Abstract
Background
Rhizobium grahamii belongs to a new phylogenetic group of rhizobia together with Rhizobium mesoamericanum and other species. R. grahamii has a broad-host-range that includes Leucaena leucocephala and Phaseolus vulgaris, although it is a poor competitor for P. vulgaris nodulation in the presence of Rhizobium etli or Rhizobium phaseoli strains. This work analyzed the genome sequence and transfer properties of R. grahamii plasmids.
Results
Genome sequence was obtained from R. grahamii CCGE502 type strain isolated from Dalea leporina in Mexico. The CCGE502 genome comprises one chromosome and two extrachromosomal replicons (ERs), pRgrCCGE502a and pRgrCCGE502b. Additionally, a plasmid integrated in the CCGE502 chromosome was found. The genomic comparison of ERs from this group showed that gene content is more variable than average nucleotide identity (ANI). Well conserved nod and nif genes were found in R. grahamii and R. mesoamericanum with some differences. R. phaseoli Ch24-10 genes expressed in bacterial cells in roots were found to be conserved in pRgrCCGE502b. Regarding conjugative transfer we were unable to transfer the R. grahamii CCGE502 symbiotic plasmid and its megaplasmid to other rhizobial hosts but we could transfer the symbiotic plasmid to Agrobacterium tumefaciens with transfer dependent on homoserine lactones.
Conclusion
Variable degrees of nucleotide identity and gene content conservation were found among the different R. grahamii CCGE502 replicons in comparison to R. mesoamericanum genomes. The extrachromosomal replicons from R. grahamii were more similar to those found in phylogenetically related Rhizobium species. However, limited similarities of R. grahamii CCGE502 symbiotic plasmid and megaplasmid were observed in other more distant Rhizobium species. The set of conserved genes in R. grahamii comprises some of those that are highly expressed in R. phaseoli on plant roots, suggesting that they play an important role in root colonization.
Similar content being viewed by others
Background
A large proportion of Rhizobium, Sinorhizobium and Agrobacterium genomes is located in extrachromosomal replicons (ERs) [1]. ERs play adaptive roles in soil bacteria [1, 2] and are enriched in particular classes of genes involved in pathogenesis, symbiosis, metabolism and antibiotic resistance. Two types of ERs have been recognized, chromids [3] and plasmids. The term chromid has been recently proposed to refer to extrachromosomal elements that carry “essential” genes and have similar G + C content and codon usage as chromosomes [3]. Nodulation and nitrogen fixation genes are located on symbiotic plasmids (pSyms) in Rhizobium, Sinorhizobium, Burkholderia and in some Mesorhizobium species [1, 4] but in some cases these genes may reside in chromids. pSyms determine the symbiotic capacities in rhizobia and may be transferred among bacteria. The term symbiovar refers to host specificity. A single symbiovar may be present in different rhizobial species while a single species may exhibit different symbiovars [5]. Well conserved pSyms have been found respectively in rhizobia nodulating Phaseolus vulgaris corresponding to symbiovars (sv) tropici or phaseoli [6, 7], and we wondered if conserved pSyms are a rule or an exception in rhizobia [8]. An “acaciella” symbiotic plasmid seems to be contained in the related Ensifer (also named Sinorhizobium) species, E. mexicanum and E. chiapanecum[9]. Symbiovar mimosae is found in the related species Rhizobium etli and Rhizobium phaseoli and symbiovar meliloti is the most widespread found in several Ensifer or Mesorhizobium species [5].
A novel phylogenetic group in rhizobia is now recognized for Rhizobium grahamii, Rhizobium mesoamericanum[10], Rhizobium endophyticum[11], Rhizobium sp. OR191 [12], Rhizobium sp. LPU83 [13], Rhizobium tibeticum[14] and Rhizobium sp. CF122 [15]. R. grahamii, R. mesoamericanum, Rhizobium sp. OR191 and Rhizobium sp. LPU83 are broad host range bacteria. They are capable of forming nodules on P. vulgaris although they are not fully efficient or competitive. R. endophyticum is non-symbiotic as it lacks a symbiotic plasmid [11]. R. grahamii and R. mesoamericanum are closely related species. R. grahamii strains have been isolated from nodules of Dalea leporina, Leucaena leucocephala and from Clitoria ternatea growing naturally as weeds in agricultural bean fields in central Mexico [16]; or from P. vulgaris nodules. R. mesoamericanum strains have been isolated from Mimosa pudica in Costa Rica, French Guiana and New Caledonia [17–19] and from P. vulgaris nodules in Los Tuxtlas rain forest in Mexico [10]. Seemingly, R. mesoamericanum strains were introduced to New Caledonia together with their mimosa hosts [18], maybe on seeds as described before for other rhizobia [20].
Genome sequences are available for R. grahamii, R. mesoamericanum[10, 21] and Rhizobium sp. CF122 [15]. Whole genome comparison of related species would provide clues on the divergence mechanisms involved in speciation. Numerical estimates such as average nucleotide identity (ANI) and genome conservation estimates have been found useful to globally compare genomes [22], and we use them here. In this work we present 1) an improved version of the R. grahamii CCGE502 genome, 2) a genomic comparison of ERs in related rhizobia, 3) evidence of the natural integration of an ER in the R. grahamii CCGE502 chromosome, and 4) an evaluation of the conjugative transfer ability of the R. grahamii CCGE502 symbiotic plasmid and megaplasmid to other Rhizobium species.
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this work are described in Table 1. Rhizobium and Agrobacterium tumefaciens strains were grown at 30°C on PY medium [23]. Escherichia coli cells were grown on LB medium [24] at 37°C. When required, antibiotics were added at the following concentrations (in μg ml-1): nalidixic acid (Nal) 20, spectinomycin (Sp) 75, kanamycin (Km) 15, neomycin (Nm) 60, rifampicin (Rif) 100, streptomycin (Sm) 50, gentamicin (Gm) 30.
Bacterial matings
Conjugation of E. coli and Rhizobium was done biparentally, using E. coli S17-1 as the donor [30]. Transconjugants were selected with the appropriate antibiotics. Conjugation experiments were performed on PY plates at 30°C using overnight cultures. Donors and recipients were mixed in a 1:2 ratio and incubated overnight. The mixtures were serially diluted and plated on suitable selective media. To study conjugative transfer of the R. grahamii CCGE502 pSym, it was tagged (see below).
PCR amplification and cloning
The oligonucleotides used in this study were purchased from Unidad de Síntesis Química, IBT-UNAM. PCR amplification was carried out with recombinant TaqDNA polymerase (Invitrogen) and PFU (Fermentas) as specified by the manufacturer. PCR products were purified with the High Pure PCR Purification Kit (Roche). Vectors were purified with the High Pure Plasmid Isolation Kit (Roche). T4 polynucleotide ligase was used as indicated by the manufacturer (Fermentas).
Genetic manipulations
The symbiotic plasmid pRgrCCGE502a was tagged with a Not I-cassette carrying Gm and green fluorescence protein (Gm-GFP). GFP (gfpmut3*) protein was from plasmid pJBA28 [35] that harbors a Not I cassette with a Ssp I site. Gm was from pBSL142 [36] and cloned at Ssp I site. A fragment corresponding to RGCCGE502_32801 was amplified with PFU using Fw_32801 and Rv_32801 and cloned at the Sma I site of pK18mob:sacB obtaining pMJAM01. This plasmid was digested with Not I and the Not I- (Gm-GFP) cassette was ligated to obtain pMJAM02 in E. coli S17-1 that was mated with R. grahamii CCGE502. Transconjugants were plated on PY Gm and Nm, selecting single recombinants. These colonies were checked by PCR with Fw_ext_32801 and Rv_ext_32801, combined with internal primers of the vector. Once the orientation of the insert was verified, one colony was grown to stationary phase and plated on PY sucrose and Gm. Finally the colonies obtained were checked by PCR to confirm double recombination and were named R. grahamii CCGE502a:GFP.
A traI mutant was obtained by deletion of a 428 base pair (bp) internal fragment of this gene (locus tag RGCCGE502_33766, size 621 bp). Two fragments of the gene were amplified. The first 265-bp fragment was amplified with PFU using Fw_33766_1 and Rv_33766_1. The second 272-bp fragment was amplified with Fw_33766_2 and Rv_33766_2. Fragment 1 was cloned blunt-ended in Sma I-digested pK18mob:sacB to obtain pMJAM03; and fragment 2 was cloned as a BamH I-Hind III fragment in the same vector to obtain pMJAM04 where both fragments are in the same orientation. The final construction was transformed into E. coli S17-1. The procedure to obtain the mutant in R. grahamii CCGE502 was the same as described above: first, transconjugants were plated on PY Nm, to select single recombinants which were used to perform PCR reactions to detect deleted derivative strains. External primers to verify insertions were Fw_ext_traI and Rv_ext_traI. Fragments amplified with these primers were 1500 bp and 1001 bp for wild type strain and deleted mutants, respectively. The mutant was designated R. grahamii CCGE502ΔtraI.
The symbiotic plasmid pRgrCCGE502a carrying the traI deletion was tagged by insertion of pG18mob2 [31] in the nodC gene. An internal fragment of nodC was amplified with PFU, employing Fw_nodC and Rv_nodC and cloned blunt-end in the SmaI site of pG18mob2 to obtain pMJAM05. The construction was transformed into S17-1 and transferred by mating to R. grahamii CCGE502ΔtraI. Transconjugants were verified by PCR combining Fw_ext_nodB or Rv_ext_nodC and M13 primers. The resultant strain was designated R. grahamii CCGE502ΔtraI::nodC.
Megaplasmid pRgrCCGE502b was tagged by insertion of plasmid pK18mob:sacB[32] in an intergenic region between RGCCGE502_28748 and RGCCGE502_28753. A 692-bp fragment was amplified with PFU, Fw_28753 and Rv_28753 and cloned blunt-end in the SmaI site of pK18mob:sacB to obtain pMJAM06. The construction was transformed into S17-1 and transferred by mating to R. grahamii CCGE502. Recombinants were verified by PCR combining Fw_ext_28753 or Rv_ext_28753 and M13 primers. The strain was designated R. grahamii CCGE502b:Km.
N-acyl-homoserine-lactone (AHL) detection
Autoinducers were detected by thin-layer chromatography (TLC) with the reporter plasmid pZLR4 [37] that contains the traR gene and traG::lacZ reporter fusion from pTiC58, independently cloned into the broad-host-range vector pBBR1MCS5 [38]. Extracts from R. grahamii CCGE502 and mutants were prepared from 5-ml cultures grown in PY medium. Briefly, cultures were extracted twice with equal volumes of ethyl acetate, bacteria were removed by centrifugation and supernatants evaporated to dryness. Residues from 5-ml cultures were dissolved in 50–100 μl of ethyl acetate.
Eckhardt gel analysis
This was performed as described [39], with liquid early-exponential-phase cultures in horizontal gels with sodium dodecyl sulfate in agarose.
Gap closure
Gap filling was done over the contigs of the sequence assembly AEYE01000000 [40]. Ten contigs corresponding to symbiotic plasmid pRgrCCGE502a and sixteen corresponding to megaplasmid pRgrCCGE502b were selected. A new assembly was done with Phrap assembler using the 454 pyrosequencing mate-paired reads and edited with Consed (23.0) program [41]. A total of 1920 contigs were obtained and compared with the scaffolds corresponding to pRgrCCGE502a and pRgrCCGE502b of the original assembly. Contigs that overlapped with the pRgrCCGE502a and pRgrCCGE502b scaffolds were selected and analyzed at their ends to obtain the sequence that protruded into the gap region. Those protruding sequences were edited manually to fill the scaffold gaps. The complete pRgrCCGE502a and pRgrCCGE02b sequences were aligned with Illumina reads using Consed to verify the coverage of the new molecules. In some cases these processes located small contigs (corresponding to IS or repetitive sequences) to close a gap. A final annotation of the new version AEYE02000000 was performed by the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP). The replicons gave an estimated genome size of 7,156 kbp.
Sequence comparisons
Average nucleotide identity (ANI) between sequences and sequence conservation was calculated with JSpecies software [22].
Phylogenetic inference
Multiple sequence alignments were performed with CLUSTAL_X version 1.83 [42] and manually checked with BioEdit [43]. Best-fit models of sequence evolution were selected for each gene with ProtTest 2.4, using the Akaike information criterion [44]. Maximum-likelihood phylogenies were constructed with PhyML 3 using subtree pruning and regrafting moves to improve tree topology [45]. Support for tree nodes was evaluated by the Shimodaira–Hasegawa-like approximate likelihood-ratio test implemented in PhyML.
Results
The genome of R. grahamii CCGE502 consists of three circular replicons, one chromosome and two ERs: one megaplasmid and a symbiotic plasmid. The first draft sequence [40] consisted of ten contigs for the symbiotic plasmid pRgrCCGE502a and sixteen corresponding to the megaplasmid pRgrCCGE502b. The version described in this paper is version AEYE02000000.
Chromosome
The ca. 5,400-kbp chromosome of R. grahamii CCGE502 is the largest reported to date in Rhizobium. A genomic island of ca. 1,073 kbp that may have originated from the integration of a plasmid or an Integrative and Conjugative Element (ICE) [46] may account for its large size. Interestingly, this island has 57.1% G + C content, lower than the rest of the chromosome (59.7%) and the megaplasmid pRgrCCGE502b (59.1%), and more comparable to that of the symbiotic plasmid pRgrCCGE502a (57.4%). It is not similar to any known sequenced plasmid, and has a mosaic structure with genes resembling many different bacteria. It contains a repABC operon and a complete set of genes for a type IV secretion system. According to the latest classification of plasmid transfer systems proposed by Ding et al.[47] and based on the TraA relaxase and the TraG coupling protein phylogenies, the integrated replicon contains a type IVB rhizobial plasmid secretion system. However, the transfer mechanism of this new group still remains unclear. The chromosomal island encodes proteins related to chemotaxis, DNA metabolism and ABC transporters, among others. It is interesting to note that the location of the homologous genes in other bacteria is variable, they may be in plasmids or chromosomes. A BLASTN comparison of the R. grahamii CCGE502 chromosome with those of R. mesoamericanum STM3625, Rhizobium tropici CIAT 899 and R. etli CFN42 is shown in Figure 1A. Usually, the GC skew in bacterial chromosomes shows a bias toward G over the leading strand while the bias is to C on the lagging strand and indicates the origin of replication and the ending site [48]. In the R. grahamii chromosome the distinct GC skew indicates that the genomic island is a recent insertion. In order to validate that this integration is not an artifact of the assembly, we tagged the island by the insertion of a suicide vector containing a homologous region, to transfer the island to an A. tumefaciens free plasmid, but no transfer was detected. We also performed a Southern blot using a probe directed to the genomic island and hybridized a membrane of an Eckhardt gel. A signal was observed in the wells of the gel but not in the plasmids bands (not shown). Finally we did a PCR reaction employing primers outside and inside the genomic island and obtained a product of the expected size (not shown). Except for the genomic island, the R. grahamii chromosome is conserved with other rhizobial chromosomes (Figure 1A, Additional file 1: Table S1).
Genomic comparison of R. grahamii and other rhizobia. A) Chromosomal alignment of R. grahamii and other rhizobial chromosomes. Each replicon was split in silico in 10 kbp fragments and aligned by BlastN with R. grahamii CCGE502 chromosome as a reference (internal black circle with size labels). When 70% of identity in each fragment with the reference was found, a color line was used to indicate the conservation in the genomes. The colors used are: blue for R. etli CFN42, green for R. tropici CIAT 899 and red for R. mesoamericanum STM3625. The black circle with peaks represents the G + C content, and the outside internal circle the GC skew of R. grahamii CCGE502 chromosome. Black arrows indicate the location of the genomic island. B) ANI and C) conserved DNA values between replicons of R. grahamii CCGE502 and R. mesoamericanum CCGE501 (blue) or STM3625 (red).
Megaplasmid pRgrCCGE502b
The megaplasmid of R. grahamii CCGE502 appears to conform to the definition of a chromid; it had a similar G + C content as the chromosome (59.1% and 59.7% respectively), a plasmid-type maintenance and replication systems (repABC) and a group of genes present in others chromids such as pRetCFN42e from R. etli CFN42 [3]. However we have not yet tried to cure this replicon from the bacteria. In pRetCFN42e, Landeta et al. [49] analyzed a set of genes, most of which were also present in pRgrCCGE502b such as hutUGHI for histidine degradation; pcaDCHGB for protocatechuic acid degradation; agpA, agaL1 and agaL2, involved in melobiose consumption; nadABC involved in the initial steps of NAD biosynthesis, cls responsible of cardiolipin synthesis, thiMED participating in the thiamine salvage pathway, cobFGHIJKLM involved in cobalamin biosynthesis (vitamin B12) and cyoABCDE, encoding the cytochrome O terminal oxidase. Additionally, on pRgrCCGE502b we found minCDE genes, involved in septum formation and actP for copper extrusion. Two essential genes required for growth in rich medium are present in pRetCFN42e, RHE_PE00001 and RHE_PE00024. R. grahamii showed an ortholog 68% identical to RHE_PE00001 also on pRgrCCGE502b, but RHE_PE00024 was not found in the genome. All these genes are present in single copy in each genome. Furthermore, some of the R. phaseoli Ch24-10 genes found to be highly expressed in maize or bean rhizosphere [1] were found to be conserved in pRgrCCGE502b (e.g. cyoAB, hutUGH, apgA, cls, cobG and actP).
Most of the genes analyzed that were located on pRgrCCGE502b gave high identities, between 60 and 90%, to Rhizobium sp. CF122 and some with R. mesoamericanum STM625 gene sequences [21]. CF122 was isolated from Populus deltoides rhizosphere in North Carolina [15]. The ANI values we estimated for the genomes of Rhizobium sp. CF122 and R. grahamii or R. mesoamericanum were 87.5% and 87.8%, respectively. CF122 should correspond to a species other than R. grahamii or R. mesoamericanum considering its low ANI values with the reported related species.
ANI values between the megaplasmids in the “grahamii” group was nearly 85% (Figure 1B) but the percentage of conserved DNA between these replicons was around 14% (Figure 1C). ANI values of the corresponding chromosomes were estimated to be around 86% and conserved DNA around 75% (Figure 1B and C). In comparison with the R. etli CFN42 chromid, pRetCFN42e, these values were 83.28% and 13.75% (Additional file 2: Table S2).
Symbiotic plasmid pRgrCCGE502a
Symbiosis genes were found on plasmid pRgrCCGE502a, most were located in a 108 kbp region. nodABC genes, responsible for synthesis of the Nod factor core, were located upstream of nodSUIJHPQ. NodS is an N- methyltransferase and NodU is a carbamoyltransferase responsible for adding substitutions at the C-2 and C-6 position, respectively, on the non-reducing N- acetyl-d-glucosamine of the Nod factor. nodHPQ gene products are involved in the sulfation of C-6 of the reducing terminus [50, 51] and NodIJ are involved in the export of Nod factors [52, 53]. The R. grahamii pSym also has nodEF-hsnT. NodE and NodF are involved in the synthesis of unsaturated fatty acids [54] and HsnT is an acyltransferase of non specified function. Based on the nod genes found, R. grahamii Nod factor structure was predicted as a chitin backbone of N- acetylglucosamine residues N- acylated with polyunsaturated fatty acids, N- methylated at the C-2 nonreducing terminal and carbamoylated at C-6 of the same residue. At the reducing end this Nod factor may be substituted at the C-6 position with sulfate.
The symbiotic plasmids most similar to pRgrCCGE502a were those from R. mesoamericanum strains. A comparison of nod genes revealed that R. grahamii CCGE502 and R. mesomericanum STM3625 have almost the same nodulation gene products, ranging from 69% to 99% amino acid similarity (Figure 2). Despite this similarity, some differences were observed in overall pSym gene content as well as in individual nod genes (Figure 1C, Figure 2). R. mesoamericanum STM3625 lacks nodEF-hsnT but harbors two copies of nodA and three copies of nodD, while R. grahamii only presented one nodA and two nodD gene copies. R. grahamii had two nodO and one nodM gene copies located distant to the sym cluster. They encode a Ca-binding protein that is thought to form cation-specific channels in plant membranes [55] and a glucosamine 6-phosphate synthase, respectively. R. mesoamericanum STM3625 also has two nodO and one nodM gene copies; nodO2 and nodM showed an identical genetic context, while nodO1 is found in a different genetic context.
Alignment of symbiotic plasmids of R. grahamii CCGE502 (pRgrCCGE502a) and R. mesoamericanum STM3625 (pRmeSTM3625 2). Numbers indicate nucleotide positions and arrows the open reading frames in each replicon. Red and yellow lines indicate conserved regions with the same direction. Yellow lines show conserved symbiosis regions including nif, fix and nod genes. Blue lines indicate inverted conserved regions.
In relation to nif/fix genes, a complete set of genes for nitrogen fixation were found in R. grahamii. Some repeated genes, such as nifQ and nifW were also found. nifW had not been found in other Rhizobium species. There were two copies in both R. grahamii and R. mesoamericanum STM3625. Moreover, RGCCGE502_32751 (nifW1) had 92% similarity with BNN_260005 from R. mesoamericanum strain STM3625, and RGCCGE502_33006 (nifW2) had 98% similarity with BNN_270058 from R. mesoamericanum strain STM3625. nifQ was located next to nifW genes in R. grahamii and in R. mesoamericanum STM3625. nifW has an unknown function while nifQ is implicated in the processing of molybdenum, specifically for the biosynthesis of the iron-molybdenum cofactor of the nitrogenase. There are more nif genes in R. grahamii and R. mesoamericanum than in E. meliloti or R. leguminosarum sv. viciae (Table 2).
R. grahamii and R. mesoamericanum symbiotic plasmids showed an ANI of 94.54% (Table 3). Synteny analysis showed that the pSyms of both species are the most closely related (Figure 2), while only short and fragmented similarities were observed between the pSym of R. grahamii and those of R. tropici CIAT 899 and other species. In spite of the high sequence identity of genes between R. grahamii and R. mesoamericanum, the percentage of conserved DNA was only 42% to 51% (depending on the query sequence) of the total molecule (Table 3). In contrast, pSyms of phaseoli strains Ch24-10, CIAT652 and CFN42 showed higher conservation 88 to 95% (Table 3). Also, the percentage of conserved DNA was 96% among three symbiotic plasmids belonging to sv. tropici.
Phylogenetic analysis of RepB proteins of R. grahamii CCGE502
Rhizobial plasmids have repABC operons involved in their replication and maintenance. RepA and RepB are proteins that participate in active plasmid segregation and RepC is the replication initiator protein [57]. Additional repC gene copies have been found separated from repAB and may have different evolutionary origins [58]. pRgrCCGE502a has one independent repC gene copy located at the nodulation cluster. Four repB gene copies were found, one encoded in the genomic island of CCGE502 chromosome, two in pRgrCCGE502b and one in pRgrCCGE502a (Figure 3). Megaplasmid RepB proteins from R. grahamii and R. mesoamericanum were closely related (Figure 3, filled and empty circles) as well as those of the symbiotic plasmids respectively (Figure 3, stars). RepB of R. etli pRetCFN42a (YP_471770.1) was related to the corresponding sequences from the symbiotic plasmids in the “grahamii” group (Figure 3, stars). In the symbiotic plasmids, repABC operons were located next to Mating Pair Formation (Mpf) and DNA transfer and replication (Dtr) system genes.
Maximum likelihood phylogeny of RepB proteins. LG + I + G + F was used as model of amino acid substitution. Labels indicate the replicon and the GenBank accession numbers. Squares indicate proteins with genes found in symbiotic plasmids, circles indicate RepB of R. grahamii and R. mesoamericanum megaplasmids: filled circles specify proteins encoded by genes organized in a repABC operon and empty circles specify RepB proteins encoded in a repAB operon. Stars indicate proteins of R. grahamii and R. mesoamericanum encoded in symbiotic plasmids, together with RepB of pRetCFN42a. The arrow indicates the chromosomal RepB. Numbers close to tree nodes indicate branch support evaluated by the Shimodaira–Hasegawa-like approximate likelihood-ratio test (only values higher than 50% are shown). Scale bar, 0.2 amino acid substitutions per site.
The presence of a repB gene localized in the chromosome may be considered as further evidence that this region originated from a plasmid (Figure 3, arrow). It grouped with the corresponding genes from pRL7 of R. leguminosarum sv. viciae and from pRmeSTM3625 3 of R. mesoamericanum STM3625. A phylogenetic analysis of RepC proteins revealed similar results (not shown) to those obtained with RepB phylogenies except that the chromosomal RepC protein grouped with the corresponding protein from pRetCFN42d of R. etli.
Conjugative transfer of the symbiotic plasmid and megaplasmid of R. grahamii CCGE502
The organization of the trb cluster (Mpf proteins) and tra cluster (Dtr proteins) is identical in R. grahamii CCGE502 and R. etli CFN42 (identities of 95%), only differing in that cinR is present in pRetCFN42a but absent in the symbiotic plasmid pRgrCCGE502a. The high similarity among the conjugative transfer genes could suggest a similar regulation of plasmid transfer. In R. etli CFN42, three genes present in pRetCFN42a are necessary for plasmid transfer dependent on quorum sensing: traI, N- acyl-homoserine synthase, cinR and traR, both encoding transcriptional regulators [25]. Notably, mobilization of pRetCFN42d (pSym) depends on its cointegration with pRetCFN42a [59]. R. grahamii CCGE502 has traI (RGCCGE502_33766) and traR (RGCCGE502_33821) genes in the symbiotic plasmid. A traI mutant of R. grahamii, CCGE502aΔtraI did not produce AHLs (Figure 4). As Figure 4 shows, an A. tumefaciens GMI9023 transconjugant carrying pRgrCCGE502a:GFP produced all AHLs present in R. grahamii, albeit at a highly reduced level (see below), suggesting that RGCCGE502_33766 is responsible for all the spots detected by TLC.
Thin-layer chromatogram of the AHLs produced by R. grahamii CCGE502 and derivatives. 1) R. grahamii CCGE502 wild type strain; 2) R. grahamii CCGE502aΔtraI; 3) A. tumefaciens GMI9023 (pRgrCCGE502a: GFP); 4) A. tumefaciens GMI9023 (pRgrCCGE502aΔtraI) and 5) A. tumefaciens GMI9023 (negative control). Equal amounts of sample were loaded in each lane, except at lane 3 where the sample was ten-fold concentrated.
The symbiotic plasmid of R. grahamii CCGE502a:GFP could be transferred at a frequency of ca. 10-6 transconjugants per donor cell to the plasmid-free A. tumefaciens GMI9023 strain [28], but this transfer was abolished when the traI-mutant was assessed (fewer than 3.0 × 10-1 transconjugants per donor cell). Thus, we considered that conjugative transfer of pRgrCCGE502a was regulated by quorum sensing as occurs with pRetCFN42a. Although pRgrCCGE502a could be transferred to A. tumefaciens GMI9023, transfer of this pSym to R. mesoamericanum CCGE501, R. etli CFN2001 [25], Sinorhizobium fredii GR64-4 [26], Ensifer meliloti SmA818R [27], R. phaseoli Ch24-10, Rhizobium sp. LPU83 [27] and R. endophyticum CCGE2052 [11] was tried unsuccessfully.
Due to the close relationship of RepC proteins of pRgrCCGE502a and pRetCFN42a (RGCCGE502_33751 and RHE_PA00182), we considered that they could be incompatible. Nevertheless a plasmid cured strain (without pRetCFN42a and pRetCFN42d) also was unable to act as a recipient. Furthermore, pRgrCCGE502a:GFP could not be mobilized from the A. tumefaciens transconjugants. Remobilization experiments were done either from GMI9023 (pRgrCCGE502a:GFP) or GMI9023 (pRgrCCGE502a:GFP, pRgrCCGE502b:Km) to another GMI9023 (SpR) and no transconjugants were obtained. The production of AHLs in the genomic background of A. tumefaciens is at least ten-fold lower than in R. grahamii (Figure 4) and this event may explain why pRgrCCGE502a:GFP could not be transferred from GMI9023. However A. tumefaciens overexpressing the AHLs of R. grahamii, GMI9023 (pRgrCCGE502a:GFP, pBBR1MCS2::traI) was not able to mobilize the symbiotic plasmid, indicating that additional factors are needed. Some of these factors could be encoded in the chromosome and thus they are not present when transfer is assayed from A. tumefaciens carrying the plasmids of R. grahamii as donor.
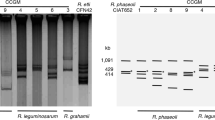
By triparental conjugation (using pRK2013 as helper) megaplasmid pRgrCCGE502b:Km was transferred to A. tumefaciens GMI9023 or GMI9023 (pRgrCCGE502a:GFP) but it could not be transferred to Rhizobium species such as R. etli CFN42. Figure 5 shows the plasmid profile of R. grahamii wild type strain and A. tumefaciens GMI9023 carrying pRgrCCGE502a or pRgrCCGE502b or both plasmids.
Plasmid profiles in Eckhardt gels. 1) R. grahamii CCGE502, 2) A. tumefaciens GMI9023, 3) A. tumefaciens GMI9023 (pRgrCCGE502a: GFP), 4) A. tumefaciens GMI9023 (pRgrCCGE502b:Km), 5) A. tumefaciens GMI9023 (pRgrCCGE502a: GFP, pRgrCCGE502b:Km), 6) R. grahamii CCGE502a: GFP and 7) R. grahamii CCGE502b:Km. Ccc DNA: closed circular chromosome of A. tumefaciens GMI9023.
Discussion and conclusions
When comparing genomes from closely related rhizobial species (e.g. R. tropici and R. rhizogenes or R. leguminosarum and R. etli), it was observed that there is a larger degree of conservation in the chromosomes than in the ERs [3, 60]. We confirmed here a high degree of conservation between the chromosomes of strains in the “grahamii” group, namely R. grahamii CCGE502, R. mesoamericanum CCGE501 and STM3625, as well as Rhizobium sp. CF122. However, in other cases a larger degree of nucleotide conservation has been observed in the symbiotic plasmids (e.g. symbiotic plasmids from the tropici or phaseoli symbiovars) than in chromosomes. In R. grahamii and R. mesoamericanum we observed the largest nucleotide identity in pSyms (ANI around 94%), but not as large as among tropici and phaseoli symbiotic plasmids with ANI of 99 or 98% respectively (Table 3). The conservation of pSyms may be explained by the lateral transfer of a successful plasmid (epidemic plasmid in terms of Souza et al.[61]) or a wandering plasmid among different rhizobial lineages [62] or from being a recently evolved replicon. In the case of the phaseoli plasmids we favored the latter explanation [4, 62–64]. Anyhow, it seems reasonable to consider that limited replicon transfer among related species would lead to an isolated evolutionary history linked to a single genomic background. The phaseoli and tropici plasmids have been found to be conjugative with a high frequency of transfer among rhizobia [65], and the “phaseoli” pSym is found in distantly related species such as R. giardinii or R. gallicum[66]. In contrast we were unable to transfer R. grahamii ERs to other rhizobia. It is worth noting that tropici symbiotic plasmids are more conserved than phaseoli ones, and both are more conserved than the grahamii group pSyms. It is tempting to suggest that genome conservation among distinct species is related to transferability. On the other hand, transfer of plasmids to novel hosts can also detonate their evolution by picking up new genetic information (that would affect the genomic content) from other genomic backgrounds. We do not know if in natural habitats or in the presence of a microbial community, the lack of transferability of R. grahamii ERs holds true. Besides, the limited conservation of pSyms among R. grahamii and R. mesoamericanum suggests that they are not frequently interchanged among these species. Transfer of the R. grahamii symbiotic plasmid to Agrobacterium was dependent on quorum sensing, a mechanism that regulates transfer of plasmids in rhizobia [25, 67] and agrobacteria [68, 69]. This lack of ER flow and existence of a genetic barrier could be due to different mechanisms, such as DNA restriction/methylation systems or to surface or entry exclusion systems. Surface exclusion at the level of formation of stable mating aggregates and entry exclusion seem to inhibit conjugation in a later step of the mating aggregate [70, 71]. Limited transfer may be due to a system similar to CRISPR/Cas, an adaptive immunity system found in Archaea and bacteria that eliminates virus or plasmids in a new host [72, 73]. These possibilities deserve further research.
Putative chromids (megaplasmids) in the grahamii group have a lower percentage of gene content conservation than the chromosomes and symbiotic plasmids, in spite of their fairly high ANI values (Figure 1B and C). Considering the conserved genomic content in chromosomes, symbiotic plasmids and putative chromids in the grahamii group, there clearly are three different degrees of conservation (Figure 1C). We suggest a layout where the rhizobial genome is a 3 gear genome with different rates of change in each of the replicon types. In animals and plants, different regions of the genome exhibit variable levels of genetic divergence between populations (reviewed in Nosil et al.[74]).
The extrachromosomal replicons of R. grahamii CCGE502 were related to those from R. mesoamericanum. An exception is the plasmid integrated in the R. grahamii chromosome for which no equivalent plasmid was found in R. mesoamericanum or in other rhizobia. However some common genes were found in the R. grahamii integrated replicon and in other Rhizobium species. ER organization plasticity was reported previously in rhizobia with the integration of plasmids or megaplasmids into the chromosome [75, 76]. This seems to have occurred in R. grahamii CCGE502 as we report here.
It is noteworthy that some of the genes highly expressed in R. phaseoli Ch24-10 when colonizing roots were found to be conserved in R. grahamii CCGE502 and do not seem to constitute a single genomic island, instead they were patchily distributed in pRgrCCGE502b. Such genes may have an important role in root colonization and seem to have been preserved during rhizobial divergence.
Availability of supporting data
The data set supporting the results of this article is available in the Treebase repository, http://treebase.org/treebase-web/search/study/summary.html?id=14994.
References
López-Guerrero MG, Ormeño-Orrillo E, Acosta JL, Mendoza-Vargas A, Rogel MA, Ramírez MA, Rosenblueth M, Martínez-Romero J, Martínez-Romero E: Rhizobial extrachromosomal replicon variability, stability and expression in natural niches. Plasmid. 2012, 68: 149-158. 10.1016/j.plasmid.2012.07.002.
Heuer H, Smalla K: Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev. 2012, 36: 1083-1104. 10.1111/j.1574-6976.2012.00337.x.
Harrison PW, Lower RP, Kim NK, Young JP: Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol. 2010, 18: 141-148. 10.1016/j.tim.2009.12.010.
Wang ET, Van Berkum P, Sui XH, Beyene D, Chen WX, Martínez-Romero E: Diversity of rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp. nov. Int J Syst Bacteriol. 1999, 49: 51-65. 10.1099/00207713-49-1-51.
Rogel MA, Ormeño-Orrillo E, Martínez Romero E: Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol. 2011, 34: 96-104. 10.1016/j.syapm.2010.11.015.
González V, Acosta JL, Santamaría RI, Bustos P, Fernández JL, Hernández González IL, Díaz R, Flores M, Palacios R, Mora J, Dávila G: Conserved symbiotic plasmid DNA sequences in the multireplicon pangenomic structure of Rhizobium etli. Appl Environ Microbiol. 2010, 76: 1604-1614. 10.1128/AEM.02039-09.
Ormeño-Orrillo E, Menna P, Almeida LG, Ollero FJ, Nicolas MF, Pains Rodrigues Ribeiro Vasconcelos AT, Megías M, Hungria M, Martínez-Romero E: Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genomics. 2012, 13: 735-10.1186/1471-2164-13-735.
Martínez E, Ormeño E, Rogel MA, González V, Acosta JL, Martínez J: 21st North American Nitrogen Fixation Conference: 13–18 June 2010. Trends in rhizobial evolution: conservation of symbiotic plasmids, exception or rule. 2010, Columbia, Missouri, U.S.A, 8-
Rincón-Rosales R, Lloret L, Ponce E, Martínez-Romero E: Rhizobia with different symbiotic efficiencies nodulate Acaciella angustissima in Mexico, including Sinorhizobium chiapanecum sp. nov. which has common symbiotic genes with Sinorhizobium mexicanum. FEMS Microbiol Ecol. 2009, 67: 103-117. 10.1111/j.1574-6941.2008.00590.x.
López-López A, Rogel-Hernández MA, Barois I, Ortiz Ceballos AI, Martínez J, Ormeño-Orrillo E, Martínez-Romero E: Rhizobium grahamii sp. nov., from nodules of Dalea leporina, Leucaena leucocephala and Clitoria ternatea, and Rhizobium mesoamericanum sp. nov., from nodules of Phaseolus vulgaris, siratro, cowpea and Mimosa pudica. Int J Syst Evol Microbiol. 2012, 62: 2264-2271. 10.1099/ijs.0.033555-0.
López-López A, Rogel MA, Ormeño-Orrillo E, Martínez-Romero J, Martínez-Romero E: Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst Appl Microbiol. 2010, 33: 322-327. 10.1016/j.syapm.2010.07.005.
Eardly BD, Young JP, Selander RK: Phylogenetic position of Rhizobium sp. strain Or 191, a symbiont of both Medicago sativa and Phaseolus vulgaris, based on partial sequences of the 16S rRNA and nifH genes. Appl Environ Microbiol. 1992, 58: 1809-1815.
Torres Tejerizo G, Del Papa MF, Draghi W, Lozano M, Giusti MÁ, Martini C, Salas ME, Salto I, Wibberg D, Szczepanowski R, Weidner S, Schlüter A, Lagares A, Pistorio M: First genomic analysis of the broad-host-range Rhizobium sp. LPU83 strain, a member of the low-genetic diversity Oregon-like Rhizobium sp. group. J Biotechnol. 2011, 155: 3-10. 10.1016/j.jbiotec.2011.01.011.
Hou BC, Wang ET, Li Y, Jia RZ, Chen WF, Gao Y, Dong RJ, Chen WX: Rhizobium tibeticum sp. nov., a symbiotic bacterium isolated from Trigonella archiducis-nicolai (Sirj.) Vassilcz. Int J Syst Evol Microbiol. 2009, 59: 3051-3057. 10.1099/ijs.0.009647-0.
Brown SD, Utturkar SM, Klingeman DM, Johnson CM, Martin SL, Land ML, Lu TY, Schadt CW, Doktycz MJ, Pelletier DA: Twenty-one genome sequences from Pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of Populus deltoides. J Bacteriol. 2012, 194: 5991-5993. 10.1128/JB.01243-12.
Martínez E, Pardo MA, Palacios R, Cevallos MA: Reiteration of nitrogen gene sequences and specificity of Rhizobium in nodulation and nitrogen fixation in Phaseolus vulgaris. J Gen Microbiol. 1985, 131: 1779-1786.
Barrett CF, Parker MA: Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl Environ Microbiol. 2006, 72: 1198-1206. 10.1128/AEM.72.2.1198-1206.2006.
Klonowska A, Chaintreuil C, Tisseyre P, Miche L, Melkonian R, Ducousso M, Laguerre G, Brunel B, Moulin L: Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol Ecol. 2012, 81: 618-635. 10.1111/j.1574-6941.2012.01393.x.
Mishra RP, Tisseyre P, Melkonian R, Chaintreuil C, Miche L, Klonowska A, González S, Bena G, Laguerre G, Moulin L: Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol Ecol. 2012, 79: 487-503. 10.1111/j.1574-6941.2011.01235.x.
Pérez-Ramírez NO, Rogel MA, Wang E, Castellanos JZ, Martínez-Romero E: Seeds of Phaseolus vulgaris bean carry Rhizobium etli. FEMS Microbiol Ecol. 1998, 26: 289-296.
Moulin L, Mornico D, Melkonian R, Klonowska A: Draft genome sequence of Rhizobium mesoamericanum STM3625, a nitrogen-fixing symbiont of Mimosa pudica isolated in French Guiana (South America). Genome Announc. 2013, 1: e00066-12.
Richter M, Rosselló-Mora R: Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009, 106: 19126-19131. 10.1073/pnas.0906412106.
Noel KD, Sanchez A, Fernández L, Leemans J, Cevallos MA: Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J Bacteriol. 1984, 158: 148-155.
Miller JH: Experiments in molecular genetics. 1972, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press
Tun-Garrido C, Bustos P, González V, Brom S: Conjugative transfer of p42a from Rhizobium etli CFN42, which is required for mobilization of the symbiotic plasmid, is regulated by quorum sensing. J Bacteriol. 2003, 185: 1681-1692. 10.1128/JB.185.5.1681-1692.2003.
Cervantes L, Bustos P, Girard L, Santamaría RI, Dávila G, Vinuesa P, Romero D, Brom S: The conjugative plasmid of a bean-nodulating Sinorhizobium fredii strain is assembled from sequences of two Rhizobium plasmids and the chromosome of a Sinorhizobium strain. BMC Microbiol. 2011, 11: 149-10.1186/1471-2180-11-149.
Torres Tejerizo G, Del Papa MF, De los Ángeles Giusti M, Draghi W, Lozano M, Lagares A, Pistorio M: Characterization of extrachromosomal replicons present in the extended host range Rhizobium sp. LPU83. Plasmid. 2010, 64: 177-185. 10.1016/j.plasmid.2010.07.004.
Rosenberg C, Hughet T: The pAtC58 plasmid of Agrobacterium tumefaciens is not essential for tumor induction. Mol Gen Genet. 1984, 196: 533-536. 10.1007/BF00436205.
Sambrook J, Fitsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. 1989, Cold Spring Harbor: Cold Spring Harbor Press
Simon R, Priefer U, Pühler A: A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biol Technol. 1983, 1: 784-791.
Kirchner O, Tauch A: Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J Biotechnol. 2003, 104: 287-299. 10.1016/S0168-1656(03)00148-2.
Schäfer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Pühler A: Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994, 145: 69-73. 10.1016/0378-1119(94)90324-7.
Figurski DH, Helinski DR: Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979, 76: 1648-1652. 10.1073/pnas.76.4.1648.
Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans AD, Jefferson RA: β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology. 1995, 141: 1691-1705. 10.1099/13500872-141-7-1691.
Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S: New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998, 64: 2240-2246.
Alexeyev MF, Shokolenko IN, Croughan TP: Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995, 160: 63-67. 10.1016/0378-1119(95)00108-I.
Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Rinehart KL, Farrand SK: Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997, 94: 6036-6041. 10.1073/pnas.94.12.6036.
Cha C, Gao P, Chen YC, Shaw PD, Farrand SK: Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant Microbe Interact. 1998, 11: 1119-1129. 10.1094/MPMI.1998.11.11.1119.
Hynes MF, McGregor NF: Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol Microbiol. 1990, 4: 567-574. 10.1111/j.1365-2958.1990.tb00625.x.
Althabegoiti MJ, Lozano L, Torres-Tejerizo G, Ormeño-Orrillo E, Rogel MA, González V, Martínez-Romero E: Genome sequence of Rhizobium grahamii CCGE502, a broad-host-range symbiont with low nodulation competitiveness in Phaseolus vulgaris. J Bacteriol. 2012, 194: 6651-6652. 10.1128/JB.01785-12.
Gordon D, Abajian C, Green P: Consed: a graphical tool for sequence finishing. Genome Res. 1998, 8: 195-202. 10.1101/gr.8.3.195.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25: 4876-4882. 10.1093/nar/25.24.4876.
Hall TA: BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999, 41: 95-98.
Abascal F, Zardoya R, Posada D: ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005, 21: 2104-2105. 10.1093/bioinformatics/bti263.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O: New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010, 59: 307-321. 10.1093/sysbio/syq010.
Wozniak RA, Waldor MK: Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010, 8: 552-563. 10.1038/nrmicro2382.
Ding H, Yip CB, Hynes MF: Genetic characterization of a novel rhizobial plasmid conjugation system in Rhizobium leguminosarum bv. viciae Strain VF39SM. J Bacteriol. 2013, 195: 328-339. 10.1128/JB.01234-12.
Bentley SD, Parkhill J: Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004, 38: 771-792. 10.1146/annurev.genet.38.072902.094318.
Landeta C, Dávalos A, Cevallos MA, Geiger O, Brom S, Romero D: Plasmids with a chromosome-like role in rhizobia. J Bacteriol. 2011, 193: 1317-1326. 10.1128/JB.01184-10.
Roché P, Debellé F, Maillet F, Lerouge P, Faucher C, Truchet G, Dénarié J, Promé JC: Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell. 1991, 67: 1131-1143. 10.1016/0092-8674(91)90290-F.
Torres Tejerizo G, Del Papa MF, Soria-Diaz ME, Draghi W, Lozano M, Giusti Mde L, Manyani H, Megías M, Gil Serrano A, Pühler A, Niehaus K, Lagares A, Pistorio M: The nodulation of alfalfa by the acid-tolerant Rhizobium sp. strain LPU83 does not require sulfated forms of lipochitooligosaccharide nodulation signals. J Bacteriol. 2011, 193: 30-39. 10.1128/JB.01009-10.
Spaink HP, Wijfjes AH, Lugtenberg BJ: Rhizobium NodI and NodJ proteins play a role in the efficiency of secretion of lipochitin oligosaccharides. J Bacteriol. 1995, 177: 6276-6281.
Cárdenas L, Domínguez J, Santana O, Quinto C: The role of the nodI and nodJ genes in the transport of Nod metabolites in Rhizobium etli. Gene. 1996, 173: 183-187. 10.1016/0378-1119(96)00166-7.
Spaink HP, Sheeley DM, van Brussel AAN, Glushka J, York WS, Tak T, Geiger O, Kennedy EP, Reinhold VN, Lugtenberg BJJ: A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature. 1991, 354: 125-130. 10.1038/354125a0.
Sutton JM, Lea EJ, Downie JA: The nodulation-signaling protein NodO from Rhizobium leguminosarum biovar viciae forms ion channels in membranes. Proc Natl Acad Sci USA. 1994, 91: 9990-9994. 10.1073/pnas.91.21.9990.
Masson-Boivin C, Giraud E, Perret X, Batut J: Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes?. Trends Microbiol. 2009, 17: 458-466. 10.1016/j.tim.2009.07.004.
Cevallos MA, Cervantes-Rivera R, Gutiérrez-Ríos RM: The repABC plasmid family. Plasmid. 2008, 60: 19-37. 10.1016/j.plasmid.2008.03.001.
Mercado-Blanco J, Olivares J: The large nonsymbiotic plasmid pRmeGR4a of Rhizobium meliloti GR4 encodes a protein involved in replication that has homology with the RepC protein of Agrobacterium plasmids. Plasmid. 1994, 32: 75-79. 10.1006/plas.1994.1046.
Brom S, García-De Los Santos A, Cervantes L, Palacios R, Romero D: Rhizobium etli symbiotic plasmid transfer, nodulation competitivity and cellular growth require interaction among different replicons. Plasmid. 2000, 44: 34-43. 10.1006/plas.2000.1469.
Crossman LC, Castillo-Ramírez S, McAnnula C, Lozano L, Vernikos GS, Acosta JL, Ghazoui ZF, Hernández-González I, Meakin G, Walker AW, Hynes MF, Young JP, Downie JA, Romero D, Johnston AW, Dávila G, Parkhill J, González V: A common genomic framework for a diverse assembly of plasmids in the symbiotic nitrogen fixing bacteria. PLoS One. 2008, 3: e2567-10.1371/journal.pone.0002567.
Souza V, Eguiarte LE: Bacteria gone native vs. bacteria gone awry?: plasmidic transfer and bacterial evolution. Proc Natl Acad Sci USA. 1997, 94: 5501-5503. 10.1073/pnas.94.11.5501.
Martínez-Romero E: Coevolution in Rhizobium-legume symbiosis?. DNA Cell Biol. 2009, 28: 361-370. 10.1089/dna.2009.0863.
Lozano L, Hernández-González I, Bustos P, Santamaría RI, Souza V, Young JP, Dávila G, González V: Evolutionary dynamics of insertion sequences in relation to the evolutionary histories of the chromosome and symbiotic plasmid genes of Rhizobium etli populations. Appl Environ Microbiol. 2010, 76: 6504-6513. 10.1128/AEM.01001-10.
Servín-Garcidueñas LE, Rogel MA, Ormeño-Orrillo E, Delgado-Salinas A, Martínez-Romero J, Sánchez F, Martínez-Romero E: Genome sequence of Rhizobium sp. strain CCGE510, a symbiont isolated from nodules of the endangered wild bean Phaseolus albescens. J Bacteriol. 2012, 194: 6310-6311. 10.1128/JB.01536-12.
Rogel MA, Hernández-Lucas I, Kuykendall LD, Balkwill DL, Martínez-Romero E: Nitrogen-fixing nodules with Ensifer adhaerens harboring Rhizobium tropici symbiotic plasmids. Appl Environ Microbiol. 2001, 67: 3264-3268. 10.1128/AEM.67.7.3264-3268.2001.
Amarger N, Macheret V, Laguerre G: Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int J Syst Bacteriol. 1997, 47: 996-1006. 10.1099/00207713-47-4-996.
He X, Chang W, Pierce DL, Seib LO, Wagner J, Fuqua C: Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J Bacteriol. 2003, 185: 809-822. 10.1128/JB.185.3.809-822.2003.
Zhang L, Murphy PJ, Kerr A, Tate ME: Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature. 1993, 362: 446-448. 10.1038/362446a0.
Piper KR, Beck von Bodman S, Farrand SK: Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993, 362: 448-450. 10.1038/362448a0.
Garcillán-Barcia MP, De la Cruz F: Why is entry exclusion an essential feature of conjugative plasmids?. Plasmid. 2008, 60: 1-18. 10.1016/j.plasmid.2008.03.002.
Pistorio M, Giusti MA, Del Papa MF, Draghi WO, Lozano MJ, Tejerizo GT, Lagares A: Conjugal properties of the Sinorhizobium meliloti plasmid mobilome. FEMS Microbiol Ecol. 2008, 65: 372-382. 10.1111/j.1574-6941.2008.00509.x.
Álvarez-Martínez CE, Christie PJ: Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009, 73: 775-808. 10.1128/MMBR.00023-09.
Van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ: CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009, 34: 401-407. 10.1016/j.tibs.2009.05.002.
Nosil P, Funk DJ, Ortiz-Barrientos D: Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009, 18: 375-402. 10.1111/j.1365-294X.2008.03946.x.
Mavingui P, Flores M, Guo X, Dávila G, Perret X, Broughton WJ, Palacios R: Dynamics of genome architecture in Rhizobium sp. strain NGR234. J Bacteriol. 2002, 184: 171-176. 10.1128/JB.184.1.171-176.2002.
Morton ER, Merritt PM, Bever JD, Fuqua C: Large deletions in the pAtC58 megaplasmid of Agrobacterium tumefaciens can confer reduced carriage cost and increased expression of virulence genes. Genome Biol Evol. 2013, 5 (7): 1353-1364. 10.1093/gbe/evt095.
Acknowledgements
This work was supported by PAPIIT IN205412 and Fundacion Produce San Luis Potosi, Mexico. We thank Dr. Susana Brom for her valuable advice on transfer assays, to SB and Dr. Michael Dunn for critically reading the manuscript and to Julio Martínez Romero, Humberto Peralta, Maria de Lourdes Girard and Yolanda Mora for technical support. G.T.T and M.J.A are members of the Research Career of CONICET and received fellowships from DGAPA, UNAM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MJA obtained the bacterial DNA and together with LL assembled and worked on the genome. Also, MJA carried out the molecular genetics experiments and wrote the manuscript. MAR assisted in laboratory experiments. EOO participated in sequence annotation, analysis and prepared some illustrations. GTT participated in design and discussion of genetics experiments. JM and coworkers performed plasmid profiles, isolated a novel R. grahamii strain, helped closing gaps and participated in discussion. EMR conceived the study, wrote and revised the manuscript. All authors approved the final manuscript.
Electronic supplementary material
12866_2013_2179_MOESM1_ESM.docx
Additional file 1: Table S1: Average nucleotide identity (ANI) and percentage of conserved DNA between chromosomes. (DOCX 24 KB)
12866_2013_2179_MOESM2_ESM.docx
Additional file 2: Table S2: Average nucleotide identity (ANI) and percentage of conserved DNA between chromids. (DOCX 25 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Althabegoiti, M.J., Ormeño-Orrillo, E., Lozano, L. et al. Characterization of Rhizobium grahamii extrachromosomal replicons and their transfer among rhizobia. BMC Microbiol 14, 6 (2014). https://doi.org/10.1186/1471-2180-14-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-14-6