Abstract
Background
Periodontitis is a bacterial infection of the periodontal tissues. The Gram-negative anaerobic bacterium Porphyromonas gingivalis is considered a major causative agent. One of the virulence factors of P. gingivalis is capsular polysaccharide (CPS). Non-encapsulated strains have been shown to be less virulent in mouse models than encapsulated strains.
Results
To examine the role of the CPS in host-pathogen interactions we constructed an insertional isogenic P. gingivalis knockout in the epimerase-coding gene epsC that is located at the end of the CPS biosynthesis locus. This mutant was subsequently shown to be non-encapsulated. K1 capsule biosynthesis could be restored by in trans expression of an intact epsC gene. We used the epsC mutant, the W83 wild type strain and the complemented mutant to challenge human gingival fibroblasts to examine the immune response by quantification of IL-1β, IL-6 and IL-8 transcription levels. For each of the cytokines significantly higher expression levels were found when fibroblasts were challenged with the epsC mutant compared to those challenged with the W83 wild type, ranging from two times higher for IL-1β to five times higher for IL-8.
Conclusions
These experiments provide the first evidence that P. gingivalis CPS acts as an interface between the pathogen and the host that may reduce the host's pro-inflammatory immune response. The higher virulence of encapsulated strains may be caused by this phenomenon which enables the bacteria to evade the immune system.
Similar content being viewed by others
Background
Porphyromonas gingivalis is a major pathogen in destructive periodontal diseases including chronic and aggressive periodontitis that are characterized by breakdown of the tooth-supporting tissues [1–3]. P. gingivalis is a black pigmented, often encapsulated, strict anaerobic, Gram negative coccobacillus that occurs in the human oral cavity.
Among the variety of virulence factors that have been described for P. gingivalis, CPS has shown to be a major factor in experimental infections. Studies in a mouse infection model have revealed that encapsulated P. gingivalis strains are more virulent than non-encapsulated strains [4–7]. Non-encapsulated strains mostly cause non-invasive, localized abscesses whereas encapsulated strains cause invasive, spreading phlegmonous infections after subcutaneous inoculation of experimental animals.
Six distinct capsular serotypes have currently been described (K1-K6) [8, 9] and a seventh serotype (K7) has been suggested by R. E. Schifferle (personal communication). Small differences in virulence have been found between capsular serotypes and strong variation in virulence has been described between strains of the same capsular serotype [10]. CPS of all serotypes has been tested for induction of immunological responses in macrophages and it has been revealed that the CPS of K1 serotype strains induces higher chemokine expression in murine peritoneal macrophages than the other serotypes [11]. These data suggest that the K1 CPS plays an important role in host-pathogen interaction. The chemical composition of the K1 CPS has been studied to a limited extent. It has been reported that the CPS of K1 (strain W50) comprises of mannuronic acid (ManA), glucuronic acid (GlcA), galacturonic acid (GalA), galactose and N-acetylglucosamine (GlcNAc), but the CPS structure has not been solved [12].
Although CPS is a major structure at the interface between the bacterial cell and the host, the exact role of P. gingivalis CPS is not yet clear. Adhesion to epithelial cells has been shown to be higher for non-encapsulated P. gingivalis and the level and mechanism of co-aggregation has been shown to be CPS dependent [5, 13, 14]. In many pathogens CPS has been found to be involved in evasion of the host immune system by circumvention of phagocytosis, opsonization and complement killing [15–17].
The aim of this study was to investigate in vitro differences in host response during infection with a wild type and an isogenic non-encapsulated mutant of a naturally encapsulated strain. The well-studied K1 serotype W83 strain was used as the wild type strain since its CPS biosynthesis locus has been described [18, 19]. An insertional mutation in PG0120 (epsC) was constructed, which yielded a non-encapsulated strain. The gene has been annotated as a UDP-GlcNAc 2-epimerase.
This epsC mutant is tested in a fibroblast infection model [20] since fibroblasts are the most abundant stromal cells in soft connective tissue of the gingiva [21] and among the first cells encountering periodontal infections by anaerobic bacteria like P. gingivalis. And above all, fibroblasts have been shown to be involved in the immune response in periodontitis [22, 23]. Human gingival fibroblasts were infected with W83 and the epsC mutant and transcription of IL-1β, IL-6 and IL-8 was determined as host response parameters. This study provides the first direct evidence that P. gingivalis CPS reduces the host immune response, thereby potentially enabling evasion of the immune system to sustain successful long-term infection.
Results
EpsC mutant construction
After transformation of the linearized plasmid pΔEpsC to P. gingivalis W83 the epsC insertional mutation was confirmed by specific PCR amplifications and agarose gel electrophoresis of the products (data not shown). Primer combinations epsC BamHI F × PG0119 R and EryF F × epsC EcoRI R (Table 1) ensured that a 1.2 Kb fragment of pΔEpsC had been integrated by double crossover at PG0120 (epsC) as expected, replacing the intact copy with the insertionally inactivated copy (Figure 1).
Schematic representation of the knockout strategy to construct the epsC insertional mutation in W83. A. The genetic arrangement of the 3'-end of the CPS locus in the W83 wild type strain with the grey rectangles representing the genes present. B. Construct pΔepsC for insertional inactivation of epsC. The 1.2 Kb epsC was inserted into BamH I-EcoR I digested pGEX-6-p3 (oval) and interrupted by insertion of a 1.2 Kb EryF (shaded rectangle) in the single ClaI restriction site present. The dashed lines between A and B show the homologous crossover regions between the plasmid and W83 CPS locus. C. The final arrangement of the 3'-end of the P. gingivalis CPS locus after double crossover showing the insertional inactivation of epsC. Arrows represent the primers used to confirm the integrity of the epsC mutant.
To examine if the mutation had an influence on the growth characteristics of the epsC mutant both W83 and the epsC mutant were grown in brain heart infusion broth supplemented with hemin (5 μg/ml) and menadione (1 μg/ml) (BHI+H/M). Phase-contrast microscopy revealed that the mutant grows in aggregates, but no difference in growth rate was observed.
EpsC mutant characterization
The potential polar effect of the insertional inactivation on the down stream gene of epsC named hup-1 was examined. Total RNA was extracted from W83 and the epsC mutant in the early exponential phase and the hup-1 expression levels were evaluated by Real-Time PCR. No significant difference in expression of hup-1 was found between W83 and the epsC mutant (data not shown).
To show the effect of capsule-loss on the surface structure of P. gingivalis the hydrophobicity of the epsC mutant was tested by the capacity to adhere to hexadecane. While 3% of W83 cells was shown to adhere to hexadecane more than 60% of the epsC mutant cells was adhered to hexadecane. 19% of the complemented mutant cells was adhered to hexadecane (see Additional file 1).
Reactivity with the CPS-specific polyclonal rabbit antisera against P. gingivalis serotypes K1-K6 [8, 9] was examined for W83 and the epsC mutant. The epsC mutant was not recognized by any of the antisera including the K1 antiserum, whereas the wild type strain was only recognized by the K1 antiserum (Figure 2). Differences in CPS characteristics were also studied by Percoll density gradient centrifugation, which can reveal density differences between encapsulated and non-encapsulted bacteroides strains [24]. Percoll density gradient centrifugation analyses of W83 and the epsC mutant showed that the density of the mutant had been changed (Figure 3). Where W83 mostly settled at the 20-30% interface, the epsC mutant settled at the 50-60% interface. Note that the appearance of W83 is diffuse and not restricted to the 20-30% interface. The mutant settles as a compact and granulous layer.
Double immunodiffusion analysis of autoclaved supernatants of P. gingivalis strains. Samples of W83, the epsC mutant and the complemented mutant were tested against the K1-specific antiserum (central well). Note that the white precipitate indicating recognition of CPS with the antiserum is absent in case of the epsC mutant, whereas the intact epsC copy restores the wild-type K1 antiserum recognition in the complemented mutant.
Percoll density gradient centrifugation of W83 and epsC mutant. 1 ml of a OD690 = 4 suspension of overnight-grown P. gingivalis was layered on top of a stepwise Percoll gradient (10-80%) and centrifuged at 8000 × g for one hour. The gradient is visualized using fuchsine-stained layers in the marker (M).W83 reproducibly settles in the interfaces of 10-20%, 20-30% and 30-40% where most of the bacterial material is found in the 20-30% interface. The epsC mutant settles as a distinct, granulous band at the 50-60% interface.
To conclusively examine the absence of CPS in the epsC mutant, light microscopy was performed using India ink in combination with fuchsine staining (Figure 4). The negative India ink staining allows direct visualization of the capsule, appearing as a light halo surrounding the P. gingivalis cell. Fuchsine is used to stain the cell body. The halos around the W83 wild type strain are clearly visible in the phase contrast microscopic picture, whereas halos are absent around the epsC mutant. The intact epsC gene in trans under control of the CP25 promoter rescues the wild-type phenotype enabling the complemented mutant to produce a K1 capsule again (Figures 2 and 4).
Negative capsule staining of fuchsine-stained P. gingivalis cells with India Ink. Phase contrast microscopic picture at a 1000× magnification of (A) W83 wild type strain, (B) epsC mutant and (C) the complemented epsC mutant in an India ink preparation which reveals the capsule as a white halo (arrow). The inset shows an extra six times magnification.
Fibroblast response to P. gingivalis challenge
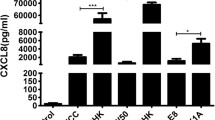
To study the effect of the epsC deletion on the host immune response six hour infection studies of human gingival fibroblasts with W83 and the epsC mutant were performed. Figure 5 shows IL-1β, IL-6 and IL-8 expression of infected gingival fibroblasts relative to the non-infected negative control which is set to 1 and normalized against expression of housekeeping gene GAPDH.
Relative expression of IL-1β , IL-6 and IL-8 genes in human gingival fibroblasts (HGF1) infected with P. gingivalis W83 and the epsC mutant. After a 6-hour challenge with P. gingivalis cells at MOI 1000:1 or 10.000:1 as indicated on the Y-axis, the expression levels of IL-1β, IL-6 and IL-8 in human gingival fibroblasts were measured using RT-PCR and represented as a relative value compared to a non-infected control sample which is set to a value of 1. Significant differences p < 0.01 are indicated by an asterisk.
At multiplicity of infection (MOI) 1000:1 of both strains a small induction of the tested genes could be detected compared to the non-infected control, but significant induction for all three genes was found when MOI 10.000:1 was used for infection.At MOI 1000:1 IL-6 and IL-8 expression showed a significantly higher induction (150-fold and 37-fold induction respectively) in the cells challenged with the epsC mutant when compared to the wild-type (6-fold and 2-fold induction respectively), IL-1β did not show a difference compared to the wild-type. However, when gingival fibroblasts were challenged with MOI 10.000 bacteria all three tested genes showed a significantly higher induction in the cells challenged with the epsC mutant than with W83 (figure 5). When fibroblasts were challenged with the complemented mutant the response was almost completely restored to wild-type levels (see Additional file 2).
Sedimentation of the epsC mutant in comparison to W83 was analyzed in the same buffer as used in the infection experiments. No significant sedimentation differences were found between W83 and the epsC mutant within the 6 hours needed for infection of the fibroblasts (data not shown).
Since infections were done with viable P. gingivalis, survival of the bacteria during the 6-hour aerobic period of infection in DMEM medium had to be ensured. Therefore a 6-hour survival experiment was performed in the 24-well plates used for the fibroblast challenge. On average 60-75% of W83, epsC mutant and complemented mutant cells survived for 6 hours in Dulbecco's modified Eagle's Medium (DMEM; Sigma Chemical Co.) supplemented with 10% fetal calf serum (FCS) (see Additional file 3).
Discussion
The aim of this paper was to understand the role of P. gingivalis CPS in the response of human gingival fibroblasts.P. gingivalis CPS has been regarded as an important virulence factor. It has been shown to induce inflammatory mediators in in vitro studies [11]. The capsule also plays an important role in shielding of immune response inducers in several bacterial species [25–27]. Since a distinct CPS biosynthesis locus in P. gingivalis has been described and shown to be functional [18, 19], studying the role of P. gingivalis CPS in the immune response by use of a mutant became feasible. For this purpose an insertional isogenic knockout in epsC, a potential capsular biosynthesis gene within the CPS biosynthesis locus present in strains of different serotypes, was constructed to prevent capsule synthesis. The homologue of this gene in Listeria monocytogenes lmo2537 has been shown to be essential for survival, and has been suggested to be involved in the maintenance of cell shape by providing a precursor of the teichoic acid linkage unit that serves as an acceptor for the main teichoic acid chain assembly [28]. Construction of the P. gingivalis epsC mutant shows that the epsC gene is not essential for P. gingivalis viability.
In the present study the mutant is shown to be non-encapsulated by double immuno-diffusion, density gradient centrifugation and India ink staining. Complementation resulted in rescue of wild-type K1 capsule biosynthesis. Although the exact role of epsC remains to be elucidated, this finding provides evidence that EpsC is essential in P. gingivalis CPS biosynthesis.
The epsC mutant was expected to have altered immunological properties. To examine the role of CPS, both the wild-type and the epsC mutant were used in an in vitro challenge of primary human gingival fibroblasts. Since the epsC mutant has altered physical properties, it was important to compare the sedimentation rate and viability of both the wild type and the mutant strain since these could have influenced the amount of living bacterial cells that are in contact with the fibroblasts. No differences were observed between the strains during the 6 hours of infection.
From the infection experiments of the gingival fibroblasts it became apparent that pro-inflammatory mediators IL-1β, IL-6 and IL-8 expression levels were up-regulated after a 6-hour challenge with both wild-type W83 and the epsC mutant in comparison to the non-infected control, especially when MOIs of 10.000:1 were used.
A challenge with the epsC mutant induced a significantly higher pro-inflammatory immune response than a challenge with the wild type W83, as shown by IL-1β, IL-6 and IL-8 gene expression. So, even though purified P. gingivalis CPS has been shown to stimulate pro-inflammatory cytokine expression in murine peritoneal macrophages [11] the absence of capsule induces extra cytokine induction when viable P. gingivalis cells were used to challenge fibroblasts.
Capsular polysaccharides of several bacteria have been implicated in down-regulation of pro-inflammatory cytokine production, including Klebsiella pneumonia [29]. Bacteroides fragilis capsular polysaccharide complex has been shown to induce IL-10 expression, a regulating cytokine which may cause suppression of the immune system [30].
An explanation of our results may be that the CPS prevents more potent immune inducers to be recognized by Toll-like receptors on the fibroblasts. It has been shown that the capsular antigen in Salmonella typhi, referred to as Vi-antigen, is able to prevent Toll-like receptor 4 recognition of LPS, thereby reducing expression of pro-inflammatory TNF-α and IL-6 [31–33]. In E. coli the capsule may cover short (10 nm) bacterial adhesins, which do not penetrate the 0.2-1.0 μm capsular layer, preventing them from being recognized by the immune system [26]. Likewise, P. gingivalis strain W83 was described as to have a small amount of short fimbriae that might be mostly covered by the CPS [34].
Another or additional explanation of our findings could be immune suppression by P. gingivalis CPS, meaning that CPS would actively modulate the immune response of the fibroblasts, leading to lower inflammatory cytokine expression levels, potentially enabling P. gingivalis to evade the immune system.
For several bacteria it has been described that capsular biosynthesis can be modulated depending on environmental conditions [35, 36]. Although presently no regulation of P. gingivalis capsule expression has been described, we can not exclude the possibility that in the in vivo situation capsule expression is regulated. However, the reduced pro-inflammatory host's immune response by the encapsulated strain may explain the documented differences between natural P. gingivalis strains in spreading. Whereas non-encapsulated strains are tackled directly by the immune system in localized abscesses, the more virulent encapsulated strains can evade this defence and cause phlegmonous infections [4–7].
Conclusions
The epimerase-coding gene epsC of P. gingivalis is essential for CPS synthesis. The absence of CPS results in increased induction of IL-1β, IL-6 and IL-8 in human gingival fibroblasts upon in vitro infection with viable P. gingivalis cells. P. gingivalis CPS acts as a functional interface between the pathogen and the host. The CPS-related reduced pro-inflammatory response can explain why natural non-encapsulated strains cause localized abscesses and encapsulated strains spreading phlegmonous infections.
Methods
Bacterial maintenance
P. gingivalis strains were grown either on 5% horse blood agar plates (Oxoid no. 2, Basingstoke, UK) supplemented with hemin (5 μg/ml) and menadione (1 μg/ml) (BA+H/M plates) or BHI+H/M, both, at 37°C in an anaerobic atmosphere of 80% N2, 10% H2, and 10% CO2. Mutants were selected in the presence of 5 μg/ml erythromycin. Complemented mutants were selected in the presence of 50 μg/ml gentamycin and 1 μg/ml tetracycline. Purity of P. gingivalis liquid and plate-grown cultures was routinely checked by gram staining and microscopic examination.
Escherichia coli DH5α was used for maintenance and construction of plasmids. DH5α was cultured in Luria-Bertani (LB) broth or on solid medium (LB broth with addition of 1.5% agar). Ampicillin (Na+ salt; 100 μg/ml) was added to the growth media to select for pUC-derived plasmids. E. coli S17-1 grown on LB supplemented with 5 μg/ml tetracycline carrying the complementation construct pT-PG0120 was used for conjugation with P. gingivalis.
Human gingival fibroblasts
The gingival fibroblasts (HGF1 and HGF2) used in this study were collected from extracted third molars of two periodontally healthy subjects with a high pro-inflammatory immunological response when challenged with P. gingivalis [20]. Donors had given written informed consent, and the study was approved by the VUmc Medical Ethical committee.
Genomic DNA isolation from P. gingivalis
Genomic DNA from P. gingivalis strains was isolated from plate-grown bacteria using the DNeasy tissue kit (Qiagen Benelux BV). The DNA concentration of all samples after purification was between 20 ng/μl and 60 ng/μl.
Generation of an insertional knockout construct for epsC
To make an insertional knockout of epsC in the W83 wild type strain we constructed plasmid pΔEpsC. Primers epsC BamHI-F and epsC EcoRI-R (see table 1 for details) were used to amplify the 1.2 Kb epsC gene from P. gingivalis W83 genomic DNA in a PCR reaction. Pfu polymerase (Fermentas, GmbH, St. Leon-Rot, Germany) was used according to the manufacturer's protocol with 100 ng of genomic DNA. The PCR program started with 95°C for 5 min and then 25 cycles of 95°C, 55°C and 72°C for 30 s, 30 s and 2.5 min respectively and was ended by one step of 72°C for 5 min. The amplified fragment was cleaned using the Qiagen PCR purification kit (Qiagen Benelux B.V.) and restricted with Bam HI and Eco RI. This restricted epsC gene fragment was ligated into Bam HI-Eco RI restricted pGEX-6p-3 plasmid to yield pGEX-PG0120.
The 1.2 Kb EryF erythromycin resistance cassettes for use in P. gingivalis was amplified from plasmid pEP4351 using primers EryF ClaI F and EryF ClaI R. and after restriction with Cla I this fragment was ligated into the Cla I-restricted pGEX-PG0120 plasmid yielding pΔEpsC. The Sca I-linearized pΔEpsC plasmid was used for insertional inactivation of epsC in P. gingivalis strain W83.
Complementation of the epsC mutant
The 120 bp artificial constitutive CP25 promoter [37] was amplified from plasmid pDM15 [38] using primers CP25 ClaI F and CP25 AscI R. The intact epsC 1.2 Kb gene was amplified from genomic DNA of P. gingivalis strain W83 using primers epsC AscI F and epsC SpeI R. After ligation of these fragments into cloning vector pJET1.2 (Fermentas, GmbH, St. Leon-Rot, Germany) the constructed expression cassette was cut out with Xho I and Hind III and ligated into the Sal I and Hind III digested pT-COW shuttle plasmid [39] to yield the complementation construct pT-PG0120.
Transformation of P. gingivalis
BHI+H/M was inoculated with P. gingivalis W83 from a 6-day-old blood agar plate. This pre-culture was anaerobically incubated at 37°C for 2 days. 2 ml of the pre-culture was used to inoculate a 100 ml culture. The next day this culture was used to inoculate 2 × 100 ml of fresh BHI+H/M to an OD690 of 0.2. After six hours of anaerobic incubation at 37°C the cells were harvested by centrifugation in mid-exponential phase. The pellet was washed two times in 20 ml EPB (10% glycerol, 1 mM MgCl2) and after that resuspended in 2 ml of EPB. Aliquots of 200 μl were stored at -80°C and used for electroporation.
200 ng of Pst I digested pΔEpsC was added to 200 μl of W83 P. gingivalis cells. The mixture was transferred to a 2 mm electroporation cuvette and electroporated using an Electro Cell Manipulator 600 (BTX Instrument Division, Holliston, MA, USA; 25 μF, 2.5 kV, 186 Ω). 1 ml of BHI+H/M was added immediately after the pulse. The cells were left for recovery anaerobically at 37°C for 18 hours. The suspension was plated on BA+H/M plates with 5 μg/ml erythromycin for selection of the transformants. The authenticity of the insertional knockout epsC mutants was verified using primer combinations epsC BamHI F × PG0119 R and EryF ClaI F × epsC EcoRI R. Furthermore, using Real-Time PCR, the expression of the downstream gene hup-1 in both W83 and the epsC mutant was monitored using primers hup-1 F and hup-1 R to exclude polar effects. W83 and the epsC mutant were grown till early exponential phase. The cell pellets were collected by centrifugation and resuspended in RLT buffer (Qiagen, Benelux B. V.). The cells were disrupted using a Fast Prep Cell Disrupter (Bio 101, Thermo electron corporation, Milford, USA) and centrifuged, the total RNA was extracted from the supernatant according to the manufacturer's protocol of Qiagen RNeasy® mini kit (Qiagen Benelux B.V.). The residual contaminating genomic DNA was removed by Turbo DNA-free™ kit (Ambion, Austin, USA). mRNA was then reverse transcribed using the Fermentas first-strand cDNA synthesis kit (Fermentas GmbH, St. Leon-Rot, Germany) according to the manufacturer's protocol.
The synthesized cDNA was further analyzed using Real-Time PCR with gene-specific primers on an ABI Prism 7000 Sequence Detecting System (Applied Biosystems, Nieuwerkerk a/d lJssel, The Netherlands). Gene expression was normalized to the expression of glucokinase (glk), amplified with primers glk F and glk R [40]. The relative hup-1 expression levels of W83 from three independent experiments were compared in duplicate to those of the epsC mutant.
Conjugation of P. gingivalis
To complement the epsC mutant, plasmid pT-PG0120 was transferred into the mutant by conjugation following a protocol described earlier [41], with slight modifications. For selection of P. gingivalis after the over-night conjugation we used 50 μg/ml of gentamycin in our blood agar plates instead of 150 μg/ml. Integrity of the trans-conjugants was confirmed by colony PCR and plasmid isolation combined with restriction analysis using a plasmid isolation kit (Qiagen Benelux B.V.).
Percoll density gradient centrifugation
Percoll density gradients were in principle prepared as described by Patrick and Reid [24]. In short, a 9:1 stock solution of Percoll (Pharmacia, Biotech AB, Uppsala, Sweden) was prepared with 1.5 M NaCl. Solutions containing 80, 70, 60, 50, 40, 30, 20 and 10% Percoll in 0.15 NaCl were prepared from the stock. In an open top 14 ml polycarbonate tube (Kontron instruments, Milan, Italy) 1.5 ml of each of the solutions was carefully layered on top of the previous starting with 80%. 1 ml of an anaerobically grown over night culture of wild type and the epsC mutant concentrated to an OD690 of 4 in PBS was added to the top of the 10% layer and centrifuged for one hour at 8000 × g at 20°C in a Centrikon TST 41.14 rotor (Kontron instruments, Milan, Italy) using a Centrikon T-1170 (Kontron instruments, Milan, Italy) centrifuge.
Hydrophobicty of P. gingivalis
W83, the epsC mutant and the complemented mutant were grown 18 hours in BHI+H/M. The bacteria were washed twice in PBS after which the OD600 was set to 0.5. After addition of 150 μl n-hexadecane to 3 ml of this suspension the mix was vortexed 30 seconds, rested for 5 seconds and vortexed for 25 seconds. After exactly 10 minutes incubation at room temperature a sample was taken to measure the OD600 of the aqueous phase. The percentage of bacteria adhered to hexadecane was calculated by the formula: (OD600 before-OD600 after)/OD600 before × 100%. The presented data in Additional figure 1 were collected from two experiments using triplicate measurements.
P. gingivalis serotyping
Serotyping of P. gingivalis was based on the detection of the six described K-antigens [8, 9]. In short, serotype-specific, polyclonal antisera were obtained after immunization of rabbits with whole bacterial cells of the six P. gingivalis type strains [42]. Bacterial antigens for double immunodiffusion tests were prepared as described previously [8]. Immunodiffusion was carried out in 1% agarose (Sigma Chemical Co., St. Louis, MO, type 1, low EEO) in 50 mM Tris-HCl buffer (pH 8.6). 10 μl antiserum and 10 μl of antigen were loaded and allowed to diffuse and precipitate for 48 hours at room temperature.
India ink negative staining
P. gingivalis cells were taken from 4 day-old plates and resuspended in 1 ml of PBS. On a glass slide 10 μl of this suspension was mixed with 10 μl of India ink (Talens, Apeldoorn, The Netherlands) and using another glass slide a thin film was made. The film was air-dried. A drop of 0.2% fuchsine was carefully added onto the film and removed after 2 minutes by decanting. Then the film was air-dried. Pictures were taken with a Leica DC500 camera on a Zeiss Axioskop using phase-contrast.
Growth curve
Pre-cultures of W83 and the epsC mutant were grown anaerobically for 18 hours in BHI+H/M at 37°C. The pre-cultures were diluted to an OD690 of 0.05 in duplo in fresh BHI+H/M and incubated anaerobically at 37°C. Every few hours the OD690 was measured and a sample was taken for cfu-counts.
Sedimentation of P. gingivalis
W83 and the epsC mutant were grown anaerobically for 18 hours in BHI+H/M at 37°C. After 3 wash steps in phosphate buffered saline (PBS) the OD690 was standardized to 5 in DMEM with 10% FCS. 10 ml of this culture was added to 40 ml DMEM with 10% FCS in a 100 ml flask to set the OD690 to 1. The cultures were incubated standing still at 37°C for six hours. At regular time intervals, a 200 μl sample was taken 0.5 cm from the liquid surface and the decrease of the OD690 values was determined as a measure for sedimentation.
Survival of P. gingivalis
W83, the epsC mutant and the complemented mutant were grown anaerobically for 18 hours in BHI+H/M at 37°C. After 2 wash steps in phosphate buffered saline (PBS) the pellets were resuspended in DMEM with 10% FCS to an OD690 of 0.05 as used in fibroblast infections at MOI 10.000:1. 500 μl of these suspensions was incubated at 37°C in a humidified atmosphere of 5% CO2 in air. Samples for cfu-counts were taken at t = 0 hours, t = 3 hours and t = 6 hours and dilutions were plated on BA+H/M plates.
Infection of gingival fibroblasts with P. gingivalis
Bacteria were grown overnight for 18 hours in BHI+H/M. The bacterial cells were washed three times in PBS and then used to infect gingival fibroblasts at MOIs of 1000:1 and 10.000:1 (bacteria cells: fibroblasts) in a total volume of 500 μl DMEM with 10% FCS in 24-well plates. The plates were incubated for 6 hours at 37°C in a humidified atmosphere of 5% CO2 in air. The cells were washed twice with cold PBS. Then 350 μl lysis buffer (1% β-mercapthanol in RLT buffer) was added to the cells according to the protocol of Qiagen RNeasy® mini kit (Qiagen Benelux B.V.) after which the plate was stored at -80°C for later use.
RNA isolation and reverse transcription
mRNA was isolated from the gingival fibroblast lysates according to the manufacturer's protocol of Qiagen RNeasy® mini kit (Qiagen Benelux B.V.). The mRNA concentrations of the samples were determined using the Nanodrop ND_1000 (Isogen Life Science). mRNA was reverse transcribed using the Fermentas first-strand cDNA synthesis kit (Fermentas GmbH, St. Leon-Rot, Germany) according to the manufacturer's protocol.
Real-Time PCR
cDNA synthesized from mRNA isolated from gingival fibroblasts after infection with P. gingivalis was analyzed in quadruple using Real-Time PCR with gene-specific primers on a ABI Prism 7000 Sequence Detecting System (Applied Biosystems, Nieuwerkerk a/d lJssel, The Netherlands). Reactions were performed with 2 ng cDNA in a total volume of 8 μl containing SYBR Green PCR Master Mix (Applied Biosystems) and 0.99 pM of each primer. After activation of the AmpliTaq Gold DNA polymerase for 10 minutes at 94°C, 40 cycles were run of a two step PCR consisting of a denaturation step at 95°C for 30 seconds and annealing and extension step at 60°C for 1 minute. Predicted product sizes were in the 100-200 bp range. Subsequently the PCR products were subjected to melting curve analysis to test if any unspecific PCR products were generated. The PCR reactions of the different amplicons had equal efficiencies. Samples were normalized for the expression of housekeeping gene GAPDH, which is not affected by the experimental conditions, by calculating the Δ Ct (Ct housekeeping gene - Ct gene of interest) and expression of the different genes is expressed as 2-(ΔCt). Fold increase in gene expression (induction) was expressed by 2 -(ΔΔCt), wherein ΔΔCt = ΔCtchallenged- average Ct-value non-challenged.
Statistical analysis
Differences in gene induction between multiple groups were tested by one-way analysis of variance (ANOVA) and Bonferroni's Multiple Comparison Test. Tests were performed with GraphPad Prism version 4.00 for Windows, GraphPad Software, San Diego California USA. Differences were considered significant at p < 0.01.
References
Lafaurie GI, Contreras A, Baron A, Botero J, Mayorga-Fayad I, Jaramillo A, Giraldo A, Gonzalez F, Mantilla S, Botero A: Demographic, clinical, and microbial aspects of chronic and aggressive periodontitis in Colombia: a multicenter study. J Periodontol. 2007, 78 (4): 629-639. 10.1902/jop.2007.060187.
Haffajee AD, Socransky SS: Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994, 5: 78-111. 10.1111/j.1600-0757.1994.tb00020.x.
Page RC, Offenbacher S, Schroeder HE, Seymour GJ, Kornman KS: Advances in the pathogenesis of periodontitis: summary of developments, clinical implications and future directions. Periodontol 2000. 1997, 14: 216-248. 10.1111/j.1600-0757.1997.tb00199.x.
Grenier D, Mayrand D: Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J Clin Microbiol. 1987, 25 (4): 738-740.
Laine ML, Appelmelk BJ, van Winkelhoff AJ: Prevalence and distribution of six capsular serotypes of Porphyromonas gingivalis in periodontitis patients. J Dent Res. 1997, 76 (12): 1840-1844. 10.1177/00220345970760120601.
Neiders ME, Chen PB, Suido H, Reynolds HS, Zambon JJ, Shlossman M, Genco RJ: Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989, 24 (3): 192-198. 10.1111/j.1600-0765.1989.tb02005.x.
van Steenbergen TJ, Delemarre FG, Namavar F, de Graaff J: Differences in virulence within the species Bacteroides gingivalis. Antonie Van Leeuwenhoek. 1987, 53 (4): 233-244. 10.1007/BF00393930.
Laine ML, Appelmelk BJ, van Winkelhoff AJ: Novel polysaccharide capsular serotypes in Porphyromonas gingivalis. J Periodontal Res. 1996, 31 (4): 278-284. 10.1111/j.1600-0765.1996.tb00494.x.
van Winkelhoff AJ, Appelmelk BJ, Kippuw N, de Graaff J: K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol Immunol. 1993, 8 (5): 259-265. 10.1111/j.1399-302X.1993.tb00571.x.
Laine ML, van Winkelhoff AJ: Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998, 13 (5): 322-325. 10.1111/j.1399-302X.1998.tb00714.x.
d'Empaire G, Baer MT, Gibson FC: K1 serotype capsular polysaccharide of Porphyromonas gingivalis elicits chemokine production from murine macrophages that facilitates cell migration. Infect Immun. 2006, 74 (11): 6236-43. 10.1128/IAI.00519-06.
Farquharson SI, Germaine GR, Gray GR: Isolation and characterization of the cell-surface polysaccharides of Porphyromonas gingivalis ATCC 53978. Oral Microbiol Immunol. 2000, 15 (3): 151-157. 10.1034/j.1399-302x.2000.150302.x.
Davey ME, Duncan MJ: Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol. 2006, 188 (15): 5510-5523. 10.1128/JB.01685-05.
Rosen G, Sela MN: Coaggregation of Porphyromonas gingivalis and Fusobacterium nucleatum PK 1594 is mediated by capsular polysaccharide and lipopolysaccharide. FEMS Microbiol Lett. 2006, 256 (2): 304-310. 10.1111/j.1574-6968.2006.00131.x.
Domenico P, Salo RJ, Cross AS, Cunha BA: Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect Immun. 1994, 62 (10): 4495-4499.
Noel GJ, Hoiseth SK, Edelson PJ: Type b capsule inhibits ingestion of Haemophilus influenzae by murine macrophages: studies with isogenic encapsulated and unencapsulated strains. The Journal of infectious diseases. 1992, 166 (1): 178-182.
Glynn AA, Howard CJ: The sensitivity to complement of strains of Escherichia coli related to their K antigens. Immunology. 1970, 18 (3): 331-346.
Aduse-Opoku J, Slaney JM, Hashim A, Gallagher A, Gallagher RP, Rangarajan M, Boutaga K, Laine ML, van Winkelhoff AJ, Curtis MA: Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect Immun. 2006, 74 (1): 449-460. 10.1128/IAI.74.1.449-460.2006.
Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, Duncan MJ: Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004, 186 (16): 5473-5479. 10.1128/JB.186.16.5473-5479.2004.
Scheres N, Laine ML, de Vries TJ, Everts V, van Winkelhoff AJ: Gingival and periodontal ligament fibroblasts differ in their inflammatory response to viable Porphyromonas gingivalis. J Periodontal Res. 2009,
Schroeder HE, Munzel-Pedrazzoli S, Page R: Correlated morphometric and biochemical analysis of gingival tissue in early chronic gingivitis in man. Archives of oral biology. 1973, 18 (7): 899-923. 10.1016/0003-9969(73)90060-5.
Lekic PC, Pender N, McCulloch CA: Is fibroblast heterogeneity relevant to the health, diseases, and treatments of periodontal tissues?. Crit Rev Oral Biol Med. 1997, 8 (3): 253-268. 10.1177/10454411970080030201.
Nagasawa T, Kobayashi H, Kiji M, Aramaki M, Mahanonda R, Kojima T, Murakami Y, Saito M, Morotome Y, Ishikawa I: LPS-stimulated human gingival fibroblasts inhibit the differentiation of monocytes into osteoclasts through the production of osteoprotegerin. Clinical and experimental immunology. 2002, 130 (2): 338-344. 10.1046/j.1365-2249.2002.01990.x.
Patrick S, Reid JH: Separation of capsulate and non-capsulate Bacteroides fragilis on a discontinuous density gradient. J Med Microbiol. 1983, 16: 239-241. 10.1099/00222615-16-2-239.
Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, Rossetti CA, Lawhon SD, Chu H, Lau T, Bevins CL: The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect Immun. 2007, 75 (9): 4342-4350. 10.1128/IAI.01571-06.
Schembri MA, Dalsgaard D, Klemm P: Capsule shields the function of short bacterial adhesins. J Bacteriol. 2004, 186 (5): 1249-1257. 10.1128/JB.186.5.1249-1257.2004.
Shifrin Y, Peleg A, Ilan O, Nadler C, Kobi S, Baruch K, Yerushalmi G, Berdichevsky T, Altuvia S, Elgrably-Weiss M: Transient shielding of intimin and the type III secretion system of enterohemorrhagic and enteropathogenic Escherichia coli by a group 4 capsule. J Bacteriol. 2008, 190 (14): 5063-5074. 10.1128/JB.00440-08.
Dubail I, Bigot A, Lazarevic V, Soldo B, Euphrasie D, Dupuis M, Charbit A: Identification of an essential gene of Listeria monocytogenes involved in teichoic acid biogenesis. J Bacteriol. 2006, 188 (18): 6580-6591. 10.1128/JB.00771-06.
Yoshida K, Matsumoto T, Tateda K, Uchida K, Tsujimoto S, Yamaguchi K: Induction of interleukin-10 and down-regulation of cytokine production by Klebsiella pneumoniae capsule in mice with pulmonary infection. Journal of medical microbiology. 2001, 50 (5): 456-461.
Gibson FC, Tzianabos AO, Onderdonk AB: The capsular polysaccharide complex of Bacteroides fragilis induces cytokine production from human and murine phagocytic cells. Infect Immun. 1996, 64 (3): 1065-1069.
Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Baumler AJ: The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun. 2005, 73 (6): 3367-3374. 10.1128/IAI.73.6.3367-3374.2005.
Vann WF, Daines DA, Murkin AS, Tanner ME, Chaffin DO, Rubens CE, Vionnet J, Silver RP: The NeuC protein of Escherichia coli K1 is a UDP N-acetylglucosamine 2-epimerase. J Bacteriol. 2004, 186 (3): 706-712. 10.1128/JB.186.3.706-712.2004.
Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, Baumler AJ: The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cellular microbiology. 2008, 10 (4): 876-890. 10.1111/j.1462-5822.2007.01090.x.
Sojar HT, Hamada N, Genco RJ: Isolation and characterization of fimbriae from a sparsely fimbriated strain of Porphyromonas gingivalis. Applied and environmental microbiology. 1997, 63 (6): 2318-2323.
Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE: Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001, 414 (6863): 555-558. 10.1038/35107092.
Tran QT, Gomez G, Khare S, Lawhon SD, Raffatellu M, Baumler AJ, Ajithdoss D, Dhavala S, Adams LG: The Salmonella enterica serotype Typhi Vi capsular antigen is expressed after entering the ileal mucosa. Infect Immun. 2010, 78 (1): 527-35. 10.1128/IAI.00972-09.
Jensen PR, Hammer K: The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Applied and environmental microbiology. 1998, 64 (1): 82-87.
Deng DM, Liu MJ, ten Cate JM, Crielaard W: The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res. 2007, 86 (7): 606-610. 10.1177/154405910708600705.
Gardner RG, Russell JB, Wilson DB, Wang GR, Shoemaker NB: Use of a modified Bacteroides -Prevotella shuttle vector to transfer a reconstructed beta-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Applied and environmental microbiology. 1996, 62 (1): 196-202.
Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, Kolenbrander PE: Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol. 2006, 188 (7): 2454-2462. 10.1128/JB.188.7.2454-2462.2006.
Belanger M, Rodrigues P, Progulske-Fox A: Genetic manipulation of Porphyromonas gingivalis. Current protocols in microbiology. 2007, Chapter 13 (Unit13C): 12-
van Winkelhoff AJ, Kippuw N, de Graaff J: Serological characterization of black-pigmented Bacteroides endodontalis. Infect Immun. 1986, 51 (3): 972-974.
Acknowledgements
We would like to thank Jeffrey Kroon for his excellent work on the transcriptional analysis of the P. gingivalis genes.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
JB performed the cloning work, mutant construction, hydrophobicity test, density gradient centrifugation, negative staining, serotyping and drafted the manuscript. NBEI made the growth curves and did the sedimentation assay. NS and NBEI together performed the fibroblast infection experiments, the transcription analyses and statistical analyses. DMD analyzed the strains using Real-Time PCR and performed part of the statistical analysis. ML, AJvW and WC were involved in the study design, supervision and helped to draft the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12866_2009_961_MOESM1_ESM.PNG
Additional file 1: Hydrophobicity of P. gingivalis strains. Percentage of bacterial cells adhered to hexadecane after extensive vortexing and 10 minutes incubation. 3.4%, 61% and 19% of the cells was adhered to hexadecane for W83, the epsC mutant and the complemented mutant respectively, indicating increased hydrophobicity for the epsC mutant. The data are the averages of two experiments comprised of triplicate measurements. The bars show the standard deviations. (PNG 8 KB)
12866_2009_961_MOESM2_ESM.PNG
Additional file 2: Effect of complementation of the epsC mutant on the immune response mutant of human gingival fibroblasts (HGF2). After a 6-hour challenge with P. gingivalis cells at MOI 10.000:1, the expression levels of IL-1β, IL-6 and IL-8 in human gingival fibroblasts were measured using RT-PCR and if possible represented as a relative value compared to a non-infected control sample which is set to a value of 1. Relative IL-1β expression could not be calculated as IL-1β was not detected in the non-infected control. Complementation almost restored the wild-type situation for IL-1β (83%), IL-6 (83%) and IL-8 (77%). (PNG 10 KB)
12866_2009_961_MOESM3_ESM.PNG
Additional file 3: Six hour survival of W83, the epsC mutant and the complemented mutant under aerobic experimental conditions. Survival of W83, the epsC mutant and the complemented mutant in 0.5 ml DMEM + 10% FCS under humidified 5% CO2 conditions was determined by cfu-counts on BA + H/M plates. Survival of 67%, 60 and 73% was found for each strain respectively. Error bars represent the standard deviations of triplicate measurements. (PNG 10 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Brunner, J., Scheres, N., El Idrissi, N.B. et al. The capsule of Porphyromonas gingivalis reduces the immune response of human gingival fibroblasts. BMC Microbiol 10, 5 (2010). https://doi.org/10.1186/1471-2180-10-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-10-5