Abstract
Background
An extremely low level methicillin resistant Staphylococcus aureus (MRSA) belonging to ST45, circulates among intravenous drug users in the Zurich area. This clone can be misinterpreted as an MSSA by phenotypic oxacillin resistance tests, although it carries a staphylococcal cassette chromosome mec (SCCmec) element encoding a functional mecA gene and it produces PBP2a.
Results
This clone carried a new 45.7-kb element, termed SCCmecN1, containing a class B mec complex (mecA- ΔmecR1::IS1272), a truncated Tn4003 harbouring the dfrA gene, and a fusB1 gene, conferring methicillin, trimethoprim and low level fusidic acid resistance, respectively. In addition to the two insertion site sequences (ISS) framing the SCCmec, a third ISS (ISS*) was identified within the element. SCCmecN1 also harboured two distinct ccrAB complexes belonging to the class 4 subtype, both of which were shown to be active and to be able to excise the SCCmecN1 or parts thereof. Slight variations in the SmaI-PFGE pattern of the clinical MRSA isolates belonging to this clone were traced back to differences in the sizes of the SCCmec J2 regions and/or to a 6.4-kb deletion extending from ISS* to the right end ISS. This latter deletion led to a variant right SCCmec-chromosomal junction site. MRSA clones carrying the shorter SCCmec with the 6.4-kb deletion were usually ciprofloxacin resistant, while strains with the complete SCCmecN1 were co-trimoxazole resistant or had no additional resistances. This suggested that the genetic backbone of the host S. aureus, although identical by PFGE pattern, had at some stage diverged with one branch acquiring a sulfonomide resistance mutation and the other ciprofloxacin resistance.
Conclusion
This description of the structure and variations of SCCmecN1 will allow for quicker and easier molecular detection of this clone and monitoring of its spread.
Similar content being viewed by others
Background
Injection drug user (IDU) populations throughout certain areas of Europe and North America have become major risk groups associated with the epidemic spread of methicillin-resistant Staphylococcus aureus (MRSA) [1–5]. The transmission of MRSA clones through both community- and healthcare-associated routes is responsible for the high incidence of soft tissue infections and increases in severe infections such as endocarditis and bacteremia in IDUs [6–8]. Such a clonal dispersal led to MRSA becoming endemic in the Zurich IDU population, where in 2003 24% of all MRSA isolates collected at the University hospital of Zurich belonged to a single, so called "drug clone" [9]. Dissemination of this clone to IDU populations in other, geographically distinct regions of Switzerland has also been recently reported, indicating that it has a capacity for spreading and colonizing new populations [10].
Clinical detection of MRSA can be complicated due to vast strain-to-strain differences in the expression of methicillin resistance. Difficulties arise from strains expressing low-level but heterogenous resistance, that upon exposure to β-lactams segregate highly resistant subpopulations resulting in therapy failure [11]. Misdiagnosis of such strains with very low, phenotypically susceptible, minimum inhibitory concentrations (MICs) is a major problem necessitating the use of molecular detection methods.
Epidemiological classification of MRSA strains is important for monitoring their prevalence and spread, and relies on molecular typing of both their core genomic background and the type of staphylococcal cassette chromosome mec (SCCmec) they harbour. SCCmec is the chromosomally integrated resistance element which carries the mecA gene, encoding the alternate penicillin-binding protein PBP2a, which confers methicillin resistance. There are currently six main types of SCCmec, differentiated according to their combinations of mec complex, containing the mecA gene and various portions of its regulatory genes mecR1 and mecI, and ccr complex containing recombinases specific for the chromosomal integration and excision of the SCCmec. Further sub-typing is based on the presence of certain additional genes or resistance determinants within the J (so called junkyard) regions J1, J2 and J3 of the element [12]. A number of non mecA-encoding SCC elements, sharing some common features with various SCCmec s, have also been discovered in methicillin sensitive Staphylococcus aureus (MSSA) or coagulase-negative staphylococcal strains [13–18].
Identification of the Zurich drug clone was based on a characteristic pulsed field gel SmaI restriction pattern and the presence of a unique, previously uncharacterised SCCmec element which was termed SCCmecN1. In addition to methicillin resistance, all drug clone isolates were shown to be resistant to trimethoprim and most were resistant to sulfamethoxazole or to ciprofloxacin. MLST typing revealed that the representative isolate of this clone, MRSA CHE482, belonged to sequence type ST45, a genotype that has been associated with epidemic MSSA and low level oxacillin resistant MRSA, which seem to have high colonization and circulation capacities [19].
All the drug clone isolates have low oxacillin resistance levels, with MICs between 0.5 and 4 μg ml-1, which can make them difficult to detect by phenotypic tests. Except for the detection of the mecA gene, genotypic tests, which rely on identifying known features of SCCmec s [20, 21] or SCCmec-chromosomal junctions [22] (X. Schneider, unpublished), have also failed to identify this clone [9].
To facilitate accurate molecular identification of this clone this manuscript presents a detailed description of the novel SCCmecN1 and describes the SCCmec variability observed so far between different isolates.
Results and Discussion
Mapping of SCCmecN1
The size of SCCmecN1 in CHE482 was estimated to be 45.7-kb, based on a series of overlapping long range PCR products amplified with primers shown in Figure 1 and listed in Table 1. This is larger than the community-associated SCCmec type IV (21–25-kb), type V (27.6-kb) and type VI (approximately 22-kb) elements, falling within the range of the classical hospital associated SCCmec types I-III which range in size from 34–67 kb [23]. Loci of interest within SCCmecN1 were then further mapped and sequenced.
Schematic organisation of the SCCmecN1 of strain CHE482. Primers are indicated by black arrows. Regions coloured in blue represent the core chromosome. Red stars indicate the excision sites of the partially cured strain CHE482Δ. Orange arrows indicate the ISS sites and the internal ISS*. The ccrAB4-1 and ccrAB4-2 complexes are highlighted in light and dark green, respectively. The truncated Tn4003 in orange comprises IS431-orf140-dfrA-thyE-IS431. The class B mec complex in yellow includes mecA- ΔmecR1-IS1272. The fusidic acid resistance gene fusB1 is shown in pink. Dotted lines indicate regions of variability. The region absent in ZH4 and ZH43 is indicated by a dashed line. JL: junction left; JR: junction right. Positions of SCCmec regions J1 and J3 are shown. A: Entire SCCmecN1 in CHE482. B: Partially cured variant CHE482Δ.
mec complex typing
SCCmec typing [20] results suggested that the mec complex did not contain mecI and PCR using primers spanning IS1272 and ΔmecR1 and sequencing over the gene junction then confirmed the presence of a class B mec complex (mecA-ΔmecR1-IS1272).
ccr typing
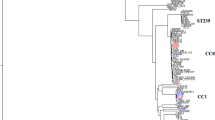
No ccr complex could be detected using the multiplex PCR for ccr types 1 to 3 as described by Ito et al. [21], however with ccr type 4-specific primers C1 and C2, a weak amplificate was produced [9]. Further sequence analysis revealed that this SCCmec contained two complete ccrAB loci which are both similar to ccrAB4 from strain HDE288, a pediatric clone isolated in Portugal carrying a type VI SCCmec element [24, 25]. Therefore specific primers to identify the drug clone ccrAB4-1/-2 genes were designed (primers 27 and 28). One of the loci, ccrAB4-1, was located at the usual ccrAB position downstream of the mec complex at the border of the J1 region. The other recombinase complex, ccrAB4-2 was located within the J3 region (Figure 1). Sequence alignments of ccrAB4-1CHE482, ccrAB4-2CHE482, ccrAB4HDE288 and ccrAB4ATCC12228 genes showed that all four loci were different, with ccrA4 genes sharing between 85.2% and 89.4% similarity with each other and ccrB4 sequences sharing between 94.3% and 92.9% similarity. Nucleotide sequence similarities of these four ccrA4 genes and ccrA genes from complex types 1–3, and of the four ccrB4 genes with ccrB genes from complexes 1–3, are shown by phylogenetic tree (Figure 2). For these alignments the sequence of ccrB4HDE288 was adjusted because the database sequence is truncated as the result of an adenine deletion at nt position 1325; leaving it 99-aa shorter than ccrB4-1CHE482 and 100-aa shorter than ccrB4-2CHE482. By adding back this adenine we could compare the whole length sequences.
Phylogenetic relatedness of selected ccrA and ccrB nucleotide sequences. The following genes were used: ccrA1 and ccrB1* from strain NCTC10442 [DDBJ:AB033763]; ccrA2 and ccrB2 from MRSA252 [EMBL:BX571856], MW2 [NCBI:NC_003923], SCCmec IIE [EMBL:AJ810120]; ccrA3 and ccrB3 of 85–3907 [DDBJ:AB047088]; ccrA4 and ccrB4* from HDE288 [GenBank:AF411935]; ccrA4 and ccrB4 of ATCC12228 [GeneBank:AE015929]; ccrC in WIS [DDBJ:AB121219]. The ccrB genes from NCTC10442 (ccrB1*) and HDE288 (ccrB4*) are truncated; for comparison we have reconstituted them by adding an adenine at the site of the frameshifts. The evolutionary relationships are shown by the length of the branches and the scale of the tree indicates the number of nucleotide substitutions per 100 bases. Alignment was done using ClustalW and tree constructed with Multalign, Lasergene 6.0. Type 4 ccrA genes are framed in red and and type 4 ccrB genes in green.
The phylogenetic trees show that ccrAB4-1CHE482, ccrAB4-2CHE482, ccrAB4HDE288 and ccrAB4ATCC12228 (S. epidermidis) form a distinct ccrAB4 cluster. The presence of two complete ccrAB4 loci in the CHE482 SCCmec indicated that SCCmecN1 had been composed from at least two different complete or partial SCC elements. Other such mosaic or composite SCC elements have been described previously [13, 26, 27], however this is the first SCCmec found to contain two copies of the same ccr complex.
Due to the presence of both a class B mec complex and ccrAB4, the CHE482 SCCmec would be most closely related to SCCmec type VI. However, due to a number of unique features, including the presence of a second ccr locus and additional antibiotic resistance determinants, it appears to be a distinct subtype of this group that we are provisionally calling SCCmecN1.
Additional resistance determinants
In addition to mecA, the resistance genes dfrA and fusB1, encoding trimethoprim and fusidic acid resistance, respectively, were also found on SCCmecN1. The dfrA gene shared 100% nucleotide identity with dfrA from pSK1 (X13290.1), which confers high level trimethoprim resistance [28]. dfrA is carried on Tn4003, a generally plasmid-encoded composite transposon with the genetic organisation IS431-rep-IS431-orf140-dfrA-thyE-IS431 [28]. We speculate that Tn4003 had jumped into the SCCmec-associated IS431, hypothesised to be a hotspot for the integration of resistance determinants [29, 30] (Figure 1). However, it had lost the rep gene (replication protein) and one of its flanking IS431 elements, leaving Tn4003 truncated (IS431-orf140-dfrA-thyE-IS431).
The fusB1 gene, found within the J1 region of SCCmecN1, was identical to the hypothetical fusidic acid resistance gene SAS0043 from the methicillin-susceptible strain MSSA476 [14], located on the 22.8-kb SCC-like element SCC476, in MSSA476. The SCC476 ccrAB genes, however, are most similar to the type 1 ccr complex from S. hominis. The fusB1 gene in CHE482 conferred only low level fusidic acid resistance of 6 μg ml-1.
SCCmec boundaries
The boundaries of the SCCmec element were sequenced using the primers 2, 3, 13 and 14 and compared to reference sequences of SCC476 from MSSA476 and SCCmec type II from N315 (Figure 3). SCCmecN1 had integrated at the same position within the attBSCC sequence at the 3' end of orfX as all previously described SCCmec and SCC elements. The ends of SCCmecN1 contained the characteristic direct and degenerate-inverted repeats found at the ends of SCCmec types I-IV and SCC476. Integration site sequences (ISS) with the consensus sequence 5'-(GA NGC NTATCATAA NT N)-3 [23] were present at both boundaries. A third ISS sequence (ISS*) was also identified about 6.4-kb upstream from the right end junction.
Chromosome-SCCmec junction sequences. The borders of strain CHE482 were aligned with the sequences of the three drug clones, ZH4, ZH43, ZH81, the cured drug clone, strain N315 (type II SCCmec), MSSA476 (SCC476)and ISS*. Light green boxes indicate the orfX region and its integration sequence site, ISS. Yellow shading highlights differences between the SCCmec ends. Direct and indirect repeats are indicated with arrows. The red letters represent the similarity of the inverted repeats. attBSCC** and ISS** consensus sequences were taken from Ito et al. [23].
Drug clone variability
Analysis of PFGE profiles of all drug clone isolates characterised by Qi et al. in 2003 [9] revealed that there were small variations in size in the 208-kb SmaI band containing mecA. Therefore the SCCmec of CHE482 and a selection of three other drug clone strains (ZH4, ZH43 and ZH81, Table 2) were cured using plasmid pSR3-1. The SmaI band carrying SCCmec was slightly larger in CHE482 and ZH81 than in ZH4 and ZH43 before curing, whereas after curing the resulting patterns were identical in all four strains (Figure 4A). This indicated that there was variability, presumably within the SCCmec.
PFGE restriction analysis. A: SmaI restriction patterns of four drug clone isolates CHE482, ZH4, ZH43, ZH81 and their corresponding cured strains ME21, ME135, ME138, ME141. The SmaI fragments carrying SCCmec are indicated by filled triangles, and the corresponding fragments of the cured strain are indicated by open triangles. Southern hybridisations using a mecA and a dfrA probe are shown below. B: PFGE patterns of the drug clone CHE482 (filled triangle), its cured strain ME21 (open triangle) and the partially cured strain CHE482Δ (red triangle).
SCCmecN1 variation
Using the long range overlapping PCR products, variations in the SCCmec elements of strains CHE482, ZH4, ZH43 and ZH81, were compared. Fragment sizes between primers 6 and 4 varied by 100 to 200 bp, and between primers 7 and 9 from 1000 to 1500 bp. The variation in the hypervariable region between the mec complex and ΔTn4003 (primers 6 and 4) could be due to different numbers of direct repeats units (dru) [31] as found in SCCmec VT compared to the WIS SCCmec V [32]. Amplification between fusB1 and the right SCCmec junction produced a 6.4-kb larger end fragment from CHE482/ZH81 than from ZH4/ZH43.
Sequencing in from the right junctions showed that the ends of CHE482/ZH81 were identical to each other with no significant similarity to any database sequences (data not shown) but they were different to those of ZH4/ZH43 (Figure 3). The latter sequences of ZH4/ZH43 were identical to the end of SCC476. In contrast, the left end chromosome-SCCmec junction sequences were identical in all drug clones analysed.
Antibiotic resistance variability
SCCmec variability also appeared to correlate with other strain differences. Strains ZH4 and ZH43 which had identical SCCmec s, were also both ciprofloxacin resistant; meanwhile CHE482 and ZH81, which share identical SCCmec s, were ciprofloxacin susceptible (Table 2). There was also variation in fusidic acid resistance levels. Most strains had relatively low fusidic acid resistance. ZH43, however, was highly resistant and resistance was not lost upon curing (Table 3), therefore resistance in this strain was probably additionally caused by a mutation in the chromosomal elongation factor G, EF-G (fusA) [33]. Since both ciprofloxacin and sulfamethoxazole resistance are chromosomal, the SCCmec variants found in the Zurich drug clones are very likely associated with different, closely related genetic backgrounds.
ccr activity
CHE482 was cured using either pME21 (ccrAB4-1) or pME22 (ccrAB4-2). Resulting isolates were screened for methicillin, trimethoprim and fusidic acid susceptibility and for amplification of a PCR product spanning the former SCCmec-chromosomal junctions (primers 1 and 14, Figure 1). Both ccrAB4-1 and ccrAB4-2 were functional and able to excise SCCmec even though their ccrA and ccrB amino acid sequences differed by 11.3% and 4.6%, respectively. This is consistent with the finding that several different ccrAB loci from type IV SCCmec s were all shown to be active, despite varying up to 3.7% in their amino acid sequences [34].
Excision variants
CHE482 was cured using pSR3-1 [35], the resulting strain ME21 was sensitive to oxacillin, fusidic acid and trimethoprim (Figure 1, Table 3). During curing experiments with pME21 and pME22 we discovered that there were also many strains that had not been completely cured. One set of cured CHE482 variants maintained a fragment of 6.4-kb, and sequencing confirmed that this fragment was the portion between the ISS at the right junction and ISS*. This indicated that excision of the main SCCmec fragment containing all three resistance determinants had occurred through recombination between the ISS at the left junction and ISS*.
ccrAB4-1 and its predicted promoter were also cloned into the E. coli-S. aureus shuttle vector pAW17 to produce the recombinant plasmid pME15. Attempts to cure CHE482 of its SCCmec element using pME15 resulted in another partially cured set of variants which had maintained fusidic acid resistance but lost oxacillin and trimethoprim resistance (CHE482Δ, Figure 1B). Analysis of these strains by PFGE, showed that the SCCmec-containing band had become smaller but not to the same extent as the completely cured strain ME21 (Figure 4B). These results indicated that only a portion of the SCCmec, containing mecA and dfrA, had been lost. PCR mapping identified the location of the missing portion and sequencing over the excision sites revealed that excision was likely to be mediated by homologous recombination across regions of high nucleotide sequence similarity surrounding the two ccr loci, as no additional ISS sequences were found. It appeared that recombination between the two ccr regions resulted in the deletion of a 22-kb fragment containing ccrAB4-1, the class B mec complex and ΔTn4003. This recombination left an SCC-like element of 23.7-kb, which contained one ccrAB complex (ccrAB4-2) and the fusidic acid resistance determinant (Figure 1B). This truncated SCC is similar in size to the MSSA476 SCC476 which also contains fusB1 and a ccrAB locus, although in SCC476 the ccr genes are most similar to ccrAB type 1.
Therefore we have identified three possible excision variants, two resulting from the presence of three ISS, as has been seen in SCCmec type IV strains [26], and the third variant caused by recombination between regions of high sequence similarity.
Conclusion
The general structure of SCCmecN1 (ccrAB4-2, dfrA, class B mec complex, ccrAB4-1, fusB1) was distinctly different from already published SCCmec types. Several regions of variability were found between different clinical drug clone isolates, especially in the right-end region where the presence or absence of a DNA fragment framed by ISS sequences was detected. Nevertheless this clone can now be identified by its resistance profile and its combination of class B mec complex and ccrAB4 complex sequences, thus allowing easier epidemiological identification.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains and plasmids are listed in Table 2. The four clinical MRSA isolates CHE482, ZH4, ZH43 and ZH81 were clones associated with intravenous drug users in the Zurich area. Apart from the type strain CHE482, strains were selected from the epidemiological study in 2003 based on their PFGE patterns and resistance profiles (Table 2) [9]. Growth was at 37°C in Luria Bertani broth (Difco Laboratories, Detroit, MI, USA). Strains harbouring the temperature-sensitive plasmids pME21 or pME22 were propagated at 30°C in the presence of 10 μg ml-1 tetracycline and those with plasmid pME15 were grown at 37°C in the presence of 50 μg ml-1 kanamycin.
Susceptibility testing
The minimal inhibitory concentrations (MIC) of antibiotics were determined by Etest on Mueller-Hinton agar plates (Difco Laboratories, Detroit, MI, USA) according to the manufacturer's instructions (AB Biodisk, Solna, Sweden). Disc diffusion of oxacillin, cefoxitin, fusidic acid and trimethoprim/sulfamethoxazole were done according to CLSI [36] on Mueller Hinton agar plates. Penicillinase production of cefoxitin-induced cells was assayed by nitrocefin hydrolysis and PBP2a production by the MRSA screen agglutination test from Denka Seiken (Tokyo, Japan) [37].
SCCmec typing
SCCmec types I through IV, ccr types 1 to 3, and ccr type 4 from the pediatric clone HDE288, were identified by PCR as described by [20, 21], and [38], respectively. A specific PCR was established to identify the drug clone ccrAB4-1/-2CHE482 using primer pair 27 and 28 (Table 1).
Localization of dfrA and mecA
SmaI digested chromosomal DNA was separated by pulsed field gel electrophoresis, PFGE [39] and hybridised sequentially [40] with a mecA (primer pair 18 and 19) and a dfrA probe (primer pair 20 and 21) (Table 1).
Cloning of the ccrAB genes of CHE482
Each of the two ccrAB complexes identified in strain CHE482, including their own promoter, were cloned into the BamHI site of the temperature-sensitive plasmid pYT3, using primers 22 and 24 for ccrAB4-1CHE482 and primers 25 and 26 for ccrAB4-2CHE482. The resulting plasmids pME21 and pME22, respectively, were electroporated into RN4220 and then transduced by Φ80α into the MRSA clinical isolates to be cured of SCCmec. The ccrAB4-1CHE482 was also cloned into the E. coli-S. aureus shuttle vector pAW17, using the primers 22 and 23 (Table 1). The resulting plasmid pME15 was then electroporated into RN4220 and transduced into the MRSA to be cured of SCCmec.
Curing of SCCmec
Curing of SCCmec was done by the method of Katayama [35] using either the temperature-sensitive plasmid pSR3-1 containing ccrAB type 2 recombinase genes, or plasmids pME21 (ccrAB4-1) or pME22 (ccrAB4-2). Curing with plasmid pME15 (ccrAB4-1) was done by transducing the plasmid into the clinical MRSA isolates, selecting for kanamycin resistant transductants at 37°C, which were then pooled and plated on LB agar containing kanamycin, grown overnight, and screened by replica plating for loss of oxacillin resistance on 1 μg ml-1 oxacillin.
Sequence analysis
Sequencing was performed with an ABI PRISM BigDye Terminator Cycle sequencing reaction kit (US Biochemicals, Cleveland, Ohio) and an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, California). Sequence assembly was accomplished using the DNAStar sequence analysis package (Lasergene 6.0).
Sequencing of the SCCmec-chromosome junctions of four drug clones was done by direct chromosomal sequencing [41] of the original and cured clones using primer 2. This nucleotide sequence provided the template for the design of primers 14, 13 and 3 (Table 1) over the chromosome-SCCmec junctions.
Mapping of SCCmec
To estimate the size of the CHE482 SCCmec, long range PCR amplification of six main fragments was performed using the polymerase TaKaRa Ex Taq (TAKARA BIO INC., Shiga, Japan). PCR was done as described by the manufacturer's recommendation. Primer pairs utilised were: 1 and 5; 4 and 6; 4 and 8; 7 and 9; 10 and 11 and 12 and 14 (Table 1). Amplified PCR fragments were run against molecular weight markers (Marker II, Fermentas International, Ontario, CA; 1 kb+, Invitrogen, Carlsbad, CA) on a 0.5% agarose gel.
Nucleotide sequence accession number
The sequences of ccrAB4-1CHE482 and ccrAB4-2CHE482 have been deposited in the GenBank (NCBI) database under accession number [GenBank: EF126185] and [GenBank:EF126186] respectively.
References
Markowitz N, Pohlod DJ, Saravolatz LD, Quinn EL: In vitro susceptibility patterns of methicillin-resistant and-susceptible Staphylococcus aureus strains in a population of parenteral drug abusers from 1972 to 1981. Antimicrob Agents Chemother. 1983, 23: 450-457.
Fleisch F, Zbinden R, Vanoli C, Ruef C: Epidemic spread of a single clone of methicillin-resistant Staphylococcus aureus among injection drug users in Zurich, Switzerland. Clin Infect Dis. 2001, 32: 581-586. 10.1086/318716.
Charlebois ED, Perdreau-Remington F, Kreiswirth B, Bangsberg DR, Ciccarone D, Diep BA, Ng VL, K C, Edlin BR, Chambers HF: Origins of community strains of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2004, 39: 47-54. 10.1086/421090.
Huang H, Flynn NM, King JH, Monchaud C, Morita M, Cohen SH: Comparisons of community-associated methicillin-resistant Staphylococcus aureus (MRSA) and hospital-associated MSRA infections in Sacramento, California. J Clin Microbiol. 2006, 44: 2423-2427. 10.1128/JCM.00254-06.
Gilbert M, MacDonald J, Gregson D, Siushansian J, Zhang K, Elsayed S, Laupland K, Louie T, Hope K, Mulvey M, Gillespie J, Nielsen D, Wheeler V, Louie M, Honish A, Keays G, Conly J: Outbreak in Alberta of community-acquired (USA300) methicillin-resistant Staphylococcus aureus in people with a history of drug use, homelessness or incarceration. Can Med Assoc J. 2006, 175: 149-154. 10.1503/cmaj.051565.
Ebright JR, Pieper B: Skin and soft tissue infections in injection drug users. Infect Dis Clin North Am. 2002, 16: 697-712. 10.1016/S0891-5520(02)00017-X.
Bassetti S, Wolfisberg L, Jaussi B, Frei R, Kuntze MF, Battegay M, Widmer AF: Carriage of Staphylococcus aureus among injection drug users: Lower prevalence in an injection heroin maintenance program than in an oral methadone program. Infect Control Hosp Epidemiol. 2004, 25: 133-137. 10.1086/502364.
Ruotsalainen E, Sammalkorpi K, Laine J, Huotari K, Sarna S, Valtonen V, Jarvinen A: Clinical manifestations and outcome in Staphylococcus aureus endocarditis among injection drug users and nonaddicts: A prospective study of 74 patients. BMC Infect Dis. 2006, 6: 137-10.1186/1471-2334-6-137.
Qi W, Ender M, O'Brien F, Imhof A, Ruef C, McCallum N, Berger-Bachi B: Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Zurich, Switzerland (2003): Prevalence of type IVSCCmec and a new SCCmec element associated with isolates from intravenous drug users. J Clin Microbiol. 2005, 43: 5164-5170. 10.1128/JCM.43.10.5164-5170.2005.
Fleisch F, Oechslin EC, Gujer AR, Ritzler E, Imhof A, Ruef C, Reinhart WH: Transregional spread of a single clone of methicillin-resistant Staphylococcus aureus between groups of drug users in Switzerland. Infection. 2005, 33: 273-277. 10.1007/s15010-005-4149-6.
Witte W, Pasemann B, Cuny C: Detection of low-level oxacillin resistance in mecA-positive Staphylococcus aureus. Clin Microbiol Infect. 2007, 13: 408-412. 10.1111/j.1469-0691.2006.01671.x.
Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K: Combination of multiplex PCRs for SCCmec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2006, 51: 264-274. 10.1128/AAC.00165-06.
Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS: Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: A possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob Agents Chemother. 2004, 48: 1823-1836. 10.1128/AAC.48.5.1823-1836.2004.
Holden MTG, Feil EJ, Lindsay JA, Peacock SJ, Day NPJ, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J: Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance. PNAS. 2004, 101: 9786-9791. 10.1073/pnas.0402521101.
Luong TT, Ouyang S, Bush K, Lee CY: Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J Bacteriol. 2002, 184: 3623-3629. 10.1128/JB.184.13.3623-3629.2002.
Katayama Y, Takeuchi F, Ito T, Ma XX, Ui-Mizutani Y, Kobayashi I, Hiramatsu K: Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J Bacteriol. 2003, 185: 2711-2722. 10.1128/JB.185.9.2711-2722.2003.
Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K: Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: A proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006, 50: 1001-1012. 10.1128/AAC.50.3.1001-1012.2006.
Kuroda M, Yamashita A, Hirakawa H, Kumano M, Morikawa K, Higashide M, Maruyama A, Inose Y, Matoba K, Toh H, Kuhara S, Hattori M, Ohta T: Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. PNAS. 2005, 102: 13272-13277. 10.1073/pnas.0502950102.
Wannet WJB, Spalburg E, Heck MEOC, Pluister GN, Willems RJL, de Neeling AJ: Widespread dissemination in the Netherlands of the epidemic Berlin methicillin-resistant Staphylococcus aureus clone with low-level resistance to oxacillin. J Clin Microbiol. 2004, 42: 3077-3082. 10.1128/JCM.42.7.3077-3082.2004.
Oliveira DC, de Lencastre H: Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002, 46: 2155-2161. 10.1128/AAC.46.7.2155-2161.2002.
Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K: Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001, 45: 1323-1336. 10.1128/AAC.45.5.1323-1336.2001.
Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, van Belkum A, Ouellette M, Roy PH, Bergeron MG: New real-time PCR assay for rapid detection of methicillin resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J Clin Microbiol. 2004, 42: 1875-1884. 10.1128/JCM.42.5.1875-1884.2004.
Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K: Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004, 48: 2637-2651. 10.1128/AAC.48.7.2637-2651.2004.
Oliveira DC, Tomasz A, de Lencastre H: The evolution of pandemic clones of methicillin resistant Staphylococcus aureus: Identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist. 2001, 7: 349-361. 10.1089/10766290152773365.
Oliveira DC, Milheirico C, de Lencastre H: Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006, 50: 3457-3459. 10.1128/AAC.00629-06.
Jansen WTM, Beitsma MM, Koeman CJ, van Wamel WJB, Verhoef J, Fluit AC: Novel mobile variants of staphylococcal cassette chromosome mec in Staphylococcus aureus. Antimicrob Agents Chemother. 2006, 50: 2072-2078. 10.1128/AAC.01539-05.
Heusser R, Ender M, Berger-Bachi B, McCallum N: Mosaic staphylococcal cassette chomosome mec (SCCmec) containing two recombinase loci and a new mec complex, B2. Antimicrob Agents Chemother. 2006
Rouch DA, Messerotti LJ, Loo LS, Jackson CA, Skurray RA: Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol. 1989, 3: 161-175. 10.1111/j.1365-2958.1989.tb01805.x.
Hanssen AM, Ericson Sollid JU: SCCmec in staphylococci: Genes on the move. FEMS Immunol Med Microbiol. 2006, 46: 8-20. 10.1111/j.1574-695X.2005.00009.x.
Berger-Bachi B: Genetics of methicilline resistance in Staphylococcus aureus. J Antimicrob Chemother. 1989, 23: 671-680. 10.1093/jac/23.5.671.
Ryffel C, Bucher R, Kayser FH, Berger-Bachi B: The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol. 1991, 173: 7416-7422.
Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS: Successful multiresistant community-associated methicillin-resistant S taphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J Clin Microbiol. 2005, 43: 4719-4730. 10.1128/JCM.43.9.4719-4730.2005.
Besier S, Ludwig A, Brade V, Wichelhaus TA: Compensatory adaptation to the loss of biological fitness associated with acquisition of fusidic acid resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2005, 49: 1426-1431. 10.1128/AAC.49.4.1426-1431.2005.
Noto MJ, Archer GL: A subset of Staphylococcus aureus strains harboring staphylococcal cassette chromosome mec (SCCmec) type IV is deficient in ccrAB-mediated SCCmec excision. Antimicrob Agents Chemother. 2006, 50: 2782-2788. 10.1128/AAC.00032-06.
Katayama Y, Ito T, Hiramatsu K: A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000, 44: 1549-1555. 10.1128/AAC.44.6.1549-1555.2000.
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: 15th informational supplement. CLSI/NCCLS document M100-S15 Clinical and Laboratory Standards Institute, Wayne, Pa. 2005
Zbinden R, Ritzler M, Ritzler E, Berger-Bachi B: Detection of penicillin-binding protein 2a by rapid slide latex agglutination test in coagulase-negative staphylococci. J Clin Microbiol. 2001, 39: 412-412. 10.1128/JCM.39.1.412.2001.
Lim TT, Chong FN, O'Brien FG, Grubb WB: Are all community methicillin-resistant Staphylococcus aureus related? A comparison of their mec regions. Pathology. 2003, 35: 336-343. 10.1080/0031302031000150498.
Wada A, Katayama Y, Hiramatsu K, Yokota T: Southern hybridization analysis of the mec A deletion from methicillin-resistant Staphylococcus aureus. Biochem Biophys Res Commun. 1991, 176: 1319-1325. 10.1016/0006-291X(91)90430-F.
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K: Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY. 2004
Wada A: An improved method for purifying bacterial genomic DNAs for direct sequencing by capillary automated sequencer. Tech Tips Online. 2001
Oliveira DC, Wu SW, de Lencastre H: Genetic organization of the downstream region of the mec A element in methicillin-resistant Staphylococcus aureus isolates carrying different polymorphisms of this region. Antimicrob Agents Chemother. 2000, 44: 1906-1910. 10.1128/AAC.44.7.1906-1910.2000.
Sa-Leao R, Santos Sanches I, Dias D, Peres I, Barros RM, de Lencastre H: Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: Relics of a formerly widely disseminated strain?. J Clin Microbiol. 1999, 37: 1913-1920.
Rossi J, Bischoff M, Wada A, Berger-Bachi B: MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob Agents Chemother. 2003, 47: 2558-2564. 10.1128/AAC.47.8.2558-2564.2003.
Acknowledgements
This study was supported by the Swiss National Science Foundation grant NF31-105390/1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
ME carried out the molecular genetic and microbiological studies and drafted the manuscript. BB conceived of the study, participated in its design and helped to draft the manuscript. NM participated in the design and coordination of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ender, M., Berger-Bächi, B. & McCallum, N. Variability in SCC mecN1 spreading among injection drug users in Zurich, Switzerland. BMC Microbiol 7, 62 (2007). https://doi.org/10.1186/1471-2180-7-62
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-7-62