Abstract
Background
Fluoroquinolones are potent antimicrobial agents used for the treatment of a wide variety of community- and nosocomial- infections. However, resistance to fluoroquinolones in Enterobacteriaceae is increasingly reported. Studies assessing the ability of fluoroquinolones to select for resistance have often used antimicrobial concentrations quite different from those actually acquired at the site of infection. The present study compared the ability to select for resistance of levofloxacin, ciprofloxacin and prulifloxacin at concentrations observed in vivo in twenty strains of Escherichia coli and Klebsiella spp. isolated from patients with respiratory and urinary infections. The frequencies of spontaneous single-step mutations at plasma peak and trough antibiotic concentrations were calculated. Multi-step selection of resistance was evaluated by performing 10 serial cultures on agar plates containing a linear gradient from trough to peak antimicrobial concentrations, followed by 10 subcultures on antibiotic-free agar. E. coli resistant strains selected after multi-step selection were characterized for DNA mutations by sequencing gyrA, gyrB, parC and parE genes.
Results
Frequencies of mutations for levofloxacin and ciprofloxacin were less than 10-11 at peak concentration, while for prulifloxacin they ranged from <10-11 to 10-5. The lowest number of resistant mutants after multistep selection was selected by levofloxacin followed by ciprofloxacin and prulifloxacin. Both ciprofloxacin- and prulifloxacin-resistant mutants presented mutations in gyrA and parC, while levofloxacin resistance was found associated only to mutations in gyrA.
Conclusions
Among the tested fluoroquinolones, levofloxacin was the most capable of limiting the occurrence of resistance.
Similar content being viewed by others
Background
Escherichia coli is worldwide the most frequent pathogen isolated from uncomplicated urinary tract infections (UTI) (70 - 95%) and, in bacteremia of nosocomial or community origin, it represents about the 15.5% and 42.1% of aetiologies, respectively [1]. Also Klebsiella spp., especially Klebsiella pneumoniae, are involved in uncomplicated UTI for 5% and represent 4.1% of bacteremias, the mortality of nosocomial infections being more than twice that of community-acquired infection [1, 2].
Fluoroquinolones (FQ) are potent antimicrobial agents used for the treatment of a wide variety of community- and nosocomial- infections. However, increasing resistance to FQ in E. coli isolated from community acquired UTI has been recently reported, with up to 29% of women harbouring FQ resistant E. coli, although FQ resistance rates varied significantly according to sex, age, type of urinary infection and geographic region [3–6]. Moreover, infections due to extended-spectrum beta-lactamases (ESBL) - producing Enterobacteriaceae are an emerging problem in the community since an high proportion of these microorganisms have been isolated from urine samples of women with uncomplicated UTI [7].
Ciprofloxacin use and ESBL production have been shown to be significantly correlated in a study on K. pneumoniae[8]. ESBL-producing strains have been shown to be significantly more frequent among ciprofloxacin-resistant E. coli than among ciprofloxacin-susceptible E. coli strains [9]. Moreover, prior use of FQs, an indwelling urinary catheter, and an invasive procedure within 72 hr prior to bacteremia have been identified as independent risk factors for ciprofloxacin resistance in bloodstream infections due to ESBL E. coli and Klebsiella spp. [2, 10–12].
Several studies have assessed the ability of FQs to select for resistance by subculturing bacteria at concentrations close to MICs. However, the antimicrobial concentrations used in these studies were quite different from those actually acquired at the site of infection [13–16]. For these reasons, we have recently modified the methodologies used to assess in vitro the selection for resistance by testing antimicrobial concentrations reported to occur in vivo[17]. The aim of the present study was to compare the ability of levofloxacin, ciprofloxacin and prulifloxacin to in vitro select for resistance in E. coli and Klebsiella spp. clinical isolates at peak (Cmax) and trough (Cmin) plasma concentrations.
Results
Susceptibility to fluoroquinolones
Basal MICs of E. coli strains ranged from 0.016 mg/L to 1 mg/L, from 0.004 mg/L to 0.5 mg/L and from 0.016 mg/L to 0.125 mg/L for levofloxacin, ciprofloxacin and prulifloxacin, respectively. MICs of Klebsiella spp. ranged between 0.03 mg/L and 1 mg/L, 0.016 mg/L and 0.5 mg/L, and 0.03 and 0.25 mg/L for levofloxacin, ciprofloxacin and prulifloxacin, respectively.
Frequency of mutation
Levofloxacin, 500 and 750 mg, and ciprofloxacin 500 mg limited bacterial growth with median frequencies of mutations below 10-11 at plasma Cmax. Median frequencies of mutations for prulifloxacin were generally higher than comparators ranging from 10-7 to 10-8 and from 10-8 to 10-9 at plasma Cmax in E. coli and Klebsiella spp., respectively (Table 1). Table 2 shows MIC values of the strains that were able to grow in the presence of the above mentioned concentrations of all tested antimicrobials. While no strain was able to grow at Cmax for levofloxacin and ciprofloxacin, 3 and 5 strains grew at prulifloxacin Cmax. These strains showed increments in MICs from 32 to 128 times for E. coli and from 32 to 128 times for Klebsiella spp. with respect to the basal values. Since in some instances, Cmin for all the study drugs, except for levofloxacin at 750 mg dosage, were below MIC values, some strains were able to diffusely grow on the agar plate. For these strains, in order to detect any change in bacterial susceptibility, MICs were evaluated for randomly sampled colonies (Table 2).
Multi-step selection of resistant bacteria
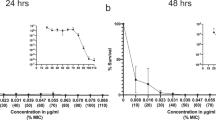
Table 3 shows the total number of strains grown after multi-step selection and MIC values after 1, 5 and 10 passages on antibiotic-gradient plates and after the subsequent 10 passages on antibiotic-free medium. After multi-step selection, a general increment in MICs was observed for all microrganisms with all tested antibiotics; no selection of resistance was observed with levofloxacin at 750 mg in E. coli and no selection of resistance was observed with levofloxacin (both doses) in Klebsiella spp.
After 10 passages on antibiotic gradient plates and 10 subcultures in antibiotic-free medium, the highest number of strains with MIC higher than the resistance breakpoint was found for ciprofloxacin and prulifloxacin both in E. coli (5 and 7 strains, respectively) and Klebsiella spp. (6 and 8 strains, respectively). Only 4 strains with MIC higher than resistance breakpoint were found with levofloxacin at 500 mg in E. coli and Klebsiella spp., whereas no resistant strains selected with the 750 mg either in E. coli or in Klebsiella spp. (Figure 1).
Multi-step selection of resistance in E. coli (A) and Klebsiella spp. (B) at plasma concentration of fluoroquinolones. 1, 5, 10 step: number of passages on antibiotic gradient agar plates. 10 step free: passages on antibiotic free agar plates. Black bars: prulifloxacin; White bars: ciprofloxacin; Dotted bars: levofloxacin.
Characterization of acquired resistance
Strains of E. coli that were selected by the multi-step assay and were able to maintain their resistance after 10 passages in antibiotic-free medium, were evaluated for acquired resistance.
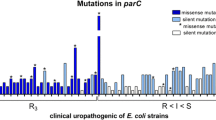
Among 16 resistant mutants, alterations in both gyrA and parC were found in 12 mutants for ciprofloxacin (n = 5) and prulifloxacin (n = 7), while only alterations in gyrA were found for levofloxacin. As reported in table 4, the 4 strains resistant to levofloxacin showed changes in Ser83Leu and Asp87Asn; while in ciprofloxacin- and prulifloxacin-resistant mutants, the mutations identified were Ser83Leu in GyrA and Ser80Ile in ParC. The same mutations were not found in the respective parent strains.
Discussion
Wild-type E. coli and K. pneumoniae clinical isolates are susceptible to quinolones, but resistance to these agents in Gram-negative bacteria has increased in recent years, probably caused by excessive and inappropriate use of these drugs [18]. Particularly, due to under-dosing and mono-therapy against moderately susceptible pathogens, FQ resistance has developed among common pathogens, like E. coli and Klebsiella spp., mainly conferred by ESBLs and AmpC enzymes [19]. ESBL production has been reported to be two times more common in infected patients who received ciprofloxacin than in those who did not (15% vs 7.4%) [8].
In a study performed over 5 years in Croatia on changes in susceptibility of E. coli from UTI, Moeal et al have shown a statistically significant change in antimicrobial resistance over that period only for ciprofloxacin [20]. This has been hypothesized to be related to the inappropriate use of quinolones for humans as well as in veterinary medicine [21]. Prolonged use (> 20 days) of low dose (250 mg twice a day) of the more potent fluoroquinolones such as ciprofloxacin or levofloxacin, has been shown to be the most significant risk factor for acquisition of resistance [22, 23]. Strategies to counteract bacterial resistances include use of the appropriate dosages of these molecules for the correct indication and/or use of synergistic combinations, particularly in the more complicated infections.
Results of this study indicate that levofloxacin presents the lowest frequencies of mutations at plasma Cmax (<10-11) and a lower propensity than prulifloxacin or ciprofloxacin to select in vitro for resistance. In regard to genetic characterization of resistance, only alterations in gyrA were found for levofloxacin, however, alterations in gyrA and parC were found for ciprofloxacin and prulifloxacin. Point mutations within DNA gyrase are known to cause a reduction in the affinity of the enzyme for FQs, decreasing the susceptibility of bacteria to these molecules. Topoisomerase IV is the second target for FQ in the absence of susceptible gyrase. Therefore, multiple mutations in gyrA and/or parC are required for high level FQ resistance in E. coli[23, 24]. In our study, both ciprofloxacin and prulifloxacin resistant mutants presented mutations in gyrA and parC, while levofloxacin resistance was found associated only with mutations in gyrA. These results seem to indicate that levofloxacin resistance at a concentration observed during treatment might develop more slowly and might be lower than resistance to the other FQs tested in the present study.
However, this study did not evaluated other mechanisms other than the target enzyme that might be involved in the observed resistant strains, including decreased intracellular drug accumulation as a result of alterations in the outer membrane proteins of the wall cell, or active efflux of the drug mediated by a number of efflux pumps.
As far as FQ resistance in Klebsiella spp. is concerned, plasmid-mediated quinolone resistance mechanisms associated with the qnr gene and the aac(6')-Ib-cr gene in ESBL producing strains have been described [25, 26]. The first encodes target protection proteins of the pent peptide repeat family and seems to be associated with low level quinolone resistance, while the aac(6')-Ib-cr gene encodes a variant of the common aminoglycoside acetyltransferase which is able to reduce the activity of some FQ, thus enhancing the selection of chromosomal mutations [25]. Although in the present study the presence of plasmid-mediated resistance was not investigated, it can not be excluded that these genes might be involved in selection of resistance observed after serial exposure to fluoroquinolones.
In a previous study, we have shown that combinations of a fluoroquinolone with a beta-lactam may both provide improved antimicrobial activity and limit the occurrence of resistance in ESBL-producing E. coli clinical isolates [27]. Therefore, the use of combination therapy could be an attractive strategy to limit occurrence of resistance.
Conclusions
In conclusion, among the tested fluoroquinolones, levofloxacin was the most able to limit occurrence of resistance in vitro. However, in order to limit the occurrence of resistance, appropriate dosages of fluoroquinolones should be respected in the therapy of infections caused by Enterobacteriaceae, as well as use of synergistic combinations in the most complicated infections.
Methods
Strains
Twenty clinical isolates of E. coli and Klebsiella spp., collected from patients presenting with community infections in 2005 at L. Sacco Hospital, Milan, were included into the study. Susceptibility to the drugs under evaluation was considered as a pre-requisite for the study. One isolate per patient was used in order to avoid inclusion of the same strain. All isolates were stored at -80°C in brain-heart infusion broth containing 10% (w/v) glycerol until use.
Antibiotics
Levofloxacin (sanofi-aventis, S.p.A. Milan, Italy); ciprofloxacin (Bayer Italia, S.p.A., Milan, Italy), and prulifloxacin (Aziende Chimiche Riunite Angelini Francesco ACRAF S.p.A, S. Palomba-Pomezia, Italy) were used to prepare stock solutions at concentrations of 5120 mg/L. Plasma maximum and minimum concentrations (Cmax, Cmin) of each antimicrobial studied were chosen from those obtained at steady state in previously published studies after oral administration [28–31]. Thus, the Cmax were as following: levofloxacin 500 mg (5.29 mg/L); levofloxacin 750 mg (11.98 mg/L); ciprofloxacin 500 mg (2.11 mg/L); prulifloxacin 600 mg (2 mg/L) [28–31]. The tested plasma Cmin were respectively: 0.60 mg/L for levofloxacin 500 mg; 1.69 mg/L for levofloxacin 750 mg; 0.08 mg/L for ciprofloxacin 500 mg; 0.10 mg/L for prulifloxacin 600 mg [28–31].
Determination of MIC
Antibiotic susceptibilities to the study drugs were determined by the microdilution broth assay in accordance with CLSI approved standards [32]. Since no CLSI breakpoints for prulifloxacin against E. coli and Klebsiella spp. were available, reduced susceptibility to this agent was defined as a MIC ≥ 4 mg/L [32]. Resistance to levofloxacin and ciprofloxacin was defined by MIC values ≥ 8 and 4 mg/L, respectively [33].
Frequency of mutation
Colonies from an overnight culture in Mueller Hinton agar were resuspended in brain heart infusion (BHI) broth at a load of about 1010 CFU/mL. An aliquot of 100 μL from the bacterial suspension was spread onto Mueller Hinton agar plates containing antibiotics at plasma Cmax and Cmin, as reported above. After incubation for 72 h, the frequency of mutation was calculated from the ratio between colonies grown on antibiotic-containing plates and the initial inoculum, determined by plating 100 μL of bacterial suspension, after proper dilution, onto Mueller Hinton agar plates. Five colonies from each antibiotic containing plate were randomly selected and their MIC for the corresponding antibiotic was determined as described above. When MIC was higher than the tested concentration, as occurred for Cmin for some strains, so that colony counts was not possible because of extensive growth on plate surface, frequency of mutation was not calculated, but the MIC was equally determined.
Multi-step selection of resistant bacteria
The ability to select for antibiotic resistance was evaluated by performing serial subcultures on Mueller Hinton agar plates, containing a gradient ranging from Cmax to Cmin. Gradients were prepared in Petri dishes, which were poured with two layers of agar, as described elsewhere [34]. The bottom layer consisted of Mueller Hinton agar containing the antibiotic at Cmin, which was allowed to harden with the plate slanted sufficiently to cover the entire bottom. The top layer, added to the dish in the normal position, contained antibiotics at Cmax.
An inoculum of 1010 CFU/mL of each strain was homogenously spread onto each plate and incubated for 48 hrs at 37°C. After incubation, colonies grown at the highest drug concentration were sampled, checked for purity, and re-plated on a new antibiotic-containing agar plates. A total of 10 consecutive passages on antibiotic containing plates were followed by 10 passages on antibiotic-free plates in order to evaluate stability of acquired resistance. MIC values were determined after 1, 5 and 10 passages on antibiotic containing plates and after 5 and 10 passages in antibiotic free medium in order to evaluate stability of acquired resistance. Acquisition of resistance was defined as a MIC value higher than resistance breakpoint.
Characterization of acquired resistance
To determine whether E. coli mutants that had acquired stable resistance to quinolones had alterations in topoisomerase IV or DNA gyrase, parC, parE, gyrA, and gyrB were amplified by PCR and sequenced as described previously [35].
Amplification products were purified with the QIAquick PCR purification kit (Qiagen Inc., Milan Italy) using the manufacturer's instructions. Sequencing was performed on an ABI PRISM 310 genetic analyzer (Applied Biosystems, Monza, Italy).
Only mutations known to be associated with resistance to fluoroquinolones were considered (Ser83, Asp87 and Ala93 in GyrA, Ser80 and Glu84 in ParC) [36].
References
Luzzaro F, Viganò EF, Fossato D, Grossi A, Sala A, Sturla C, Saudelli M, Toniolo A, AMCLI Lombardia Hospital Infectious Study Group: Prevalence and drug susceptibility of pathogens causing bloodstream infections in northern Italy: a two years study in 16 hospitals. Eur J Clin Microbiol Infect Dis. 2002, 21: 849-855.
Kang CI, Kim SH, Bang JW, Kim HB, Kim NJ, Kim EC, Oh MD, Choe KW: Community-acquired versus nosocomial Klebsiella pneumoniae bacteriemia: clinical features, treatment outcomes, and clinical implication of antimicrobial resistance. J Korean Med Sci. 2006, 21: 816-822. 10.3346/jkms.2006.21.5.816.
Gobernado M, Valdes L, Alos JI, García Rey C, Dal-Ré Saavedra R, García de Lomas J: Quinolone resistance in female outpatient urinary tract isolates of Escherichia coli: age-related differences. Rev Esp Quimioterap. 2007, 20: 206-210.
Andreu A, Alos JI, Gobernado M, Marco F, de la Rosa M, García-Rodríguez JA, García-Rodríguez JA, Grupo Cooperativo Español para el Estudio de la Sensibilidad Antimicrobiana de los Patógenos Urinarios: Etiology and antimicrobial susceptibility among uropathogens causing community-acquired urinary tract infections: a nationwide surveillance study. Enferm Infec Microbiol Clin. 2005, 23: 4-9. 10.1157/13070401.
Aypack C, Altunsoy A, Duzgun N: Empiric antibiotic therapy in acute uncomplicated infections and fluoroquinolones resistance: a prospective observational study. Ann Clin Microbiol Antimicrob. 2009, 24: 27-10.1186/1476-0711-8-27.
McIsaac WJ, Mazzulli T, Permaul J, Moineddin R, Low DE: Community-acquired antibiotic resistance in urinary isolates from adult women in Canada. Can J Infect Dis Med Microbiol. 2006, 17: 337-340.
Muratani T, Matsumoto T: Urinary tract infection caused by fluoroquinolone- and cephem-resistant Enterobacteriaceae. Int J Antimicrob Agents. 2006, 28 (suppl 1): 10-13. 10.1016/j.ijantimicag.2006.05.009.
Arslan H, Azap OK, Ergonul O, Timurkaynak F, Urinary Tract Infection Study Group: Risk factors for ciprofloxacin resistance among Escherichia coli strains isolated from community-acquired urinary tract infections in Turkey. J Antimicrob Chemother. 2005, 56: 914-918. 10.1093/jac/dki344.
Tolun V, Kucukbasmaci O, Torumkuney-Akbulut D, Catal C, Anğ-Küçüker M, Anğ O: Relationship between ciprofloxacin resistance and extended-spectrum beta-lactamase production in Escherichia coli and Klebsiella pneumoniae strains. Clin Microbiol Infect. 2004, 10: 72-75. 10.1111/j.1469-0691.2004.00723.x.
Lin CY, Huang SH, Chen TC, Lu PL, Lin WR, Chen YH: Risk factors of ciprofloxacin resistance in urinary Escherichia coli isolates. J Microbiol Immunol Infect. 2008, 41: 325-331.
Killgore KM, March KL, Guglielmo BJ: Risk factors for community-acquired ciprofloxacin resistant Escherichia coli urinary tract infection. Ann Pharmacother. 2004, 38: 1148-1152. 10.1345/aph.1D622.
Pena C, Albareda JM, Pallares R, Pujol M, Tubau F, Ariza J: Relationship between quinolones use and emergence of ciprofloxacin-resistant Escherichia coli in bloodstream infections. Antimicrob Agents Chemother. 1995, 39: 520-524.
Cebríán L, Rodríguez JC, Escribiano I, Royo SG: Evaluation of several fluoroquinolones and beta-lactams in terms of their capability to restrict the selection of fluoroquinolone-resistant Salmonella: in vitro models. APMIS. 2007, 15: 1376-1382. 10.1111/j.1600-0463.2007.00799.x.
Kim MJ, Yun HJ, Kang JW, Kim S, Kwak JH, Choi EC: In vitro development of resistance to a novel fluoroquinolone, DW286, in methicillin-resistant Staphylococcus aureus clinical isolates. J Antimicrob Chemother. 2003, 51: 1011-1016. 10.1093/jac/dkg162.
Browne FA, Clark C, Bozdogan B, Dewasse BE, Jacobs MR, Appelbaum PC: Single and multi-step resistance selection study in Streptococcus pneumoniae comparing ceftriaxone with levofloxacin, gatifloxacin and moxifloxacin. Int J Antimicrob Agents. 2002, 20: 93-99. 10.1016/S0924-8579(02)00120-6.
Sierra JM, Cabeza JG, Ruiz Chaler M, Montero T, Hernandez J, Mensa J, Llagostera M, Vila J: The selection of resistance to and the mutagenicity of different fluoroquinolones in Staphylococcus aureus and Streptococcus pneumoniae. Clin Microbiol Infect. 2005, 11: 750-758. 10.1111/j.1469-0691.2005.01211.x.
Drago L, Nicola L, De Vecchi E: A comparative in-vitro evaluation of resistance selection after exposure to teicoplanin, vancomycin, linezolid and quinupristin-dalfopristin in Staphylococcus aureus and Enterococcus spp. Clin Microbiol Infect. 2008, 14: 608-611. 10.1111/j.1469-0691.2008.01993.x.
Nseir S, Ader F, Marquette CH, Durocher A: Impact of fluoroquinolone use on multidrug-resistant bacteria emergence. Pathol Biol (Paris). 2005, 53: 470-475.
Denton M: Enterobacteriaceae. Int J Antimicrob Agents. 2007, 29 (suppl 3): 9-12. 10.1016/S0924-8579(07)72174-X.
Barisić Z, Borzić E, Kraljević KS, Carev M, Zoranić V, Kaliterna V: Rise in ciprofloxacin resistance in Escherichia coli from urinary tract infections from 1999-2004. Int J Antimicrob Agents. 2005, 25: 550-551. 10.1016/j.ijantimicag.2005.04.001.
Morales RA, McDowell RM: Risk assessment and economic analysis for managing risks to human health from pathogenic microorganisms in the food supply. J Food Prot. 1998, 61: 1567-1570.
Chenia HY, Pillay B, Pillay D: Analysis of the mechanisms of fluoroquinolone resistance in urinary tract pathogens. J Antimicrob Chemother. 2006, 58: 1274-1278. 10.1093/jac/dkl404.
Ruiz J: Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003, 51: 1109-1117. 10.1093/jac/dkg222.
Lautenbach E, Fishman NO, Metlay JP, Mao X, Bilker WB, Tolomeo P, Nachamkin I: Phenotypic and genotypic characterization of fecal Escherichia coli isolates with decreased susceptibility to fluoroquinolones: results from a large hospital-based surveillance initiative. J Infect Dis. 2006, 194: 79-85. 10.1086/503046.
Wang M, Sahm DF, Jacoby GA, Hooper DC: Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agent Chemother. 2004, 48: 1295-1299. 10.1128/AAC.48.4.1295-1299.2004.
Ambrozic Avgustin J, Keber R, Zerjavic K, Orazem T, Grabnar M: Emergence of the quinolone resistance-mediating gene aac(6')-Ib-cr in extended-spectrum-β-lactamase-producing Klebsiella isolates collected in Slovenia. Antimicrob Agent Chemother. 2007, 51: 4171-4173. 10.1128/AAC.01480-06.
Drago L, De Vecchi E, Nicola L, Legnani D, Lombardi A, Gismondo MR: In vitro synergy and selection of resistance by fluoroquinolones plus amikacin or beta-lactams against extended-spectrum beta-lactamase-producing Escherichia coli. J Chemother. 2005, 17: 46-53.
Gotfried MH, Danziger LH, Rodvold KA: Steady-state plasma and intrapulmonary concentrations of levofloxacin and ciprofloxacin in healthy adult subjects. Chest. 2001, 119: 1114-1122. 10.1378/chest.119.4.1114.
Capitano B, Mattoes HM, Shore E, O'Brien A, Braman S, Sutherland C, Nicolau DP: Steady state intrapulmonary concentrations of moxifloxacin, levofloxacin, and azithromycin in older adults. Chest. 2004, 125: 965-973. 10.1378/chest.125.3.965.
Keam SJ, Perry CM: Prulifloxacin. Drugs. 2004, 64: 2221-2234. 10.2165/00003495-200464190-00005.
Picollo R, Brion N, Gualano V, Millérioux L, Marchetti M, Rosignoli MT, Dionisio P: Pharmacokinetics and tolerability of prulifloxacin after single oral administration. Arzneimittelforschung. 2003, 53: 201-205.
Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing; Fifteenth Informational Supplement. M100-S15. 2005, Wayne (PA) CLSI
Matera MG: Pharmacologic characteristics of prulifloxacin. Pulm Pharmacol Ther. 2006, 19 (suppl 1): 20-29. 10.1016/j.pupt.2005.09.009.
De Vecchi E, Nicola L, Ossola F, Drago L: In vitro selection of resistance in Streptococcus pneumoniae at in vivo fluoroquinolone concentrations. J Antimicrob Chemother. 2009, 63: 721-727. 10.1093/jac/dkp020.
Cattoir V, Lesprit P, Lascols C, Denamur E, Legrand P, Soussy CJ, Cambau E: In vivo selection during ofloxacin therapy of Escherichia coli with combined topoisomerase mutations that confer high resistance to ofloxacin but susceptibility to nalidixic acid. J Antimicrob Chemother. 2006, 58: 1054-1057. 10.1093/jac/dkl361.
Chang TM, Lu PL, Li HH, Chang CY, Chen TC, Chang LL: Characterization of fluoroquinolone resistance mechanisms and their correlation with the degree of resistance to clinically used fluoroquinolones among Escherichia coli isolates. J Chemother. 2007, 19: 488-494.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
This work was supported by an unrestricted grant from sanofi-aventis. L. Drago has acted as a speaker for sanofi-aventis.
Authors' contributions
LD participated in designing the study, data analysis and in the writing of the paper. LN performed all experiments and participated in data collection and analysis. RM participated in writing of the paper. EDV participated in designing the study, data analysis and in the writing of the paper. All authors read and approved the final manuscript.
Lorenzo Drago, Lucia Nicola and Elena De Vecchi contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Drago, L., Nicola, L., Mattina, R. et al. In vitro selection of resistance in Escherichia coli and Klebsiella spp. at in vivo fluoroquinolone concentrations. BMC Microbiol 10, 119 (2010). https://doi.org/10.1186/1471-2180-10-119
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-10-119