Abstract
Background
Previous research has identified the potential for the existence of two separate lineages of Escherichia coli O157:H7. Clinical isolates tended to cluster primarily within one of these two lineages. To determine if there are virulence related genes differentially expressed between the two lineages we chose to utilize microarray technology to perform an initial screening.
Results
Using a 610 gene microarray, designed against the E. coli O157 EDL 933 transcriptome, targeting primarily virulence systems, we chose 3 representative Lineage I isolates (LI groups mostly clinical isolates) and 3 representative Lineage II isolates (LII groups mostly bovine isolates). Using standard dye swap experimental designs, statistically different expression (P < 0.05) of 73 genes between the two lineages was revealed. Result highlights indicate that under in vitro anaerobic growth conditions, there is up-regulation of stx2b, ure D, curli (csg AFEG), and stress related genes (hsl J, csp G, ibp B, ibp A) in Lineage I, which may contribute to enhanced virulence or transmission potential. Lineage II exhibits significant up-regulation of type III secretion apparatus, LPS, and flagella related transcripts.
Conclusion
These results give insight into comparative regulation of virulence genes as well as providing directions for future research. Ultimately, evaluating the expression of key virulence factors among different E. coli O157 isolates has inherent value and the interpretation of such expression data will continue to evolve as our understanding of virulence, pathogenesis and transmission improves.
Similar content being viewed by others
Background
Kim et al., [1] utilized octamer-based genome scanning to evaluate genome diversity among E. coli O157 isolates. Based upon this genetic fingerprinting method they noted two distinct lineages of this pathogen, one of which tended to cluster the majority of human isolates utilized in their study, and the second which grouped together isolates primarily of bovine origin. They suggested that one of these lineages (Lineage II) may not efficiently transmit to humans from bovine sources. Pradel et al. [2] also found that there were distinct lineages among isolates derived from patients with hemolytic-uremic syndrome (HUS) when evaluated genetically using a combination of stx2-RFLP (restriction fragment length polymorphism analyses), stx2 variant, and plasmid profile analyses. They also suggested that there may be a separate lineage, which was more virulent for humans, along with a lineage, which may not be as pathogenic. Yang et al. [3] utilized a lineage-specific polymorphism assay consisting of 6 genetic markers and found that they could differentiate two lineages of E. coli O157 indicating that the occurrence of these two lineages may be widespread. Barkocy-Gallagher [4] using Xba1 RFLP analysis also found distinct clusters of E. coli O157, including a cluster where most isolates lacked flagella and stx1 genes, leading them to suggest the potential for the existence of clustered isolates having differential abilities to cause disease.
The expression of several virulence factors in relation to the existence of two lineages of EHECs have been evaluated as well. McNally et al. [5] found clear differences in the expression of locus of enterocyte effacement (LEE)-encoded factors between different strains. It was found that, EspD, when used as an indicator of LEE expression, was expressed at higher concentrations in the majority of strains that were of human origin (15 of 20) compared with only a few (4 of 20) isolates that were of bovine origin (P < 0.001). They concluded that a subset of E. coli O157 isolates (stx+ eae+) in cattle were capable of causing severe disease in humans. Another study evaluating gene expression conducted by Richie et al., [6] found that HUS derived isolates expressed higher concentrations of stx2 than bovine derived isolates.
Based upon the proposed existence of a less pathogenic lineage of E. coli O157, it has been postulated that much of the Class I recall of millions of pounds of meat annually [7] might be greatly reduced. However, even if a separate lineage of E. coli O157 (conclusively proven not to cause disease in humans) were identified and concrete methods for differentiating this lineage developed, it would still be unlikely (because of liability issues) to have the suggested impact on the meat industry. Yet, the study of genetic differences between two lineages of this pathogen that possess different virulence or transmission potential could still have wide ranging and significant economic or scientific benefits. For example, if a specific lineage could be more readily eradicated during the farm to fork process, based upon their genetic differences, this might indirectly have the originally intended effect of reducing the volume of Class I recalls. In addition, from a purely scientific standpoint, clues as to why certain isolates may be more pathogenic or more easily transmitted, based upon genetic differences, is of obvious importance in the study of virulence.
Results and discussion
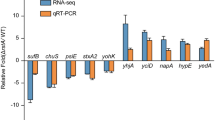
Microarray analyses, validated by quantitative PCR, showed that, of the 610 genes on the array, 179 genes were consistently and differentially regulated between the two lineages. Of these 179 regulated genes, 73 transcripts showed statistically significant (p < 0.05) differences in expression of greater than 1.2 fold (Table 1 and Table 2) between each member of the two lineages. Table 1 shows those transcripts whose expression was greater (P < 0.05) in each of the LI isolates. Three heat shock and one cold shock protein transcripts were the most upregulated in the LI isolates compared to the LII. In LII isolates cyoE, hscA, and fimbrial subunit 1 were most highly upregulated compared to LI. Table 2 shows those transcripts whose expression was statistically higher (P < 0.05) in each of the LII isolates. Six transcripts that exhibited enough expression difference to be evaluated by CT using quantitative PCR were chosen at random from these 73 and Q-PCR performed as a validation method. These included ure D, cyoE, hscA, nrfB [see Additional file 1], chap4, and stx 2B. Results of Q-PCR were found to agree in each instance with the results of the microarray experiment. Supplementary dataset 1 [see Additional file 1] provides a listing of the 106 genes that were shown to be consistently up-regulated or down-regulated as part of the microarray experiment, but which did not fully meet the stringent selection and statistical requirements additional supplemental dataset 2 provides all the genes on the array.
Results of the microarray experiments showed that the LI isolates express higher transcription of ureD (Table 2), as well as ureA, ureB, ureC (supplemental data), compared to LII. In addition stx2B (Table 2) and stx2A (supplemental data) transcripts are detected in higher abundance in Lineage 1. Lineage I also exhibits up-regulation of key fimbria related transcripts, especially fliC, fliT, and fliP. Other attachment related transcripts csgA, csgF, csgE, and csgG (curli) were also up-regulated, which could also be highly significant in promotion of pathogenesis [8–12]. When using all of the regulated genes as a single data set for Gene Ontology [13] based analyses, it was found that, up-regulation of genes associated with regulation of urease activity, GTP binding, metabolism, nitrogen metabolism and regulation of transcription were statistically (p < 0.05) more represented in LI isolates (Table 3). In LII isolates peptidase activity, transferase activity, and DNA binding activity were statistically more represented (p < 0.05). These differences could point to a fundamental difference in the environmental response and control networks of these lineages that promotes survival and differential expression of virulence attributes in response to specific environments and hosts. These types of control networks could be the key to understanding differential virulence or transmission potential if such a phenomenon could be proven to exist within the O157 serogroup.
Stx2
The role of stx2 in pathogenesis is well accepted [14–18] and up-regulation of constitutive stx2 expression in the hypothesized more pathogenic LI isolates may not be a surprising finding. The up-regulation of stx2B and stx2A [see Additional file 1] transcripts is accompanied by up-regulation of regulatory genes associated with Stx2 expression. A complicated network of interactions between the oraA (recX), dinI, lexA, umuD, SSB, recA, psiB and possibly other unidentified proteins, act in the regulation of RecA function. The role of recA as part of an SOS response is to cleave repressors that in addition to the SOS response ultimately lead to Stx2 production [19]. OraA (also known as recX) and dinl are coregulators (competing regulators) of recA [20] and both were up-regulated in LI isolates along with the stx2 subunit transcripts. OraA is thought to be co-transcribed with recA during SOS response [21]. RecA specific oligos were not included in the array but we might expect that being co-transcribed along with oraA that it would likely be up-regulated in LI as well. PsiB, (supplemental) is also up-regulated in LI and thought to prevent ssDNA from inducing an SOS response by inhibiting activation of recA protein [22]. PsiB, is found on many conjugative plasmids near the origin of conjugative transfer and has anti-recombinase activities [23]. Expression of the dinI protein of E. coli inhibits both the co-protease and recombinase activities of recA in vivo [24]. Yet, in spite of all of the regulators of SOS response in LI isolates, we still observe a significant up-regulation of stx2a and stx2b transcripts which have been shown to be expressed as part of an SOS response [25–27].
With up-regulation of dinl, psiB, oraA and also with the up-regulation of stx2a and stx2b and various other genes related to stress response it could be an indication that LI isolates do have differentially regulated pathways the enhance its toxin expression potential. It does appear that the current LI isolates have a modified regulatory system response, which significantly promotes Stx2 toxin production compared to LII isolates. We have also considered that LII isolates may have mutations affecting the integrity of the stx2 prophage's late regulatory transcripts shown to encode stx2 [28–31]. Future work looking at the actual Stx2 toxin levels as well as evaluation of the structural integrity of the Stx2 phage in these 6 isolates via sequencing or PCR would be a beneficial follow up to this research. We have performed Stx2b specific ELISA and quantitative PCR analysis of 20 additional LI and 20 additional LII isolates as part of a follow-up study, and found that the LI isolates have statistically (p < 0.05) higher transcription rates and protein concentrations under these same conditions (data not shown). If these LII isolates have a defective toxin production system this could be a strong indication that they lack one of the key virulence factors contributing to the pathogenicity of O157 [15, 16, 32–34].
Urease
Enterohemorrhagic E. coli has been shown to be highly adaptable to various extreme environments (water, heat, freezing, acid, desiccation, hypo- and hyperosmotic, disinfectants etc) which contributes greatly to its success as a pathogen [35–46]. To succeed as a enteric pathogen with a low infectious dose [47–49], E. coli O157 must be able to survive passage through the acidic environment of the stomach if they are to cause gastrointestinal disease [50]. As an indication of their evolutionary focused ability for surviving acidic environments they possess 3 acid resistance pathways [51] and urease could act as an additional system to modify anion concentrations. Therefore the up-regulation of urease in LI isolates is of interest in spite of recent work indicating that E. coli O157 has only rarely been shown to exhibit urease activity [52–55]. As an example, a previous study noted that lack of urease activity in EHEC strains is often due to a base substitution in the ureD gene causing an early termination of the transcript [54]. Urease expression and activity be condition, host, or environment specific and could be expressed only in specific environments to beneficially modify internal and/or surrounding anion concentrations, enabling EHEC to survive acidic conditions and contributing to its low infectious dose. Thus, environmental (bovine) isolates may not possess or have sufficient selective pressure for maintenance of detectable levels of urease transcript expression under the conditions evaluated.
Previous research by Heimer et al [52] suggests regulation of the urease operon is through fur (not differentially regulated) and an unknown trans-acting factor. It was hypothesized that this transacting factor is missing in E. coli O157:H7 strain EDL933 (atcc # 43895) though other O157 strains (IN1 and MO28) have been shown to possess some urease activity. However, none of the isolates showed differential regulation of fur which may be an indication that the LI isolates may be differentially expressing this proposed transacting factor, which is promoting up-regulation of the urease operons under the current growth conditions. It is likely that based upon previous evaluation that there is some low level urease activity that is not evident in E. coli O157 strains using conventional methods such as Christensen agar [56]. We have begun investigations of the effects of pH, different laboratory media, anoxia, nickel supplementation, and cytosolic specific urease based acid resistance assays on the ability to detect urease activity in O157 isolates.
Curli
Several factors related to attachment are up-regulated in LI isolates. These include curli fibers, type III secretion apparatus genes. This suggests that LI isolates have constitutive up-regulation of many genes that are involved in intimate attachment. It was reported that curli fibers are infrequently expressed during in vitro growth of E. coli O157:H7 [8] and that strains containing variations at the csgD promoter region, which induced expression of curli, are associated with increased virulence in mice and increased invasion of HEp-2 cells [57]. In this experiment there was significant up-regulation of csgA and csgD as well as some evidence for up-regulation of the both csg operons [see Additional file 1] in the LI isolates compared to LII, yet genes involved in regulation of curli operons do not correspond to this observation. RpoS has been shown to interact with hns (neither differentially regulated) to derepress csgAB expression [58]. Further contradicting the increased expression of curli operons in LI, ompR is up-regulated in LII. Increased ompR expression has also been associated with increased curli production yet a single point mutation, in ompR [59]. Future work should likely evaluate whether curli fibres are actually being produced and assembled under these in vitro conditions in LI isolates.
Virulence gene regulation
One of the more interesting of the up-regulated genes in LI is rfaH. Originally, discovered as a primary regulator of LPS-core synthesis in Salmonella enterica and E. coli [60, 61], RfaH is noted as a primary virulence regulator of E coli that functions as a transcriptional anti-terminator [62, 63] in long operons. These operons include those encoding the F-factor, O-antigens, different capsules, hemin uptake receptor, alpha-hemolysin, and CNF-1 [64–73]. Inactivation of rfaH in uropathogenic E. coli has be shown to inhibit pathogenicity completely [74]. RfaH mutants have been shown to have reduced ability to survive/grow in the presence of bile salts [75]. The up-regulation of rfaH in LI isolates may be an important avenue to pursue as a means to explain their hypothesized enhanced virulence.
LEE
LII isolates showed an increased expression of toxB which is known to promote expression of genes encoded by locus of enterocyte effacement (LEE). Indeed, several of the esp (A, B, P) showed slight cumulative up-regulation. In addition, most of the etp genes involved in the type II transport system were also up-regulated. The type II secretion system was recently noted as also being involved in intimate attachment through secretion of stcE [76]. These results showing upregulation of such an important virulence factor in LII isolates points out two key features that are of importance in this manuscript. The first is that these results as intended can help with identification of isolates which may serve as good regulatory models for providing additional insight into virulence expression. In addition, these results are obviously counter to the overall hypothesis that LI is either more virulent or has more potential for transmission and therefore serve as a caution for the interpretation of results. Thus, as with all microarray studies care must be taken in interpretation of the results, yet negative results or results counter to the hypothesis should not be ignored.
LPS, fimbria, and Flagella
LII isolates also show notable up-regulation of genes involved in a number of systems that are noted as virulence factors. Of interest in LII is the comparative up-regulation of LPS, fimbria (FimH), capsule, and flagella related genes (Table 1 and supplement). Considering that the isolates were grown under anaerobic conditions the increase in LPS and flagella related transcripts represents what may be a typical K-12 like E. coli response to anoxic conditions [77] in the LII isolates, while the LI seem to be lacking this common profile. The hypothesized decrease virulence of LII may be partially explained by the more pronounced regulation of certain virulence factors by LI. Another interesting aspect that is related to the expression of genes associated with motility and the results seen here is the hypothesis proposed by Monday et al. [78], which is related to a competitive interaction between different type III secretion systems. According to this hypothesis there could be a competitive interaction between the type III secretion systems associated with flagellar export and assembly and the type III secretion system that mediates the injection of virulence factors (LEE). Thus, because O157 has multiple type III systems there is the potential for these systems to interfere with one another. This competition could ultimately affect the expression of motility and/or virulence factors. Thus, because there is an increase in LEE expression as well as motility genes in LII isolates it may be a result of an interaction of the type III regulatory networks in these isolates.
In proper proportions type 1 fimbriae and the LPS of uropathogenic E. coli are known to operate together to induce apoptosis in human neutrophils [79]. The cooperative effects of these virulence attributes may function as a mechanism by which E. coli induces infections of the urinary tract. However, if LPS is over produced, excess LPS is likely to be secreted by bacteria into their environment, which may have the opposite effect. In fact, it has been documented that if significant amounts of LPS is released from non-adherent bacteria this has an anti-apoptotic effect on neutrophils, suggesting that LPS can also serve as an important regulator of neutrophil survival in tissue [79]. Up regulation of LPS by LII isolates compared to LI isolates, if this excess LPS were shed from the bacteria, maintained in the cytoplasm, or deposited in excess onto the membrane might also be toxic to the bacteria inhibiting its own growth and interaction with its environment [80]. Overproduction of LPS could also alter bacterial cell morphology by accumulation in the bacterial cytosol, which could also potentially prevent pathogenesis. Previous work [81] and [82] demonstrated that E. coli O157 exhibiting reduced production of O157 LPS side chains displayed an increased binding to tissue culture cells. It was concluded that the presence of the O157 polysaccharide has the potential to interfere with the adherence and its expression is not required to produce the attaching and effacing lesions. Excess LPS may act to mask adhesive structures present on the bacterial surface. It is also possible that the physicochemical properties of the cell such as surface charge or hydrophobicity may be altered by lack of or excess LPS. These hypothetical interactions and the effects of LPS expression on pathogenesis are again a highly interesting topic for future research.
Conclusion
It has been hypothesized by various researchers that a less pathogenic lineage of E. coli O157 exists. Geared toward finding evidence that might direct research toward genetic mechanisms that support the hypothesis of differential virulence or transmission potential we evaluated representatives from these two lineages in a preliminary study. The results highlight several of the more important virulence factors as being differentially regulated, as well as various regulatory networks that may provide useful insight and targets for future research. Key virulence factors were shown to be upregulated in LI, especially those that have been suggested to promote virulence and transmission potential. However, other contradictory findings were also uncovered in which several virulence factors more associated with colonization and pathogenesis were also upregulated in LII isolates. Many previous studies describing regulatory mechanisms are supported by the results of this study, providing some additional insight into the control of virulence genes. Though the hypotheses considered as part of this research is still far from conclusive, the results do provide a valuable foundation that will direct future research. Ultimately, evaluating the expression of key virulence factors among different E. coli O157 isolates is valuable beyond the reasoning discussed within the confines of this report, and the interpretation of such expression data will continue as the understanding of virulence, pathogenesis and transmission evolves.
All cells have stress response pathways that help to maintain homeostasis, however it appears that these two lineages of O157 may have diverged just enough that their regulatory pathways are geared for different purposes, ultimately promoting survival in different environments and hosts. It is not clear yet, though research is ongoing, whether LII isolates have lower transmission potential or lower virulence or indeed whether there is enough divergence between the two lineages to consider them as separate. One hypothesis presented in the literature and also supported by the data presented is that LII strains may be more co-evolved as a symbiont of cattle, which promotes its long-term survival in this specific reservoir. For instance, stx2 expression may not be as beneficial in colonization of a bovine host as it has been noted that intestinal receptors for Shiga toxin are found in humans but not cattle [83], while LEE island expression may be very important [84]. Popular theories of pathogen evolution suggest that as a pathogen evolves within finite populations, the pathogen tends to become less virulent (attenuation) to the host thereby promoting though various mechanisms of evolution its own transmission and survival among the populations [85]. This may be exemplified by the differential expression of stress response genes, which could prime or maintain an isolate of E. coli O157 in a genetic state that is able to rapidly respond to conditions the isolate might encounter during transmission from animal to human hosts, through the farm to fork process, thereby increasing its transmission potential.
Methods
Bacterial isolates and growth conditions
A working set of lineage (20 LI and 20 LII) isolates as described in Kim et al. [1] were obtained from A. Benson (University of Nebraska, Dept. Of Food Science and Technology, Lincoln, Neb.). LI isolates 43895, fda518, frik533 and LII isolates ne037, frik2000, frik1985 were chosen at random and utilized in the current analyses. Isolates were grown on LB agar under anaerobic conditions for 12 hours. Previous growth studies noted that these 6 isolates displayed similar growth curves, OD600, and concentration (data not shown). Stationary phase was selected to ensure that all isolates and cultures were at the same stage of growth. Isolates were of the Stx2vha genotype and all exhibited typical O157 phenotype characteristics including acid tolerance, lack of sorbitol fermentation, lack of glucuronidase activity and beta hemolysis on tryptose blood agar (Difco, Sparks, MD) with washed, defibrinated sheep blood (Oxoid, Lenexa, KS). All isolates also displayed the same phenotypes using API20 (bioMerieux, Durham, NC).
Microarray design
Using the transcriptome of E. coli O157:H7 EDL933 an oligonucleotide microarray (~50mer) was designed. Based upon funding available we were able to choose 610 genes [see Additional file 2] including 10 negative control genes derived from pig sequences, which were selected based upon their being associated with virulence or with regulation of virulence genes. Specifications of oligos were based upon various design characteristics such as temperature of melting, 3' location, specificity, lack of repeat nucleotides, etc. [86]. Oligos were synthesized and normalized in concentration by Integrated DNA Technologies Inc. (Coralville, IA). Oligos were resuspended in Epoxide Slide Spotting Solution and printed onto Epoxide Coated Slides (Corning Inc., Corning, NY). Each array consisted of duplicate elements and each slide contained a duplicate array.
Microarray protocol
All procedures were performed according to respective manufacturer protocols. Colonies were resuspended immediately in RNAprotect Bacteria Reagent (Qiagen Inc., Valencia, CA) after they were harvested. Total RNA was extracted using RNeasy Protect Bacteria Mini Kit (Qiagen Inc.) and DNA removed using RNase-Free DNase Set (Qiagen Inc.). RNA was quantified using a nanodrop ND-1000 device (NanoDrop Technologies, Wilmington, DE) and quality confirmed by electrophoresis. RNA was labeled with either CyDye3-dCTP or CyDye5-dCTP (Amersham Biosciences) using the LabelStar kit (Qiagen Inc.) and Random nonamers (Sigma-Aldrich Inc., St. Louis, MO). Labeled cDNA was hybridized to the microarray using Universal Hybridization Solution (Corning Inc.).
Microarray analysis
Each microarray experiment was performed in duplicate and each experiment also had a corresponding dye swap for an added technical replication. As an example of a dye swap design LI is labeled with cy3 and LII is labeled with cy5 in one array and in the second array LI is labeled with cy5 and LII labeled with cy3. Dye swaps are not biological replicates but provide technical replication that accounts for different dye incorporation rates. Images were captured using a Genepix 4000B (Molecular Devices Corporation, Union City, CA) laser scanner and images processed using GenePix 6.0 software (Molecular Devices Corporation). Analysis was performed using Acuity 4.0 software as well as GeneSpring 11.0 software (Agilent Technologies, Palo Alto, CA). Results were compared between the two software packages to assure conformity of results. Slides were normalized using standard settings (ratio based so that the mean of the ratio of means, of all features, were equal to 1.0). All ratios less than 0.1 and greater than 10.0 were excluded, as well as bad, low signal, absent, or unfound features. To obtain our final data provided in Table 1 and Table 2 we required that all arrays, duplicate elements on each array, and these same features on the dye-swap experiments (after mathematical conversion x' = -x) to provide agreement, show significant relevance at the p < 0.05 level, and exhibit at least 1.2 fold regulation. A supplemental dataset was derived for those genes that showed a tendency to be differentially expressed. Usually, the lack of inclusion into the stringent dataset was only based upon the quality of the signal in one of the array or dye swap comparisons. Therefore, these results are provided for information and discussion purposes.
Quantitative PCR
The results of the array were validated using quantitative PCR. Subsets of the regulated genes were chosen at random and primers designed using Primer Select 2.0 software (Applied Biosystems, Foster City, CA). RNA was quantified using NanoDrop system and then using QuantiTect SYBR Green RT-PCR kit (Qiagen Inc.) relative CT was determined with 16s as a control gene, using ABI 7500 Real Time-PCR system (Applied Biosystems).
Functional analysis
HT-GO-FAT software was used to perform the functional GO related analysis. Functional classifications were determined for the regulated genes using HT-GO-FAT and the LIRU8 database. Statistics for higher represented classifications were also determined using HT-GO-FAT. A dedicated Amigo database was also prepared based upon the microarray and the EDL933 transcriptome and can be found at the above URL.
Statistical analysis
Acuity 4.0 built in statistics algorithms were utilized for all statistics related to microarrays. One sample t test was used to determine the significantly regulated genes. Random samples assigned by computer generation. Standard methods were utilized for evaluation of quantitative PCR based upon target gene Ct values (number of cycles of PCR before a threshold of detection is crossed) normalized with the Ct value of an appropriate housekeeping gene (fadD) to compensate for variation in initial RNA and cDNA concentrations. The first normalization procedure provides the initial ΔCt value. The sample ΔCt values were then normalized against the smallest ΔCt value identified in the complete data set, termed ΔΔCt. Finally, the ΔΔCt value for each sample was transformed by the function 2ΔΔCT to produce the final gene expression value for each sample. This method allowed for direct comparison of relative gene expression values between isolates. Gene Ontology related statistics were calculated as described by Al-Shahrour et al [87].
USDA disclaimer
The use of trade, firm, or corporation names in this publication is for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture
References
Kim J, Nietfeldt J, Benson AK: Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci U S A. 1999, 96: 13288-13293. 10.1073/pnas.96.23.13288.
Pradel N, Boukhors K, Bertin Y, Forestier C, Martin C, Livrelli V: Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France. Appl Environ Microbiol. 2001, 67: 2460-2468. 10.1128/AEM.67.6.2460-2468.2001.
Yang Z, Kovar J, Kim J, Nietfeldt J, Smith DR, Moxley RA, Olson ME, Fey PD, Benson AK: Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl Environ Microbiol. 2004, 70: 6846-6854. 10.1128/AEM.70.11.6846-6854.2004.
Barkocy-Gallagher GA, Arthur TM, Siragusa GR, Keen JE, Elder RO, Laegreid WW, Koohmaraie M: Genotypic analyses of Escherichia coli O157:H7 and O157 nonmotile isolates recovered from beef cattle and carcasses at processing plants in the Midwestern states of the United States. Appl Environ Microbiol. 2001, 67: 3810-3818. 10.1128/AEM.67.9.3810-3818.2001.
McNally A, Roe AJ, Simpson S, Thomson-Carter FM, Hoey DE, Currie C, Chakraborty T, Smith DG, Gally DL: Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect Immun. 2001, 69: 5107-5114. 10.1128/IAI.69.8.5107-5114.2001.
Ritchie JM, Wagner PL, Acheson DW, Waldor MK: Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl Environ Microbiol. 2003, 69: 1059-1066. 10.1128/AEM.69.2.1059-1066.2003.
USDA ERS, M. O, N. B: Weighing Incentives for Food Safety in Meat and Poultry. Amber Waves. 2003, USDA ERS, 1: 35-42.http://www.ers.usda.gov/Amberwaves/April03/Features/WeighingIncentives.htm
Uhlich GA, Keen JE, Elder RO: Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol. 2001, 67: 2367-2370. 10.1128/AEM.67.5.2367-2370.2001.
Ryu JH, Kim H, Frank JF, Beuchat LR: Attachment and biofilm formation on stainless steel by Escherichia coli O157:H7 as affected by curli production. Lett Appl Microbiol. 2004, 39: 359-362. 10.1111/j.1472-765X.2004.01591.x.
Ryu JH, Beuchat LR: Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl Environ Microbiol. 2005, 71: 247-254. 10.1128/AEM.71.1.247-254.2005.
Kim SH, Kim YH: Escherichia coli O157:H7 adherence to HEp-2 cells is implicated with curli expression and outer membrane integrity. J Vet Sci. 2004, 5: 119-124.
Cookson AL, Cooley WA, Woodward MJ: The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int J Med Microbiol. 2002, 292: 195-205. 10.1078/1438-4221-00203.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000, 25: 25-29. 10.1038/75556.
Werber D, Fruth A, Buchholz U, Prager R, Kramer MH, Ammon A, Tschape H: Strong association between shiga toxin-producing Escherichia coli O157 and virulence genes stx2 and eae as possible explanation for predominance of serogroup O157 in patients with haemolytic uraemic syndrome. Eur J Clin Microbiol Infect Dis. 2003, 22: 726-730. 10.1007/s10096-003-1025-0.
Ritchie JM, Thorpe CM, Rogers AB, Waldor MK: Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect Immun. 2003, 71: 7129-7139. 10.1128/IAI.71.12.7129-7139.2003.
Ludwig K, Sarkim V, Bitzan M, Karmali MA, Bobrowski C, Ruder H, Laufs R, Sobottka I, Petric M, Karch H, Muller-Wiefel DE: Shiga toxin-producing Escherichia coli infection and antibodies against Stx2 and Stx1 in household contacts of children with enteropathic hemolytic-uremic syndrome. J Clin Microbiol. 2002, 40: 1773-1782. 10.1128/JCM.40.5.1773-1782.2002.
Kimura T, Tani S, Motoki M, Matsumoto Y: Role of Shiga toxin 2 (Stx2)-binding protein, human serum amyloid P component (HuSAP), in Shiga toxin-producing Escherichia coli infections: assumption from in vitro and in vivo study using HuSAP and anti-Stx2 humanized monoclonal antibody TMA-15. Biochem Biophys Res Commun. 2003, 305: 1057-1060. 10.1016/S0006-291X(03)00901-X.
Bonnet R, Souweine B, Gauthier G, Rich C, Livrelli V, Sirot J, Joly B, Forestier C: Non-O157:H7 Stx2-producing Escherichia coli strains associated with sporadic cases of hemolytic-uremic syndrome in adults. J Clin Microbiol. 1998, 36: 1777-1780.
Fuchs S, Muhldorfer I, Donohue-Rolfe A, Kerenyi M, Emody L, Alexiev R, Nenkov P, Hacker J: Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb Pathog. 1999, 27: 13-23. 10.1006/mpat.1999.0279.
Lusetti SL, Drees JC, Stohl EA, Seifert HS, Cox MM: The DinI and RecX proteins are competing modulators of RecA function. J Biol Chem. 2004, 279: 55073-55079. 10.1074/jbc.M410371200.
Pages V, Koffel-Schwartz N, Fuchs RP: recX, a new SOS gene that is co-transcribed with the recA gene in Escherichia coli. DNA Repair (Amst). 2003, 2: 273-284. 10.1016/S1568-7864(02)00217-3.
Shen G, Xu C, Hu R, Jain MR, Nair S, Lin W, Yang CS, Chan JY, Kong AN: Comparison of (-)-Epigallocatechin-3-Gallate Elicited Liver and Small Intestine Gene Expression Profiles Between C57BL/6J Mice and C57BL/6J/Nrf2 (-/-) Mice. Pharm Res. 2005
Bagdasarian M, Bailone A, Angulo JF, Scholz P, Bagdasarian M, Devoret R: PsiB, and anti-SOS protein, is transiently expressed by the F sex factor during its transmission to an Escherichia coli K-12 recipient. Mol Microbiol. 1992, 6: 885-893.
Yasuda T, Morimatsu K, Kato R, Usukura J, Takahashi M, Ohmori H: Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. EMBO J. 2001, 20: 1192-1202. 10.1093/emboj/20.5.1192.
Tyler JS, Mills MJ, Friedman DI: The operator and early promoter region of the Shiga toxin type 2-encoding bacteriophage 933W and control of toxin expression. J Bacteriol. 2004, 186: 7670-7679. 10.1128/JB.186.22.7670-7679.2004.
Plunkett GIII, Rose DJ, Durfee TJ, Blattner FR: Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J Bacteriol. 1999, 181: 1767-1778.
Koudelka AP, Hufnagel LA, Koudelka GB: Purification and characterization of the repressor of the shiga toxin-encoding bacteriophage 933W: DNA binding, gene regulation, and autocleavage. J Bacteriol. 2004, 186: 7659-7669. 10.1128/JB.186.22.7659-7669.2004.
Muniesa M, Blanco JE, De SM, Serra-Moreno R, Blanch AR, Jofre J: Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology. 2004, 150: 2959-2971. 10.1099/mic.0.27188-0.
Miyamoto H, Nakai W, Yajima N, Fujibayashi A, Higuchi T, Sato K, Matsushiro A: Sequence analysis of Stx2-converting phage VT2-Sa shows a great divergence in early regulation and replication regions. DNA Res. 1999, 6: 235-240. 10.1093/dnares/6.4.235.
Iyoda S, Tamura K, Itoh K, Izumiya H, Ueno N, Nagata K, Togo M, Terajima J, Watanabe H: Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol Lett. 2000, 191: 7-10.
Blanch AR, Garcia-Aljaro C, Muniesa M, Jofre J: Detection, enumeration and isolation of strains carrying the stx2 gene from urban sewage. Water Sci Technol. 2003, 47: 109-116.
Mukherjee J, Chios K, Fishwild D, Hudson D, O'Donnell S, Rich SM, Donohue-Rolfe A, Tzipori S: Human Stx2-specific monoclonal antibodies prevent systemic complications of Escherichia coli O157:H7 infection. Infect Immun. 2002, 70: 612-619. 10.1128/IAI.70.2.612-619.2002.
Fraser ME, Fujinaga M, Cherney MM, Melton-Celsa AR, Twiddy EM, O'Brien AD, James MN: Structure of shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J Biol Chem. 2004, 279: 27511-27517. 10.1074/jbc.M401939200.
Donohue-Rolfe A, Kondova I, Oswald S, Hutto D, Tzipori S: Escherichia coli O157:H7 strains that express Shiga toxin (Stx) 2 alone are more neurotropic for gnotobiotic piglets than are isotypes producing only Stx1 or both Stx1 and Stx2. J Infect Dis. 2000, 181: 1825-1829. 10.1086/315421.
Yang SE, Chou CC: Growth and survival of Escherichia coli O157:H7 and Listeria monocytogenes in egg products held at different temperatures. J Food Prot. 2000, 63: 907-911.
Williams RC, Sumner SS, Golden DA: Survival of Escherichia coli O157:H7 and Salmonella in apple cider and orange juice as affected by ozone and treatment temperature. J Food Prot. 2004, 67: 2381-2386.
Whiting RC, Golden MH: Variation among Escherichia coli O157:H7 strains relative to their growth, survival, thermal inactivation, and toxin production in broth. Int J Food Microbiol. 2002, 75: 127-133. 10.1016/S0168-1605(02)00003-X.
Uyttendaele M, Jozwik E, Tutenel A, De ZL, Uradzinski J, Pierard D, Debevere J: Effect of acid resistance of Escherichia coli O157:H7 on efficacy of buffered lactic acid to decontaminate chilled beef tissue and effect of modified atmosphere packaging on survival of Escherichia coli O157:H7 on red meat. J Food Prot. 2001, 64: 1661-1666.
Uyttendaele M, Taverniers I, Debevere J: Effect of stress induced by suboptimal growth factors on survival of Escherichia coli O157:H7. Int J Food Microbiol. 2001, 66: 31-37. 10.1016/S0168-1605(00)00509-2.
Sage JR, Ingham SC: Survival of Escherichia coli O157:H7 after freezing and thawing in ground beef patties. J Food Prot. 1998, 61: 1181-1183.
Ryu JH, Beuchat LR: Influence of acid tolerance responses on survival, growth, and thermal cross-protection of Escherichia coli O157:H7 in acidified media and fruit juices. Int J Food Microbiol. 1998, 45: 185-193. 10.1016/S0168-1605(98)00165-2.
Riordan DC, Duffy G, Sheridan JJ, Whiting RC, Blair IS, McDowell DA: Effects of acid adaptation, product pH, and heating on survival of Escherichia coli O157:H7 in pepperoni. Appl Environ Microbiol. 2000, 66: 1726-1729. 10.1128/AEM.66.4.1726-1729.2000.
Nicholson FA, Groves SJ, Chambers BJ: Pathogen survival during livestock manure storage and following land application. Bioresour Technol. 2005, 96: 135-143. 10.1016/j.biortech.2004.02.030.
McGee P, Bolton DJ, Sheridan JJ, Earley B, Kelly G, Leonard N: Survival of Escherichia coli O157:H7 in farm water: its role as a vector in the transmission of the organism within herds. J Appl Microbiol. 2002, 93: 706-713. 10.1046/j.1365-2672.2002.01752.x.
McClure PJ, Hall S: Survival of Escherichia coli in foods. Symp Ser Soc Appl Microbiol. 2000, 61S-70S.
Maule A: Survival of verocytotoxigenic Escherichia coli O157 in soil, water and on surfaces. Symp Ser Soc Appl Microbiol. 2000, 71S-78S.
Tuttle J, Gomez T, Doyle MP, Wells JG, Zhao T, Tauxe RV, Griffin PM: Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol Infect. 1999, 122: 185-192. 10.1017/S0950268898001976.
Cornick NA, Helgerson AF: Transmission and infectious dose of Escherichia coli O157:H7 in swine. Appl Environ Microbiol. 2004, 70: 5331-5335. 10.1128/AEM.70.9.5331-5335.2004.
Besser TE, Richards BL, Rice DH, Hancock DD: Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol Infect. 2001, 127: 555-560. 10.1017/S095026880100615X.
Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW: Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996, 62: 3094-3100.
Price SB, Wright JC, DeGraves FJ, Castanie-Cornet MP, Foster JW: Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl Environ Microbiol. 2004, 70: 4792-4799. 10.1128/AEM.70.8.4792-4799.2004.
Heimer SR, Welch RA, Perna NT, Posfai G, Evans PS, Kaper JB, Blattner FR, Mobley HL: Urease of enterohemorrhagic Escherichia coli: evidence for regulation by fur and a trans-acting factor. Infect Immun. 2002, 70: 1027-1031. 10.1128/IAI.70.2.1027-1031.2002.
Nakano M, Iida T, Ohnishi M, Kurokawa K, Takahashi A, Tsukamoto T, Yasunaga T, Hayashi T, Honda T: Association of the urease gene with enterohemorrhagic Escherichia coli strains irrespective of their serogroups. J Clin Microbiol. 2001, 39: 4541-4543. 10.1128/JCM.39.12.4541-4543.2001.
Nakano M, Iida T, Honda T: Urease activity of enterohaemorrhagic Escherichia coli depends on a specific one-base substitution in ureD. Microbiology. 2004, 150: 3483-3489. 10.1099/mic.0.27280-0.
Friedrich AW, Kock R, Bielaszewska M, Zhang W, Karch H, Mathys W: Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J Clin Microbiol. 2005, 43: 546-550. 10.1128/JCM.43.2.546-550.2005.
Christensen WB: Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella types. J Bacteriol. 1946, 52: 461-466.
Uhlich GA, Keen JE, Elder RO: Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect Immun. 2002, 70: 395-399. 10.1128/IAI.70.1.395-399.2002.
Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S: The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993, 7: 523-536.
Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P: Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998, 180: 2442-2449.
Lindberg AA, Hellerqvist CG: Rough mutants of Salmonella typhimurium: immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J Gen Microbiol. 1980, 116: 25-32.
Creeger ES, Schulte T, Rothfield LI: Regulation of membrane glycosyltransferases by the sfrB and rfaH genes of Escherichia coli and Salmonella typhimurium. J Biol Chem. 1984, 259: 3064-3069.
Bailey MJ, Hughes C, Koronakis V: In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol Gen Genet. 2000, 262: 1052-1059. 10.1007/PL00008648.
Artsimovitch I, Landick R: The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002, 109: 193-203. 10.1016/S0092-8674(02)00724-9.
Wang L, Jensen S, Hallman R, Reeves PR: Expression of the O antigen gene cluster is regulated by RfaH through the JUMPstart sequence. FEMS Microbiol Lett. 1998, 165: 201-206.
Stevens MP, Hanfling P, Jann B, Jann K, Roberts IS: Regulation of Escherichia coli K5 capsular polysaccharide expression: evidence for involvement of RfaH in the expression of group II capsules. FEMS Microbiol Lett. 1994, 124: 93-98.
Sanderson KE, Stocker BA: Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J Bacteriol. 1981, 146: 535-541.
Nagy G, Dobrindt U, Kupfer M, Emody L, Karch H, Hacker J: Expression of hemin receptor molecule ChuA is influenced by RfaH in uropathogenic Escherichia coli strain 536. Infect Immun. 2001, 69: 1924-1928. 10.1128/IAI.69.3.1924-1928.2001.
Leeds JA, Welch RA: RfaH enhances elongation of Escherichia coli hlyCABD mRNA. J Bacteriol. 1996, 178: 1850-1857.
Landraud L, Gibert M, Popoff MR, Boquet P, Gauthier M: Expression of cnf1 by Escherichia coli J96 involves a large upstream DNA region including the hlyCABD operon, and is regulated by the RfaH protein. Mol Microbiol. 2003, 47: 1653-1667. 10.1046/j.1365-2958.2003.03391.x.
Bailey MJ, Koronakis V, Schmoll T, Hughes C: Escherichia coli HlyT protein, a transcriptional activator of haemolysin synthesis and secretion, is encoded by the rfaH (sfrB) locus required for expression of sex factor and lipopolysaccharide genes. Mol Microbiol. 1992, 6: 1003-1012.
Marolda CL, Valvano MA: Promoter region of the Escherichia coli O7-specific lipopolysaccharide gene cluster: structural and functional characterization of an upstream untranslated mRNA sequence. J Bacteriol. 1998, 180: 3070-3079.
Rahn A, Whitfield C: Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol Microbiol. 2003, 47: 1045-1060. 10.1046/j.1365-2958.2003.03354.x.
Clarke BR, Pearce R, Roberts IS: Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J Bacteriol. 1999, 181: 2279-2285.
Nagy G, Dobrindt U, Schneider G, Khan AS, Hacker J, Emody L: Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect Immun. 2002, 70: 4406-4413. 10.1128/IAI.70.8.4406-4413.2002.
Nagy G, Dobrindt U, Grozdanov L, Hacker J, Emody L: Transcriptional regulation through RfaH contributes to intestinal colonization by Escherichia coli. FEMS Microbiol Lett. 2005, 244: 173-180. 10.1016/j.femsle.2005.01.038.
Grys TE, Siegel MB, Lathem WW, Welch RA: The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect Immun. 2005, 73: 1295-1303. 10.1128/IAI.73.3.1295-1303.2005.
Landini P, Zehnder AJ: The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol. 2002, 184: 1522-1529. 10.1128/JB.184.6.1522-1529.2002.
Monday SR, Minnich SA, Feng PC: A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H- strains. J Bacteriol. 2004, 186: 2319-2327. 10.1128/JB.186.8.2319-2327.2004.
Blomgran R, Zheng L, Stendahl O: Uropathogenic Escherichia coli triggers oxygen-dependent apoptosis in human neutrophils through the cooperative effect of type 1 fimbriae and lipopolysaccharide. Infect Immun. 2004, 72: 4570-4578. 10.1128/IAI.72.8.4570-4578.2004.
Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su LH, Fierke CA, Jackman JE, Raetz CR, Coleman J, Tomoyasu T, Matsuzawa H: Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol. 1999, 31: 833-844. 10.1046/j.1365-2958.1999.01221.x.
Bilge SS, Vary JCJ, Dowell SF, Tarr PI: Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996, 64: 4795-4801.
Cockerill FIII, Beebakhee G, Soni R, Sherman P: Polysaccharide side chains are not required for attaching and effacing adhesion of Escherichia coli O157:H7. Infect Immun. 1996, 64: 3196-3200.
Pruimboom-Brees IM, Morgan TW, Ackermann MR, Nystrom ED, Samuel JE, Cornick NA, Moon HW: Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc Natl Acad Sci U S A. 2000, 97: 10325-10329. 10.1073/pnas.190329997.
an-Nystrom EA, Bosworth BT, Moon HW, O'Brien AD: Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun. 1998, 66: 4560-4563.
Nowak MA, Sasaki A, Taylor C, Fudenberg D: Emergence of cooperation and evolutionary stability in finite populations. Nature. 2004, 428: 646-650. 10.1038/nature02414.
Charbonnier Y, Gettler B, Francois P, Bento M, Renzoni A, Vaudaux P, Schlegel W, Schrenzel J: A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics. 2005, 6: 95-10.1186/1471-2164-6-95.
Al-Shahrour F, az-Uriarte R, Dopazo J: FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004, 20: 578-580. 10.1093/bioinformatics/btg455.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
SD designed the microarray, conceived of the project and wrote the manuscript, HI performed the laboratory experiments.
Electronic supplementary material
12866_2006_243_MOESM1_ESM.xls
Additional File 1: Regulated Genes p < 0.2 and > 0.05, This file and dataset contains 105 genes that failed a criteria for inclusion in the primary dataset and their significance test was not less the p = 0.05. (XLS 28 KB)
12866_2006_243_MOESM2_ESM.xls
Additional File 2: All genes contained in the Escherichia coli O157:H7 virulence array, This file provides a list of all of the genes contained in the 610 gene virulence gene O157:H7 array. (XLS 42 KB)
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Dowd, S.E., Ishizaki, H. Microarray based comparison of two Escherichia coli O157:H7 lineages. BMC Microbiol 6, 30 (2006). https://doi.org/10.1186/1471-2180-6-30
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-6-30