Abstract
Background
Enveloped viruses utilize cellular membranes to bud from infected cells. The process of virion assembly and budding is often facilitated by the presence of certain conserved motifs within viral proteins in conjunction with cellular factors. We hence examined the West Nile Virus (WNV) Envelope protein for the presence of any such motifs and their functional characterization.
Results
We identified conserved 461PXAP464 and 349YCYL352 motifs in the WNV envelope glycoprotein bearing resemblance to retroviral late domains. Disruptive mutations of PXAP to LAAL and of the highly conserved Cys350 in the YCYL motif, led to a severe reduction in WNV particle production. Similar motifs in case of retroviruses are known to interact with components of host sorting machinery like PXAP with Tsg101 and YXXL with Alix. However, in the case of WNV, siRNA mediated depletion of Alix or Tsg101 did not have an effect on WNV release. Molecular modeling suggested that while the 461PXAP464 motif is surface accessible and could potentially interact with cellular proteins required for WNV assembly, the 349YCYL352 motif was found to be internal with Cys350 important for protein folding via disulphide bonding.
Conclusions
The conserved 461PXAP464 and 349YCYL352 motifs in the WNV envelope are indispensable for WNV particle production. Although these motifs bear sequence similarity to retroviral late domains and are essential for WNV assembly, they are functionally distinct suggesting that they are not the typical late domain like motifs of retroviruses and may play a role other than Alix/Tsg101 utilization/dependence.
Similar content being viewed by others
Background
West Nile Virus (WNV) is a single stranded positive sense RNA virus of the genus Flavivirus. The 11Kb RNA genome is translated in the cytoplasm as a polyprotein and processed to yield 3 structural (Capsid C, Premembrane prM/membrane M and Envelope E) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins [1]. Co-expression of prM and E proteins alone is sufficient for production of recombinant VLPs [2] that are similar to infectious virions in antigenic properties and have been commonly used to study virus assembly and budding. Although the field of Flavivirus assembly and release remains in its infancy, recent reports have identified certain residues in the prM that are important for WNV particle secretion [3, 4]. It is known that WNV genome replication occurs in the cytoplasm in the perinuclear region and virus particles assemble and bud into the Endoplasmic Reticulum (ER) lumen. Subsequently virions are transported to the plasma membrane (PM) via the cellular secretory pathway to be released from cells by exocytosis [5–8].
Following the synthesis of viral genome and proteins, enveloped viruses utilize cellular membranes to bud from infected cells. This is often facilitated by the presence of certain conserved motifs within viral proteins and their ability to interact with the cellular processes/machinery. The best known example of this process is the interaction of retroviral late domain motifs with components of the ESCRT (Endosomal Sorting Complex Required for Transport) sorting machinery to promote budding. Three types of consensus late domain motifs have been identified thus far: (i) the PT/SAP motif recruits the ESCRT-1 component Tsg101 (Tumor Susceptibility growth factor 101) [9, 10], (ii) the YXXL late domain motif interacts with the ESCRT associated protein Alix [11, 12] and the (iii) PPXY late domain motif binds to the Nedd4 family of E3 ubiquitin ligases that are involved in cargo recruitment during Multivesicular Body (MVB) formation [13, 14]. Besides retroviruses, late domain motifs have also been identified in other enveloped viruses like rhabdoviruses (vesicular stomatitis virus, rabies virus) [15–17], filoviruses (ebola, marburg) [18–22], arenaviruses (lymphocytic choriomeningitis virus, lassa virus) [23, 24], paramyxoviruses (Nipah virus, Sendai virus) [25, 26] and DNA viruses like hepatitis B virus, vaccinia virus, herpes simplex virus-1 and Epstein Barr virus [27–33]. Amongst flaviviruses, the NS3 of Japanese encephalitis virus (JEV) has been shown to associate with Tsg101 [34] while the yellow fever virus (YFV) NS3 has been shown to interact with Alix [35] assisting in virus release. However, currently there is no information on the presence of late domains in WNV proteins.
The process of WNV budding into the lumen of the ER is topologically similar to the process of MVB biogenesis in that both occur in a direction that is away from the cytosol. MVB biogenesis is mediated by the family of ESCRT proteins namely ESCRT-0, -I, -II and -III and other associated proteins like Alix/AIP1. The membrane associated ESCRT-III complexes are finally disassembled and recycled by the ATPase Vps4. A number of enveloped viruses via the conserved late (L) domain motifs that mimic similar motifs in cellular proteins are able to recruit the ESCRT machinery to the site of virus budding [36]. Disruption of L domain motifs or their function leads to defects in the final (late) stages of virus budding characterized by the tethering of virions to the cell surface [9, 14, 36, 37]. Most data on the role of ESCRT proteins and viral late domain motifs has come from research on retroviruses that primarily bud from the plasma membrane. Although there are reports that NS3 of other Flaviviruses can interact with ESCRT components [34, 35] there are no such reports for WNV. Furthermore, it is not known whether any late domain like motifs are present in WNV structural proteins especially E protein that is essential for assembly into virus like particles [38].
Results and discussion
Identification of conserved motifs in the WNV E protein
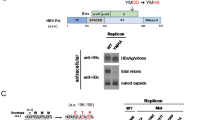
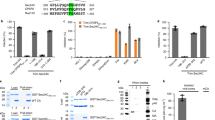
In case of Flaviviruses, the structural E protein is necessary for virus assembly and release and the production of recombinant VLPs. Hence, using sequence analysis and information based on work with other viruses we undertook this study to identify the presence of conserved motifs (a vital indicator of the functional importance) in the Flavivirus structural E proteins and determine whether they play a role in virus assembly and release. Sequence analysis of different Flavivirus structural proteins and different WNV isolates revealed the presence of conserved 461PXAP464 and 349YCYL352 motifs in the E protein (Figure 1A and B). Similar but less conserved motifs were also present in other Flaviviruses like Kunjin, JEV and St. Louis Encephalitis virus (SLE) (Figure 1B). Analysis of systematically selected WNV E protein sequences suggested that the PAAP motif was present in about 90% of the analyzed sequences while the frequency of the PSAP motif was less than 10% (Figure 1C). The YCYL motif was present in more than 95% of the WNV sequences analyzed. Table 1, depicts the occurrences of the PXAP and YCYL motifs in the protein non-redundant database (nr) database. As expected, sequence motifs that serve some biological functions, occur more often than by chance [39, 40] although it deserves mention that these motifs are maintained within the Flavivirus E proteins that themselves are highly conserved. While sequence analysis revealed the predominance of PAAP motifs over PSAP it is unclear as to what advantage the PSAP motif would render in case of WNV. From studies in HIV and that of host proteins like Hrs (Hepatocyte growth factor Receptor Substrate) it is well known that the PSAP motif is a strong binding partner of Tsg101 [41].
Sequence analysis of Flavivirus Envelope proteins. (A) Outline of WNV structural proteins C, PrM and E. (B) Presence of conserved 461PS/AAP464 and 349YCYL352 motifs in the Flavivirus envelope protein. Selected Flavivirus proteins were downloaded from NCBI [42], aligned with MAFFT [43] and the respective motif regions visualized in Jalview [44] using ClustalX-like coloring based on physicochemical properties and conservation. Virus names are shown left with NCBI GI number. (C) Frequency of YCYL and PAAP motif variants in WNV envelope. Significant protein hits (E<0.001) were first identified with Delta-BLAST [45] starting with the sequence of the envelope glycoprotein structure (PDB:2hg0) against NCBI’s non-redundant protein database restricting to West Nile virus sequences only. All hits were next aligned with MAFFT after discarding those without sequence information for the YCYL or PAAP region and removing 100% identical sequences using Jalview. The resulting set of 286 WNV sequences was analyzed for the respective motif occurrences.
Development of a rapid assay to study WNV assembly and release
We next aimed towards conducting a functional analysis to determine if WNV may utilize the above conserved motifs for virus assembly and release. To this end we developed a rapid renilla luciferase (ren-luc) based virus release assay and compared it to the classical radioimmunoprecipitation based assay (Figure 2). This would not only be a useful tool for rapid siRNA based screens or to identify potential drugs/compounds that inhibit WNV particle production but also obviate the requirement for a BSL3 facility that is necessary for working with infectious WNV. 293T cells were transfected with CprME and WNV Ren/Rep plasmids [46]. Culture supernatants were harvested 24 h post transfection and cells lysed and read for ren-luc activity (cell associated, Figure 2A and C) using the Dual Glo luciferase assay substrate (Promega). Equal volume of the harvested supernatants were then used to infect 293T cells, cells lysed and read for luciferase activity (virion-associated) 24 h post infection (Figure 2A and C). Virus release was calculated as ratio of virion associated ren-luc/(cell+virion associated ren-luc) activity. In parallel, classical radioimmunoprecipitation based virus release assay [47] was also conducted to determine the validity of the rapid assay described above (Figure 2A and B). Although, the luciferase based rapid assay also accounts for entry defects in virions, it is a convenient high throughput method for identification of general inhibitors of the virus life cycle.
Rapid assay for studying WNV assembly and release. (A) Schematic diagram delineating the steps for the rapid Ren-luc based virus release assay and comparing it to the classical radioimmunoprecipitation assay. 293T cells were transfected with WNV-CPrME along with the Ren/Rep plasmids at a ratio of 1:1 or with the pUC vector as control. (B) For radioimmunoprecipitation based assay, cells were metabolically labeled with [35S]Met-Cyst protein labeling mix (PerkinElmer) in RPMI 1640 medium supplemented with 10% FBS but devoid of Met and Cys 24 h post transfection. Following ultracentrifugation, cell and virus lysates were immunoprecipitated using anti-WNV serum, run on an SDS PAGE gel followed by fluorography. Virus release was calculated as ratio of virion associated versus cell+virion associated E protein. (C) For ren-luc based virus release assay, culture supernatants were harvested 24 h post transfection and cells lysed and read for ren-luc activity (cell associated) using the Dual Glo luciferase assay substrate (Promega). Equal volume of the harvested supernatants were then used to infect 293T cells, cells lysed and read for luciferase activity (virion-associated) 24 h post infection. Virus release was calculated as ratio of virion associated versus cell+virion associated ren-luc activity.
Tsg-5’ and Alix-V domain expression inhibits WNV assembly and release
As mentioned above, the conserved motifs identified in the WNV envelope resembled the late domain like motifs of retroviruses. We hence asked whether some of the well characterized inhibitors of ESCRT pathway previously used to study retrovirus budding would affect WNV assembly and release. To inhibit Tsg101 we utilized either Tsg-5’ expression vector that prevents HIV Gag-Tsg101 interaction or Tsg-F and TSG-3’ that have been shown to inhibit HIV release by globally disrupting the endosomal sorting machinery [48, 49]. We also used a transdominant form of Vps4 (Vps4EQ) that prevents the dissociation of ESCRT-III components at the endosomal membrane thereby inhibiting HIV-1 and Murine Leukemia Virus (MLV) budding [49–51], [52]. Similarly, the V domain of Alix (residues 364–716) which is known to bind both Equine Infectious Anemia Virus (EIAV) and HIV-1 Gag acting as a dominant-negative inhibitor of virus release [51, 53, 54] was also used. 293T cells were transfected to express CprME, WNV Ren/Rep plasmids in the presence of either control plasmid (pUC) or Tsg-F, Tsg-5’ , Tsg-3’ [49], Alix-V [53] or Vps4EQ [50] expression vectors. Virus release efficiency was then calculated by both the rapid assay and classical virus release assay. Interestingly, the expression of Tsg-5’ and Alix-V domain modestly diminished WNV release whereas no significant effect on virus release was observed on expression of Tsg-3’ Tsg-F or Vps4EQ (Figure 3A and B). While it is known that expression of Tsg-5’ affects HIV-1 release by affecting late domain function [48, 49], the precise mechanism via which Tsg-3’ , Tsg-F or Alix-V domain affect HIV release remains unknown. They could either be affecting the function of specific host proteins or universally disrupting the cell sorting machinery utilized for WNV particle production.
WNV release is inhibited on expression of Tsg-5’ and Alix V domain. 293T cells were transfected with WNV-CPrME and Ren/Rep plasmids along with control pUC or the indicated cellular protein expression constructs. Virus release was determined using the (A) classical radioimmunoprecipitation technique and (B) the rapid ren-luc based assay. Data represent mean ± SD from 3 (A) or 4 (B) independent experiments.
Mutations of the conserved PAAP and YCYL motifs in WNV envelope inhibits virus particle production
To further examine the relevance of these conserved PXAP and YCYL motifs in WNV assembly and release, we constructed mutations in the PAAP residues to either LAAL or PSAP (Figure 4A) using site directed mutagenesis. Interestingly, mutation of PAAP to LAAL caused a severe defect in virus budding, while mutation of the residues to PSAP led to virus release efficiency that was modestly better than WT (Figure 4B and C). We also mutated the YCYL domain to ACYA or AAAA. Interestingly, mutation of the above motifs to AAAA but not ACYA caused a severe defect in virus release (Figure 4B and C). It is worth mentioning that disruptive mutations to AAAA may also have other adverse side effects since the highly conserved cysteine residue may be involved in maintaining protein structure and stability.
Mutation of PAAP motif to LAAL significantly diminishes WNV release. (A) Sequence of the 461PS/AAP464 and 349YCYL352 motif bearing region and their mutagenesis strategy. 293T cells were transfected with WNV-CPrME WT or the indicated mutant DNAs along with the Ren/Rep plasmid. Virus release was determined using the (B) classical radioimmunoprecipitation technique and (C) the rapid ren-luc based assay. Pooled data (mean ± SD) from 3 (A) or 4 (B) independent experiments is shown. (D) HIV-PAAP mutant is capable of efficient release when compared to the PTAP minus mutant. 293T cells were transfected with HIV pNL4-3 WT, PTAP- or PAAP DNA. Virus release was determined 24 h post transfection after radiolabeling and immunoprecipitation with HIV-Ig.
It has previously been shown in context of HIV-1 that the PAAP motif interacts poorly with Tsg101 in in-vitro binding assays using purified proteins [9, 21, 55]. Since a large number of WNV isolates naturally bear a PAAP motif at position 461–464 instead of PTAP, we wanted to determine if a PAAP motif in the HIV p6 would permit virus release. We hence mutated the PTAP motif in HIV to PAAP and determined virus release. Although HIV-PAAP was released less efficiently than WT-HIV, it was significantly better than the PTAP deleted mutant (Figure 4D). These findings, at least in case of HIV where disruption of PT/SAP Tsg101 interaction significantly affects virus release are indicative that the PAAP motif may still be capable of binding Tsg101 albeit at a lower efficiency. Thus a PAAP motif can act as a functional late domain for HIV and hence could do the same for WNV isolates that predominantly bear PAAP motifs. Our findings are consistent with those of Demirov et al. [56] although the possibility that the PAAP motif is capable of interacting directly or indirectly with certain other host factors that favor HIV and/or WNV release cannot be ruled out.
Depletion of endogenous Alix or Tsg101 does not inhibit WNV assembly and release
Our findings that Tsg-5’ expression inhibits WNV release suggests a role for the ESCRT pathway in WNV budding. However, in other enveloped viruses that bear late domains (e.g. Gag of retroviruses, matrix of rhabdoviruses, VP40 of Ebolavirus) these motifs are located on the cytoplasmic side of the membrane and thus would be able to interact with ESCRT proteins to facilitate budding and particle release. The Flavivirus E protein on the other hand is translated into the lumen of the ER and hence these conserved motifs in WNV E protein would only be minimally exposed to the cytoplasmic side of intracellular vesicles or the plasma membrane. Hence in order to confirm the role of Tsg101 and/or Alix in WNV assembly and release we used a siRNA based approach. For this 293T cells were knocked down of endogenous Tsg101 and Alix expression using specific siRNAs and WNV release determined using both Ren-luc based and immunoprecipitation based virus release assays. As shown in Figure 5A and C, while Tsg101 depletion had no effect on WNV particle secretion, as expected, it caused a severe reduction in HIV-1 release. Alix depletion on the other hand had no effect on either HIV or WNV release (Figure 5A and C) but diminished EIAV release (Figure 5B). Thus while the conserved PXAP and YCYL motifs in WNV are important for virus assembly and release, it is most likely not due to dependence on the ESCRT component Tsg101 or the associated factor, Alix.
Depletion of endogenous Tsg101 or Alix using specific siRNA does not inhibit WNV release. 293T cells were transfected with control, Alix or Tsg101 siRNA. 24 h post transfection cells were transfected again with respective siRNAs along with (A) WT HIV-1 pNL4-3 DNA (B) WT EIAV Gag DNA or (C) WNV-CPrME plus the Ren/Rep plasmids. Virus release was determined after radiolabeling and immunoprecipitation for HIV and WNV, via western blotting for EIAV and also by the rapid ren-luc based assay for WNV. Data represent mean ± SD from 3 independent experiments (A&C). For the ren-luc based WNV assay one representative of 3 independent experiments is shown.
In the WNV E protein, the PAAP motif is surface located while the YCYL motif is deeply buried
Our siRNA mediated depletion studies above suggested that WNV may not rely on the ESCRT host cell sorting machinery for assembly and release. Thus, it is plausible that these motifs may interact with other host factors to facilitate the assembly of the virion particles. In fact our structural analysis shows that the PXAP motif is surface accessible and could participate in protein interactions with yet unidentified cellular factors (Figure 6A). In the context of the viral capsid made up of multiple envelope (E) proteins the PXAP surface motif appears to form part of the interface between the envelope subunits (Figure 6B). It also lies adjacent to the discontinuous epitope recognition site of co-crystallized neutralizing antibodies. On the other hand the YCYL motif is deeply buried and forms part of the structural core with the central cysteine participating in formation of a critical disulfide bridge (Figure 6A). This is in agreement with our findings where mutation of the YCYL motif to ACYA had little effect on virus release but mutation to AAAA severely affected budding possibly via loss of the disulphide bridging cysteine.
Crystal structure of West Nile virus envelope glycoprotein visualized with Yasara [57]. (A) Analyzed motifs on PDB:2hg0 [58] highlighted in red (PAAP) or magenta (YCYL). Structural analysis suggests that the PAAP motif is surface accessible while the YCYL motif is buried. (B) Analysis of the envelope protein in context of the assembled viral envelope PDB:3iyw [59]. Three envelope proteins are shown in gray, purple and yellow. The PAAP surface motif (red residues in black circles) appears to form part of the interface between the envelope subunits. It also lies adjacent to the discontinuous epitope recognition site of co-crystallized neutralizing antibodies (blue and green).
Conclusions
In this study, we have developed a rapid assay to study WNV assembly and release and identified conserved motifs in the viral envelope (E) that have functional relevance. These motifs bear sequence homology to late domain like motifs described in retroviruses. Experiments aimed at elucidating their role demonstrated that while expression of Tsg-5’ and Alix-V domain modestly inhibited WNV particle production, expression of Vps4EQ had no effect on WNV release. These data combined with the fact that siRNA mediated depletion of Alix or Tsg101 did not affect WNV release argues against their utilization or the ESCRT pathway by WNV. For instance, it has been documented that HSV possesses PT/SAP and YXXL motifs in several of its proteins but virus particle production is independent of Alix or Tsg101 expression [60]. Likewise, the PSAP motifs are conserved amongst the Vesiculovirus M protein without possessing L domain activity [61, 62]. However, the conserved nature of these domains in WNV and reduced virus release upon disruptive mutations argues in favor of a role in virus assembly via yet unidentified mechanism/s.
Our data are also reminiscent of the effects of Alix V domain expression versus Alix depletion on HIV particle production. While siRNA depletion of Alix does not affect HIV release, dominant negative inhibition via Alix V domain expression does [11, 53]. Moreover, it was recently demonstrated that the Alix V domain is capable of interacting with ubiquitin [51, 63, 64]. It is also known that ubiquitination plays a role in both HIV and flavivirus particle production [65, 66]. It is thus plausible that expression of the Alix V domain may alter ubiquitin dependent cellular functions thereby affecting WNV particle production. The precise mechanism behind this phenomenon with respect to HIV-1 remains to be elucidated. The fact that some WNV strains like Sarafend exhibits significant budding from the plasma membrane [67] would favor a role of ESCRT components like Alix and Tsg101 for budding.
Sequence analysis and information based on other viruses showed the presence of PXAP and YXXL conserved motifs in the E protein of Flaviviruses and different WNV strains, motifs that resemble the retroviral late domain-like motifs. It is worth mentioning that sequence analysis of a large portion of several different Flavivirus E proteins showed only 18% conservation in the amino acid residues, although the number does reflect the maximum diversity across the whole Flavivirus family [68]. This conservation was mostly seen on the inner surface of the monomers plausibly as a result of neutralizing antibody pressure. On the contrary, the PXAP and YCYL motifs were quite conserved indicating their functional relevance. Moreover, substantial changes in the consensus sequences are also found to occur in only a few areas of the E protein and may have relevance to growth in insect cells versus vertebrates [69]. Although mutational analysis confirms the importance of these domains in WNV assembly and particle formation, the role of Tsg101 and Alix in this phenomenon remains inconclusive from this study. Molecular modeling shows that the PXAP domain is present on the surface of the E protein and could potentially interact with cellular factors. On the other hand the YCYL conserved domain consisted of a conserved cysteine that is involved in disulphide bonding and protein folding. Although the YCYL motif may be critical in maintaining structure of the virus, the conservation of this motif and its functional relevance has neither been studied nor demonstrated in other Flaviviruses. Moreover, the same was not true for the PXAP domain. Interestingly, mutation of the PAAP motif to PSAP, which is an optimal binding partner for cellular sorting proteins modestly enhanced virus release. Considering the presence of only PAAP and PSAP at positions 461–464 in all the WNV sequences analyzed, the importance of this domain in virus assembly cannot be ignored. While the cellular sorting partner of PS/AAP domain in WNV could not be identified, our study opens the gate for further investigation into understanding WNV and Flavivirus assembly in general.
Further studies are needed to determine the precise mechanism via which these motifs, specially the PXAP domain, regulates WNV assembly and release and whether it functions via interaction with certain host factors or merely play a structural role in regulating virus assembly and release.
Methods
Cell culture and transfections
293T cells were cultured in DMEM supplemented with 10% FBS. All transfections were performed using Lipofactamine2000™ reagent (Invitrogen) as per the manufacturer’s instructions. In cases where transfections involved multiple DNAs, efficiency of co-transfection was carefully controlled by using an equal amount of plasmid expression vectors for each well and adjusting the total input DNA in each well to be constant by using pUC DNA.
Plasmids, antibodies, cell culture reagents, and siRNAs
The WNV CprME and Ren/Rep plasmids have been described previously [46] and were kindly provided by Dr. Ted Pierson (NIAID). Mutations in the CprME 461PAAP464 and 349YCYL352 motifs to PSAP, LAAL, ACYA and AAAA were constructed by site directed mutagenesis (Stratagene) using specific primer pairs. The full-length HIV-1 proviral clone pNL4-3 [70] and its PTAP minus derivative have been described previously [56]. The HIV PAAP mutant in the pNL4-3 backbone was constructed by site directed mutagenesis. Hemagglutinin (HA)-tagged derivatives of Tsg101-TSG-5′ and TSG-3′ in the pcGNM2 expression as well as the full-length Tsg101 expression vector (pcGNM2/TSG-F) have been previously described [49]. pEGFP-C2:VPS4A(E228Q), expressing an ATPase-deficient mutant of VPS4A fused to GFP has been previously described [50]. Expression vectors for the V domain of Alix (pcGNM2/hAlix(364–716) have been described [54]. The EIAV Gag expression vector (pPRE/GagEIAV) has been described [71].
Metabolic labeling and immunoprecipitation
The protocol for radiolabeling and immunoprecipitation of cell and virus lysates has been described in detail previously [72]. Briefly, transfected cells were starved for 30 min in RPMI medium lacking Met and Cys. Thereafter, cells were incubated for 2–3 h in RPMI medium supplemented with FBS and [35S]Met/Cys. Culture supernatants were filtered and subjected to ultracentrifugation at 100,000 x g for 45 min. Cell and virion samples were lysed in cell lysis buffer (0.5% Triton X-100, 300 mM NaCl, 50 mM Tris [pH 7.5] containing protease inhibitors [Complete; Roche]). Thereafter, they were immunoprecipitated either with HIV-Ig (Kindly provided by the NIH AIDS research and reference reagent program) or anti-WNV serum (Kindly provided by Dr. Robert B. Tesh, University of Texas Medical Branch, Galveston) coated Protein A beads. Immunoprecipitated cell lysates were washed three times in RIPA buffer and once with SDS-DOC wash (0.1% sodium dodecyl sulfate, 300 mM NaCl, 50 mM Tris [pH 7.5], 2.5 mM deoxycholic acid), resolved by SDS-PAGE followed by PhosphorImager analysis. Virus release efficiency was calculated as ratio of virion associated versus total cell plus virion associated HIV-1 Gag or WNV E protein.
Renilla based virus release assay
The overall strategy for this assay is summarized in Figure 2A. 293T cells were transfected with CprME and WNV Ren/Rep plasmids [46]. Culture supernatants were harvested 24 h post transfection and cells lysed and read for ren-luc activity using the Dual Glo luciferase assay substrate (Promega). Equal volume of the harvested supernatants were then used to infect 293T cells, cells lysed and read for luciferase activity (virion-associated) 24 h post infection. Virus release was calculated as ratio of virion associated ren-luc/(cell+virion associated ren-luc) activity. The overall strategy is summarized in Figure 2A.
Sequence analysis
Selected Flavivirus proteins were downloaded from NCBI [42]. The NCBI database was searched for sequences for complete or almost full length (>3300 amino acids) polyproteins from Flaviviruses and selected the ones with species name including West Nile Virus. If multiple sequences were available per virus name, only the longest sequence was considered. This yielded 11 different West Nile virus sequences with separate strain designations (strain name and GI numbers shown in alignment). The downloaded sequences were aligned with MAFFT [43] and the respective motif regions visualized in Jalview [44] using ClustalX-like coloring based on physicochemical properties and conservation. To systematically count the frequency of YCYL and PAAP motif variants in WNV, we first identified significant protein hits (E<0.001) with Delta-BLAST [45] starting with the sequence of the envelope glycoprotein structure (PDB:2hg0) against NCBI’s non-redundant protein database restricting to West Nile virus sequences only. Next, we aligned all hits with MAFFT [43] and discarded those without sequence information for the YCYL or PAAP region and removed 100% identical sequences using Jalview [44], leaving us with a set of 286 WNV sequences for which we calculated the respective motif occurrences.
The strain designations as listed in the alignment were taken from the NCBI taxonomy on West Nile viruses: http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=11082.
Several of these strains like Sarafend belong to the pathogenic lineage 2. These are: West Nile virus H442, West Nile virus SA381/00, West Nile virus SA93/01, West Nile virus SPU116/89. Please note that the Kunjin virus has been recognized as WNV strain which is also visible by the identical sequences in the 2 displayed patterns.
Abbreviations
- HIV:
-
Human immunodeficiency virus type 1
- WNV:
-
West Nile virus
- EIAV:
-
Equine infectious anemia virus
- JEV:
-
Japanese encephalitis virus
- YFV:
-
Yellow fever virus
- SLE:
-
St. Louis encephalitis virus
- C:
-
Capsid
- prM:
-
Pre membrane
- NS:
-
Non-structural
- VLPs:
-
Virus like particles
- ESCRT:
-
Endosomal sorting complex required for transport
- Tsg101:
-
Tumor susceptibility growth factor 101
- MVB:
-
Multi vesicular bodies
- ER:
-
Endoplasmic reticulum.
References
Brinton MA: The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu Rev Microbiol. 2002, 56: 371-402. 10.1146/annurev.micro.56.012302.160654.
Lindenbach BD, Thiel HJ, Rice CM: Flaviviridae: the viruses and their replication. 2007, Philadelphia, PA: Fields virology Lippincott William & Wilkins, 1101-1152.
Calvert AE, Huang CY, Blair CD, Roehrig JT: Mutations in the West Nile prM protein affect VLP and virion secretion in vitro. Virology. 2012, 433: 35-44. 10.1016/j.virol.2012.07.011.
Setoh YX, Prow NA, Hobson-Peters J, Lobigs M, Young PR, Khromykh AA, Hall RA: Identification of residues in West Nile virus pre-membrane protein that influence viral particle secretion and virulence. J Gen Virol. 2012, 93: 1965-1975. 10.1099/vir.0.044453-0.
Li J, Bhuvanakantham R, Howe J, Ng ML: Identifying the region influencing the cis-mode of maturation of West Nile (Sarafend) virus using chimeric infectious clones. Biochem Biophys Res Commun. 2005, 334: 714-720. 10.1016/j.bbrc.2005.06.150.
Mackenzie JM, Westaway EG: Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J Virol. 2001, 75: 10787-10799. 10.1128/JVI.75.22.10787-10799.2001.
Mason PW: Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989, 169: 354-364. 10.1016/0042-6822(89)90161-X.
Nowak T, Farber PM, Wengler G: Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989, 169: 365-376. 10.1016/0042-6822(89)90162-1.
Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al: Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001, 107: 55-65. 10.1016/S0092-8674(01)00506-2.
Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA: Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991, 88: 3195-3199. 10.1073/pnas.88.8.3195.
Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD: Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003, 100: 12414-12419. 10.1073/pnas.2133846100.
Strack B, Calistri A, Craig S, Popova E, Gottlinger HG: AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003, 114: 689-699. 10.1016/S0092-8674(03)00653-6.
Xiang Y, Cameron CE, Wills JW, Leis J: Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996, 70: 5695-5700.
Freed EO: Viral late domains. J Virol. 2002, 76: 4679-4687. 10.1128/JVI.76.10.4679-4687.2002.
Craven RC, Harty RN, Paragas J, Palese P, Wills JW: Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999, 73: 3359-3365.
Harty RN, Paragas J, Sudol M, Palese P: A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999, 73: 2921-2929.
Jayakar HR, Murti KG, Whitt MA: Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J Virol. 2000, 74: 9818-9827. 10.1128/JVI.74.21.9818-9827.2000.
Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP: A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci USA. 2000, 97: 13871-13876. 10.1073/pnas.250277297.
Kolesnikova L, Bamberg S, Berghofer B, Becker S: The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J Virol. 2004, 78: 2382-2393. 10.1128/JVI.78.5.2382-2393.2004.
Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN: Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol. 2003, 77: 1812-1819. 10.1128/JVI.77.3.1812-1819.2003.
Martin-Serrano J, Zang T, Bieniasz PD: HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001, 7: 1313-1319. 10.1038/nm1201-1313.
Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J: Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol. 2007, 81: 4895-4899. 10.1128/JVI.02829-06.
Perez M, Craven RC, de la Torre JC: The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci USA. 2003, 100: 12978-12983. 10.1073/pnas.2133782100.
Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J: Cellular factors required for Lassa virus budding. J Virol. 2006, 80: 4191-4195. 10.1128/JVI.80.8.4191-4195.2006.
Ciancanelli MJ, Basler CF: Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol. 2006, 80: 12070-12078. 10.1128/JVI.01743-06.
Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, Nagai Y, Yoshida T: AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J Virol. 2005, 79: 8933-8941. 10.1128/JVI.79.14.8933-8941.2005.
Calistri A, Sette P, Salata C, Cancellotti E, Forghieri C, Comin A, Gottlinger H, Campadelli-Fiume G, Palu G, Parolin C: Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J Virol. 2007, 81: 11468-11478. 10.1128/JVI.01364-07.
Chua HH, Lee HH, Chang SS, Lu CC, Yeh TH, Hsu TY, Cheng TH, Cheng JT, Chen MR, Tsai CH: Role of the TSG101 gene in Epstein-Barr virus late gene transcription. J Virol. 2007, 81: 2459-2471. 10.1128/JVI.02289-06.
Crump CM, Yates C, Minson T: Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol. 2007, 81: 7380-7387. 10.1128/JVI.00222-07.
Honeychurch KM, Yang G, Jordan R, Hruby DE: The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J Virol. 2007, 81: 7310-7315. 10.1128/JVI.00034-07.
Kian Chua P, Lin MH, Shih C: Potent inhibition of human Hepatitis B virus replication by a host factor Vps4. Virology. 2006, 354: 1-6. 10.1016/j.virol.2006.07.018.
Lambert C, Doring T, Prange R: Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J Virol. 2007, 81: 9050-9060. 10.1128/JVI.00479-07.
Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P: Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci USA. 2007, 104: 10205-10210. 10.1073/pnas.0704000104.
Chiou CT, Hu CC, Chen PH, Liao CL, Lin YL, Wang JJ: Association of Japanese encephalitis virus NS3 protein with microtubules and tumour susceptibility gene 101 (TSG101) protein. J Gen Virol. 2003, 84: 2795-2805. 10.1099/vir.0.19201-0.
Carpp LN, Galler R, Bonaldo MC: Interaction between the yellow fever virus nonstructural protein NS3 and the host protein Alix contributes to the release of infectious particles. Microbes Infect. 2011, 13: 85-95. 10.1016/j.micinf.2010.10.010.
Bieniasz PD: Late budding domains and host proteins in enveloped virus release. Virology. 2006, 344: 55-63. 10.1016/j.virol.2005.09.044.
Demirov DG, Freed EO: Retrovirus budding. Virus Res. 2004, 106: 87-102. 10.1016/j.virusres.2004.08.007.
Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW: N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005, 79: 13262-13274. 10.1128/JVI.79.21.13262-13274.2005.
Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ: Attributes of short linear motifs. Molecular bioSystems. 2012, 8: 268-281. 10.1039/c1mb05231d.
Ren S, Yang G, He Y, Wang Y, Li Y, Chen Z: The conservation pattern of short linear motifs is highly correlated with the function of interacting protein domains. BMC genomics. 2008, 9: 452-10.1186/1471-2164-9-452.
Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI: HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003, 162: 425-434. 10.1083/jcb.200302138.
Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al: Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012, 40: D13-D25. 10.1093/nar/gkr1184.
Katoh K, Toh H: Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008, 9: 286-298. 10.1093/bib/bbn013.
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ: Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009, 25: 1189-1191. 10.1093/bioinformatics/btp033.
Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL: Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012, 7: 12-10.1186/1745-6150-7-12.
Pierson TC, Sanchez MD, Puffer BA, Ahmed AA, Geiss BJ, Valentine LE, Altamura LA, Diamond MS, Doms RW: A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology. 2006, 346: 53-65. 10.1016/j.virol.2005.10.030.
Joshi A, Garg H, Ablan S, Freed EO, Nagashima K, Manjunath N, Shankar P: Targeting the HIV entry, assembly and release pathways for anti-HIV gene therapy. Virology. 2011, 415: 95-106. 10.1016/j.virol.2011.03.028.
Demirov DG, Ono A, Orenstein JM, Freed EO: Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002, 99: 955-960. 10.1073/pnas.032511899.
Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO: Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J Virol. 2003, 77: 6507-6519. 10.1128/JVI.77.11.6507-6519.2003.
Bishop N, Woodman P: TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001, 276: 11735-11742. 10.1074/jbc.M009863200.
Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO: Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic. 2008, 9: 1972-1983. 10.1111/j.1600-0854.2008.00817.x.
Shehu-Xhilaga M, Ablan S, Demirov DG, Chen C, Montelaro RC, Freed EO: Late domain-dependent inhibition of equine infectious anemia virus budding. J Virol. 2004, 78: 724-732. 10.1128/JVI.78.2.724-732.2004.
Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH: Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007, 14: 194-199. 10.1038/nsmb1203.
Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO: An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007, 282: 3847-3855.
Schlundt A, Sticht J, Piotukh K, Kosslick D, Jahnke N, Keller S, Schuemann M, Krause E, Freund C: Proline-rich sequence recognition: II. Proteomics analysis of Tsg101 ubiquitin-E2-like variant (UEV) interactions. Mol Cell Proteomics. 2009, 8: 2474-2486. 10.1074/mcp.M800337-MCP200.
Demirov DG, Orenstein JM, Freed EO: The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002, 76: 105-117. 10.1128/JVI.76.1.105-117.2002.
Krieger E, Koraimann G, Vriend G: Increasing the precision of comparative models with YASARA NOVA–a self-parameterizing force field. Proteins. 2002, 47: 393-402. 10.1002/prot.10104.
Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH: Crystal structure of the West Nile virus envelope glycoprotein. J Virol. 2006, 80: 11467-11474. 10.1128/JVI.01125-06.
Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG: Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc Natl Acad Sci USA. 2010, 107: 18950-18955. 10.1073/pnas.1011036107.
Pawliczek T, Crump CM: Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol. 2009, 83: 11254-11264. 10.1128/JVI.00574-09.
Irie T, Harty RN: L-domain flanking sequences are important for host interactions and efficient budding of vesicular stomatitis virus recombinants. J Virol. 2005, 79: 12617-12622. 10.1128/JVI.79.20.12617-12622.2005.
Irie T, Licata JM, Jayakar HR, Whitt MA, Bell P, Harty RN: Functional analysis of late-budding domain activity associated with the PSAP motif within the vesicular stomatitis virus M protein. J Virol. 2004, 78: 7823-7827. 10.1128/JVI.78.14.7823-7827.2004.
Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, Sundquist WI, Kopito RR: ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev Cell. 2012, 23: 1247-1254. 10.1016/j.devcel.2012.10.023.
Keren-Kaplan T, Attali I, Estrin M, Kuo LS, Farkash E, Jerabek-Willemsen M, Blutraich N, Artzi S, Peri A, Freed EO, et al: Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIX-V:ubiquitin complex and retrovirus budding. The EMBO journal. 2013, 32: 538-551. 10.1038/emboj.2013.4.
Ko A, Lee EW, Yeh JY, Yang MR, Oh W, Moon JS, Song J: MKRN1 induces degradation of West Nile virus capsid protein by functioning as an E3 ligase. J Virol. 2010, 84: 426-436. 10.1128/JVI.00725-09.
Martin-Serrano J: The role of ubiquitin in retroviral egress. Traffic. 2007, 8: 1297-1303. 10.1111/j.1600-0854.2007.00609.x.
Ng ML, Howe J, Sreenivasan V, Mulders JJ: Flavivirus West Nile (Sarafend) egress at the plasma membrane. Arch Virol. 1994, 137: 303-313. 10.1007/BF01309477.
Seligman SJ, Bucher DJ: The importance of being outer: consequences of the distinction between the outer and inner surfaces of flavivirus glycoprotein E. Trends Microbiol. 2003, 11: 108-110. 10.1016/S0966-842X(03)00005-2.
Danecek P, Lu W, Schein CH: PCP consensus sequences of flaviviruses: correlating variance with vector competence and disease phenotype. Journal of molecular biology. 2010, 396: 550-563. 10.1016/j.jmb.2009.11.070.
Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA: Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986, 59: 284-291.
Patnaik A, Chau V, Li F, Montelaro RC, Wills JW: Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J Virol. 2002, 76: 2641-2647. 10.1128/JVI.76.6.2641-2647.2002.
Freed EO, Orenstein JM, Buckler-White AJ, Martin MA: Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994, 68: 5311-5320.
Acknowledgements
We would like to thank Dr. Robert B. Tesh (University of Texas Medical Branch, Galveston) for kindly providing the WNV serum, Dr. Ted Pierson (NIAID) for the WNV constructs and the NIH AIDS research and reference reagent program for providing the HIV-Ig.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
HG and AJ designed the study, performed experiments, analyzed data and wrote the manuscript. RL, OT and SM performed sequence analysis, analyzed data and wrote the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Garg, H., Lee, R.T., Tek, N.O. et al. Identification of conserved motifs in the Westnile virus envelope essential for particle secretion. BMC Microbiol 13, 197 (2013). https://doi.org/10.1186/1471-2180-13-197
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-13-197