likely due to the fact that humans mount antibody response to SIC, a process that enhances variation by selecting escape mutants [30]. It is also reported that human anti-SIC antibodies are directed against virtually all regions of SIC that are highly polymorphic in natural population, which further strengthen the antibody mediated SIC diversification [31]. Phylogenetic analysis (Fig 1B) indicates the variant of SIC, CRS1-2, is more closely related to CRS57 [7] reported from Australia compare to SIC1.0 [6] reported from Sweden (Fig 1B). This correlation raises the possibility that crs57 may have originated from emm1-2 instead of emm1.0.
Expression of crsgene at protein level
The secreted proteins from all emm1-2 isolates were seen in SDS-PAGE (12%) and specific antisera was used for western blot analysis which confirmed the expression of CRS1-2 from all these isolates. All six emm1-2 isolates (3 throat and 3 skin) showed the CRS specific protein of 34-KDa similar to emm1.0 (Fig 1C).
Genomic location of crs1.2 gene
In emm1.0, the crs gene is located within the mga regulon whereas it is located outside the mga regulon in emm57. In this study a PCR based method [6] was applied to find the genomic location of crs1-2. The amplified product of size 1.2-kb and 2.2-kb obtained by using primer pair P1 – P2 and P3 – P4 [6] respectively indicated the genomic location of crs1-2 to be the same as for crs1.0 (Fig 2A &2B). An additional PCR based analysis, which showed a 400-bp fragment by using primer pair P5 – P6 [7] further confirmed this observation (Fig 2C). Since emm1.0 and emm1-2 shares majority of alleles, as shown by MLST analysis [17], therefore such similarity in the genomic location of crs1-2 and sic1.0 is not unexpected.
Genomic location estimation of crs 1-2. (A) 1.2-kb product amplification by primer pair P1 – P2, Lanes: M, 100-bp Ladder (NEB, USA); 1, M1 reference strain used as positive control; 2, representative clinical emm1-2 isolate; 3, sic negative strain; (-), negative control (without template) (B) 2.2-kb product amplification by primer pair P3–P4, Lanes: M, 1-kb Ladder (NEB, USA); 1, M1 reference strain used as positive control; 2, representative clinical emm1-2 isolate; 3, sic negative strain; (-), negative control(without template) (C) 400-bp product amplification by primer pair P5 – P6, Lanes: M, 100-bp Ladder (NEB, USA); 1, M1 reference strain used as positive control; 2, representative clinical emm1-2 isolate; 3, sic negative strain; (-), negative control (without template).
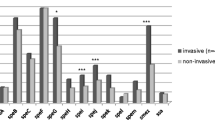
Screening of sic positive isolates for speA
To examine the presence of other virulence factors, sic positive isolates were screened for the phage encoded virulence factor speA, which is also known as a source of diversity in these GAS strains [10]. Association of the speA gene with sic positive isolates only from skin infection (Fig. 3) genetically differentiated them from throat isolates. This suggests the identification of two variants [10] of sic positive isolates, carrying the conserved crs gene. These two variants (spe A positive and negative), not only belong to different sources (throat and skin) but also to different regions of North India, and were also collected at different time periods.
Conclusion
In the present study, we found that, although emm type pattern among throat and skin isolates was different but no significant association between emm type and source (throat and skin) was observed. Out of 37 different emm types, only six emm1-2 isolates were positive for crs gene validating its highly restricted distribution. Although no intravariation was observed in the crs1-2 gene, a large number of allelic variations were observed in the crs1-2 gene in comparison to crs genes reported from other countries. This suggests the crs gene is highly divergent in comparison to drs. Moreover, variation in virulence characteristics like possession of speA in skin specific isolates not only differentiated emm1-2 isolates in two variants, but also reflects that virulence may be source specific, not type specific. The presence of conserved sic in these isolates further suggests a single allele of crs may be prevalent in the GAS isolates of Indian community, showing diverse emm type distribution in throat versus skin isolates.
Methods
Bacterial Strains
Group A streptococcus isolates (94) from throat (46 cases, comprising 36 from pharyngitis, nine RF/RHD and one Chorea) and Skin (48), were already available in the Department of Experimental Medicine and Biotechnology, Postgraduate Institute of Medical Education and Research, Chandigarh (India). Skin samples used in this study were collected during year 2000 – 2004 after obtaining ethical clearance from Institute ethics committee, Postgraduate Institute of Medical Education and Research, Chandigarh, from patients presenting with any suppurative skin lesion, wound, burn or rectum infection. However the throat isolates were collected from the throat of symptomatic patients (Pharyngitis, RF/RHD and Chorea) between the years 1995 and 2004. All these isolates were collected from hospital (Postgraduate Institute of Medical Education and Research, Chandigarh) as well as community screening (rural and urban slum) near Chandigarh after getting consent from parents.
emmtyping
emm gene sequencing was performed as previously reported [12]. DNA sequences were subjected to homology search against the bacterial DNA database http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm. Pairwise comparison of the nucleotide homology for the first 160 bases of the hyper variable region of the emm gene was conducted to designate emm type to a particular strain. Types and subtypes were designated as described earlier http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm.
Identification and Sequence analysis of crs
Published primers [5] were used for the screening of the sic (crs) gene in GAS isolates. The complete crs gene was amplified and sequenced by using a different set of published primers [29]. A Specific internal primer was designed (ACCTAAGACCGAACAATCACCA) for crs1-2 sequencing. Sequencing was carried out in an Automated DNA sequencer, model number 310, Applied Biosystems, USA. Sequence data was compared with those already deposited in the Data bank by using clustal X program [32].
Western blot analysis of CRS protein
GAS cultures were grown for 8 hrs and centrifuged at 10,000 × g for 10 mins. Supernatant proteins were precipitated with trichloroacetic acid (10% final concentration) at -20°C for approximately 20 mins. To retrieve the precipitated proteins the mixture was centrifuged at 16000 × g for 20 mins. The supernatant was discarded and the pellet was resuspended in 0.1 M NaOH. After running SDS-PAGE (12%), CRS protein was identified by using a specific antibody as described previously [7].
crsgene location in genome
The location of the crs gene on the mga regulon was mapped by PCR using primer pairs (P1 – P2, P3 – P4 and P5 – P6) that were designed from sequence flanking the crs gene [6, 7].
Screening of speA
sic positive isolates were further screened for the spe A gene as reported earlier [15].
Abbreviations
- SIC:
-
Streptococcal inhibitor of complement
- CRS:
-
closely related SIC
- DRS:
-
distantly related SIC
- RF/RHD:
-
Rheumatic fever/rheumatic heart disease.
References
Cunningham MW: Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000, 13: 470-511.
Bisno AL: Group A streptococcal infections and acute rheumatic fever. N Engl J Med. 1991, 325: 783-793.
Padmavati S: Present status of rheumatic fever and rheumatic heart disease. India. Indian Heart J. 1995, 47 (4): 395-398.
Dale JB, Chiang YC, Liu SY, Courtney HS, Hasty DL: New protective antigen of group A streptococci. J Clin Investig. 1999, 103: 1261-1268.
Hartas J, Sriprakash KS: Streptococcus pyogenes strains containing emm12 and emm55 possess a novel gene coding for distantly related SIC protein. Microb Pathog. 1999, 26 (1): 25-33.
Akesson P, Sjoholm AG, Bjorck L: Protein SIC, a novel extacellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996, 271 (2): 1081-1088.
Binks M, McMillan D, Sriprakash KS: Genomic location and variation of the gene for CRS, a complement binding protein in the M57 strains of Streptococcus pyogenes. Infect Immun. 2003, 71: 6701-6706.
Fenie-king BA, Seilly DJ, Davies A, Lachmann PJ: Streptococcal inhibitor of complement inhibits two additional components of the mucosal innate immune system: secretory leukocyte proteinase inhibitor and lysozyme. Infect Immun. 2002, 70: 4908-4916.
Frick IM, Akesson P, Rasmussen M, Schimdtchen A, Bjorck L: SIC, a Secreted Protein of Streptococcus pyogenes That Inactivates Antibacterial Peptides. J Biol Chem. 2003, 278: 16561-16566.
Hoe N, Nakashima K, Grigsby D, Pan X, Dou SJ, Niadich S, Gracia M, Kahn E, Bergmire-Sweat D, Musser JM: Rapid molecular genetic subtyping of serotype M1 group A streptococcus strain. Emerg Infect Dis. 1999, 5 (2): 254-263.
Ma X, Kikuta H, Ishiguro N, Yoshika M, Ebihara T, Murai T, Kobayanshi I, Kobayanshi K: Association of the prtf1 gene (Encoding Fibronectin Binding Protein F1) and sic gene (Encoding Streptococcal Inhibitor of Complement) with emm types of Group A Streptococci isolated from Japanese children with pharyngitis. J Clin Microbiol. 2002, 40: 3835-3837.
Sagar V, Bakshi DK, Nandi S, Ganguly NK, Kumar R, Chakraborti A: Molecular Heterogeniety among north Indian isolates of Goup A Streptococcus. Lett Appl Microbiol. 2004, 39 (1): 84-88.
Dey N, Mcmillan DJ, Yarwood PJ, Joshi RM, Kumar R, Good MF, Sriprakash KS, Vohra H: High diversity of Group A streptococcal emm types in an Indian Community: The Need to Tailor Multivalent vaccines. Clin Infect Dis. 2005, 40 (1): 46-51.
Menon T, Whatmore AM, Srivani S, Kumar MP, Anbumani N, Rajaji S: emm types of Streptococcus pyogenes in Chennai. Indian J Med Microbiol. 2001, 19 (3): 161-162.
Nandi S, Chakraborti A, Bakshi DK, Rani A, Kumar R, Ganguly NK: Association of pyogenic exotoxins genes with pharyngitis and rheumatic fever\rheumatic heart disease among Indian isolates of Streptococcus pyogenes. Lett Appl Microbiol. 2002, 35 (3): 237-41.
Sagar V, Kumar R, Ganguly NK, Menon T, Chakraborti A: Distantly related sic (drs) is less divergent then SIC. J Bacteriol. 2007, 189: 2933-2935.
MacGregor KF, Pratt BG, Kalia A, Bennett A, Bilek N, Beall B, Bessen DE: Multilocus sequence typing of Streptococcs pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J Bacteriol. 2004, 186: 4285-4294.
Tanaka D, Gyobu Y, Kodama H, Isobe J, Hosorogi S, Hiramoto Y, Karasawa T, Nakamura S: emm typing of Group A streptococcus clinical isolates: identification of dominant types for throat and skin isolates. Microbiol Immunol. 2002, 46: 419-423.
Sakota V, Fry AM, Lietman TM, Facklam RR, Li Z, Beall B: Genetically diverse group A streptococci from children in far-western Nepal share high genetic relatedness with isolates from other countries. J Clin Microbiol. 2006, 44: 2160-2166.
Kao CH, Chen PY, Huang FL, Chen CW, Chi CS, Lin YH, Shih CY, Hu BS, Li CR, Ma JS, Lau YJ, Lu KC, Yu HW: Clinical and genetic analysis of invasive and noninvasive group A streptococcal infections in central Taiwan. J Microbiol Immunol Infect. 2005, 38: 105-111.
Brandt CM, Spellerberg B, Honscha M, Truong ND, Hoevener B, Lutticken R: Typing of streptococcus pyogenes strains isolated from throat infections in the region of Achen, Germany. Infection. 2001, 29: 163-165.
McGregor KF, Bilek N, Bennett A, Kalia A, Beall B, Carapetis JR, Currie BJ, Sriprakash KS, Spratt BG, Bessen DE: Group A streptococcus streptococci from remote community have novel multilocus genotypes but share emm types and Housekeeping alleles with isolates from world wide sources. J Infect Dis. 2004, 189: 717-723.
Hu MC, Walls MA, Stroop SD, Reddish MA, Bernard B, Dale JB: Immunogenicity of 26-valent group A streptococcal vaccine. Infec Immun. 2002, 70: 2171-2177.
Dale JB, Shulman ST: Dynamic epidemiology of group A streptococcal serotypes. Lancet. 2002, 359: 889
Ho PL, Johnson DR, Yue AWY, Tsang DNC, Que TL, Beall B, Kaplan EL: Epidemiological analysis of invasive and noninvasive group A streptococcus isolates in Hong Kong. J Clin Microbiol. 2003, 41: 937-942.
Tewodros W, Kronvall G: M protein gene (emm type) analysis of group A beta hemolytic streptococci from Ethiopia reveals unique pattern. J Clin Microbiol. 2005, 43: 4369-4376.
Li Z, Sakota V, Jackson D, Franklin AR, Beall B: Array of M protein gene subtypes in 1064 recent invasive group A streptococcus isolates recoverd from active bacterial core surveillance. J Infect Dis. 2003, 188 (10): 1587-1592.
Espinosa LE, Li Z, Barreto DG, Jaimes EC, Rodriguez RS, Sakota V, Facklam RR, Beall B: M protein gene type distribution among group A streptococcal clinical isolates recovered in Mexico city, from 1991 to and Durango, Mexico from 1998 to 1999: overlap with type distribution within the United States. J Clin Microbiol. 2000, 41: 373-378.
Mejia L, Stockbauer MP, Pan XI, Cravioto A, Musser JM: Characterization of Group A streptococcus strains recovered from Maxican children with pharyngitis by automated DNA sequencing of virulence related genes: unexpectedly large variation in the gene (sic) encoding a complement inhibiting protein. J Clin Microbiol. 1997, 35: 3220-3224.
Stockbauer KE, Grigsby D, Pan X, Fu YX, Mejia LMG, Cravioto A, Musser JM: Hypervariability generated by natural selection in an extracellular complement inhibiting protein of serotype M1 strains of group A Streptococcus. Proc Natl Acad Sci. 1998, 95: 3128-3133.
Hoe NP, Kordari P, Cole R, Liu M, Palzkill T, Huang W, McLellan D, Adams GJ, Hu M, Vuopio-Varkila J, Cate TR, Pichichero ME, Edwards KM, Eskola J, Low DE, Musser JM: Human immune response to Streptococcal Inhibitor of Complement, a serotype M1 Group A Streptococcus extracellular protein involved in epidemics. J Infect Dis. 2000, 182: 1425-36.
Higgins DG, Thompson JD, Gibson TJ: Using CLUSTAL for multiple sequences alignments. Methods in Enzymology. 1996, 266: 383-402.
Acknowledgements
The financial support from ICMR, New Delhi, India is kindly acknowledged. We thank Dr Nancy Hoe, NIH, USA for providing us CRS specific antibody and Dr Martina Sanderson-Smith, University of Wollongong, AUS for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
All the authors have gone through the final manuscript. This work was a part of the Ph.D thesis of VS, done under supervision of AC (laboratory study) and RK (Field study). NKG has critically evaluated the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Sagar, V., Kumar, R., Ganguly, N.K. et al. Comparative analysis of emmtype pattern of Group A Streptococcus throat and skin isolates from India and their association with closely related SIC, a streptococcal virulence factor. BMC Microbiol 8, 150 (2008). https://doi.org/10.1186/1471-2180-8-150
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-8-150