Abstract
Background
The Archaea are highly diverse in terms of their physiology, metabolism and ecology. Presently, very few molecular characteristics are known that are uniquely shared by either all archaea or the different main groups within archaea. The evolutionary relationships among different groups within the Euryarchaeota branch are also not clearly understood.
Results
We have carried out comprehensive analyses on each open reading frame (ORFs) in the genomes of 11 archaea (3 Crenarchaeota – Aeropyrum pernix, Pyrobaculum aerophilum and Sulfolobus acidocaldarius; 8 Euryarchaeota – Pyrococcus abyssi, Methanococcus maripaludis, Methanopyrus kandleri, Methanococcoides burtonii, Halobacterium sp. NCR-1, Haloquadratum walsbyi, Thermoplasma acidophilum and Picrophilus torridus) to search for proteins that are unique to either all Archaea or for its main subgroups. These studies have identified 1448 proteins or ORFs that are distinctive characteristics of Archaea and its various subgroups and whose homologues are not found in other organisms. Six of these proteins are unique to all Archaea, 10 others are only missing in Nanoarchaeum equitans and a large number of other proteins are specific for various main groups within the Archaea (e.g. Crenarchaeota, Euryarchaeota, Sulfolobales and Desulfurococcales, Halobacteriales, Thermococci, Thermoplasmata, all methanogenic archaea or particular groups of methanogens). Of particular importance is the observation that 31 proteins are uniquely present in virtually all methanogens (including M. kandleri) and 10 additional proteins are only found in different methanogens as well as A. fulgidus. In contrast, no protein was exclusively shared by various methanogen and any of the Halobacteriales or Thermoplasmatales. These results strongly indicate that all methanogenic archaea form a monophyletic group exclusive of other archaea and that this lineage likely evolved from Archaeoglobus. In addition, 15 proteins that are uniquely shared by M. kandleri and Methanobacteriales suggest a close evolutionary relationship between them. In contrast to the phylogenomics studies, a monophyletic grouping of archaea is not supported by phylogenetic analyses based on protein sequences.
Conclusion
The identified archaea-specific proteins provide novel molecular markers or signature proteins that are distinctive characteristics of Archaea and all of its major subgroups. The species distributions of these proteins provide novel insights into the evolutionary relationships among different groups within Archaea, particularly regarding the origin of methanogenesis. Most of these proteins are of unknown function and further studies should lead to discovery of novel biochemical and physiological characteristics that are unique to either all archaea or its different subgroups.
Similar content being viewed by others
Background
Archaea are widely regarded as one of the three main domains of life [1–7], although their origin is a subject of debate [8–14]. Archaeal species were earlier believed to inhabit only extreme environments such as extremely hot, or hot and acidic, extremely saline, or very acidic or alkaline conditions [15–19]. However, recent studies provide evidence that they are widespread in different environments [3, 20]. The archaea also include methanogens, which grow under strictly anaerobic and often thermophilic conditions, and are the only organisms that derive all of their metabolic energy by reduction of CO2 by hydrogen to produce methane [21, 22]. The archaeal species branch distinctly from all other organisms in phylogenetic trees based on 16S rRNA and many other gene/protein sequences [2, 7, 23–25]. In addition, many morphological or physiological characteristics such as the presence of branched-chain ether-linked lipids in their cell membrane, lack of peptidoglycan in their cell wall, characteristic subunit pattern of RNA polymerase, presence of modified bases in tRNA, presence of a unique form of DNA polymerase, have been previously indicated as defining characteristics of archaea [1, 15]. However, as noted by Walsh and Doolittle [26], many of these features are either not shared by all archaea or they are also present in various eukaryotes or some thermophilic bacteria, indicating that they do not constitute distinctive characteristics of all Archaea.
The phylogenetic analyses of Archaea have led to their division into two major groups or phyla designated as Crenarchaeota and Euryarchaeota [1, 2, 7, 13, 27–29]. The Crenarchaeota species have also been referred to as 'Eocytes' by Lake and coworkers [30, 31]. The species from both these groups, particularly Euryarchaeota, are highly diverse in terms of their metabolism and physiology. Based on their metabolic and physiological characteristics and other unique features, five functionally distinct groups within Euryarchaeota are currently recognized: methanogens, sulfate reducers, extreme halophiles, cell wall-less archaea, and extremely thermophilic sulfur metabolizing archaea [2, 13, 32]. Some of these groups, such as methanogens, are polyphyletic in different phylogenetic trees [13, 33, 34]. However, the sets of genes or proteins that are unique to these different functional groups and distinguish them from all others remain to be identified. In the past 10 years, complete genomes of many archaeal species (29 at the time when these analyses were completed) covering all major divisions within the Archaea have been sequenced (see Table 1). Comparative analyses of these sequences provide a valuable resource for identifying different genes/proteins that are distinctive characteristics of various taxonomic and functional groups within Archaea [27, 35–37].
Whole proteins that are uniquely present in particular groups or subgroups of organisms but not found anywhere else provide valuable molecular markers for taxonomic, phylogenetic and biochemical studies. These proteins, which we refer to as signature proteins in our work, and others have called them as ORFans or conserved hypothetical proteins, are present at different phylogenetic depths, such as genus, family, order or even phylum [35, 36, 38–42]. In our recent work, a large number of such proteins that are distinctive characteristics of several groups within bacteria (viz. α-proteobacteria, ε-proteobacteria, Chlamydia and Actinobacteria), and also their subgroups, were identified [39–43]. These proteins provide not only valuable molecular markers for identifying and circumscribing species belonging to these major groups (and their subgroups) in molecular terms, but their species distribution pattern also provides useful information about the branching order within these groups. As archaea constitute a very diverse group, identification of sets of proteins that are specific for its main groups and subgroups should prove useful in terms of identifying molecular characteristics that are unique to them. Additionally, this information should also be helpful in understanding the evolutionary relationships among different groups.
Comparative studies on limited numbers of archaeal genomes have been carried out by a number of investigators using different criteria. Graham et al. [36] analyzed 9 archaeal genomes to identify signature proteins that function uniquely within the Archaea. Their definition of an archaeal signature protein required it to be present in only two different euryarchaeal species and they identified 353 archaeal signature proteins. Makarova and Koonin [27, 35] have analyzed archaeal genomes to identify core sets of genes, which are present in all archaeal species, but which are not restricted to the archaeal species. Recently, Walsh and Doolittle have analyzed prokaryotic genomes to measure dissimilarity between Archaea and Bacteria [26]. Although it was reported that 28% of the proteins from archaeal genomes are restricted to the Archaea, specific proteins that were present in different groups of archaea were not identified. Other comparative studies using different criteria have been conducted on smaller groups within archaea such as Pyrococcus, Sulfolobus and thermoacidophilic organisms (to be discussed later). However, thus far no comprehensive phylogenomics study on different archaeal genomes has been carried out using the same standard criteria to identify proteins or ORFs that are shared by all archaea or its different major lineages. In this study we have carried out comparative analyses of archaeal genomes using uniform criteria to identify proteins that are uniquely present in archaeal species at different phylogenetic depths (genus or higher) representing all major groups within the Archaea.
Results and discussion
A. Phylogenetic analyses of archaeal species
Prior to undertaking comparative studies on archaeal genomes, phylogenetic analysis of sequenced archaeal species was carried out so that the results of phylogenomics analyses could be compared with those obtained by traditional phylogenetic approaches. Phylogenetic trees for the archaeal species based on 16S rRNA as well as concatenated sequences of translation and transcription-related proteins have been published by other investigators [7, 28, 32, 44]. In the present work, we have constructed phylogenetic trees for 29 archaeal species (see Table 1) using a set of 31 universally distributed proteins that are involved in a broad range of functions [45]. The sequence of Haloquadratum walsbyi DSM 16790, which became available afterward, was not included in these studies. Phylogenetic trees based on a concatenated sequence alignment of these proteins were constructed using the neighbour-joining (NJ), maximum-likelihood (ML) and maximum-parsimony (MP) methods.
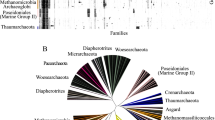
The results of these analyses are presented in Fig. 1. All three methods gave very similar tree topologies except for the branching positions of M. kandleri and Methanospirillum hungatei, which were found to be variable. Except for this, the branching pattern of the archaeal species based on our dataset is very similar to that reported by Gribaldo et al. [13, 32] based on concatenated sequences of translation and transcription-related proteins. In the tree shown, the Crenarchaeota and Euryarchaeota, the two major phyla within Archaea were clearly distinguished from each other. The phylogenetic affinity of Nanoarchaeum, which has a long-branch length, was not resolved in this or various other trees [32, 46]. Within Crenarchaeota, Pyrobaculum was indicated to be a deeper branch, and Aeropyrum branched in between the Pyrobaculum and Sulfolobus. Within Euryarchaeota, the clades corresponding to Halobacteria, Thermococci and Thermoplasmata were resolved with high bootstrap scores, but the methanogens were split into 2–3 clusters. One of these clusters that has low bootstrap score consisted of Methanobacteriales and Methanococcales with M. kandleri (Methanopyrales) branching in its vicinity [34, 47, 48]. The second cluster, with higher bootstrap score, showed a grouping of Methanomicrobiales and Methanosarcinales. These two clusters, which are separated by Thermoplasmata, Archaeoglobi and Halobacteria, have been referred to as Class I and Class II methanogens by Bapteste et al. [29].
B. Phylogenomic analyses of archaeal genomes
To search for proteins (or ORFs), which are uniquely present in either all Archaea or various subgroups of them, blast searches were performed on each open reading frame (ORF) from a total of 11 archaeal genomes (see Table 1; shaded species in Fig. 1). These genomes included 3 Crenarchaeota (viz. Aeropyrum pernix, Pyrobaculum aerophilum and Sulfolobus acidocaldarius) [49–51] and 8 divergent Euryarchaeota species covering all main functional and phylogenetic groups (see Table 1 and Fig. 1). The Euryarchaeota genomes analyzed included: Pyrococcus abyssi from extremely thermophilic sulfur metabolizing archaea [52], Methanococcus maripaludis [53] from Methanococcales, Halobacterium sp. NRC-1 and H. walsbyi from extreme halophiles [54], Thermoplasma acidophilum and Picrophilus torridus belonging to the cell wall-less archaea [19, 55], Methanococcoides burtonii from Methanosarcinales and Methanopyrus kandleri from the Methanopyrales order [56]. The chosen genomes should provide information regarding all archaeal proteins that are shared at a taxonomic level higher than a genus. The analysis of the remainder of the genomes, which was expected to provide information regarding proteins that are only unique to a given species, was not carried out.
Each ORF from these genomes was examined by means of blastp and PSI-blast searches against all available sequences from different organisms to identify proteins that are specific for only archaeal lineages. The methods and the criteria that we have used to identify proteins that are specific for either all or various subgroups of archaea are described in the Methods section. Generally, a protein was considered to be specific for a given archaeal lineage if all significant hits or alignments in the blastp and PSI-blast searches with the query protein were from the indicated group of archaeal species. In a few cases, where 1–2 isolated species from other groups also exhibited significant similarity, such proteins were retained as they provide interesting examples of lateral gene transfer (LGT) from archaea to other groups. Our analyses have identified 1448 proteins that are unique to different groups of Archaea and for which no homologues are generally found in any bacterial or eukaryotic species. Based on their specificity for different taxonomic groups, these proteins have been divided into a number of different groups (see Tables 2, 3, 4, 5, 6, 7 and Additional files). A brief description of the different subsets of archaeal-specific proteins and functional information regarding them, where known, is given below. In the description of these proteins that follows, the 'APE', 'HQ', 'Mbu', 'MK', 'MMP', 'PAB', 'PAE', 'PTO', 'Saci', 'Ta', 'VNG', and 'NEQ' part of the descriptors in proteins indicate that the original query protein sequence was from the genome of A. pernix K1, H. walsbyi DSM 16790, M. burtonii DSM 6242, M. kandleri AV19, M. maripaludis S2, P. abyssi GE5, P. aerophilum str. IM2, P. torridus DSM 9790, S. acidocaldarius DSM 639, T. acidophilum DSM 1728, Halobacterium sp. NRC-1 and N. equitans, respectively.
(a) Proteins that are specific for all Archaea
Table 2(a) shows a group of 16 proteins that are present in nearly all archaeal species but whose homologues are not found in any Bacteria or Eukaryotes with a single exception. Of these, the first 6 proteins in the left column (Table 2a) viz. PAB0063, PAB0252, PAB0316, PAB1633, PAB1716 and PAB2291, are present in all sequenced archaeal genomes. The observed E-values for these proteins from archaeal species are very low, close to 0, indicating that these proteins show very high degree of sequence conservation in various archaea. The unique presence of these proteins in all sequenced archaeal genomes indicates that these proteins could be regarded as distinctive characteristics or molecular signatures for the archaeal domain. The genes for these proteins likely evolved in a common ancestor of the Archaea and were then vertically acquired by other archaeal species. Makarova and Koonin [35] have also mentioned 6 proteins that are commonly shared by different archaea, but the identity of such proteins was not specified. These proteins are likely the same. The remaining 10 proteins in Table 2(a) are missing only in N. equitans, which is a tiny parasitic organism containing only 536 genes [57, 58]. The species distribution pattern of these proteins can be accounted for by one of the following two possibilities. First, it is possible that N. equitans is the deepest branching lineage within archaea, as has been suggested [57, 58] and the genes for these 10 proteins evolved in a common ancestor of the other archaea after its divergence (Fig. 2a). Alternatively, similar to the first 6 proteins, the genes for these 10 proteins evolved in a common ancestor of all archaea, but they were then selectively lost in N. equitans (Fig. 2b) [35, 46, 58]. Based upon our results, one cannot distinguish between these two possibilities. However, in view of the fact that the genome of N. equitans has undergone extensive genome shrinkage (only 0.49 Mb) and it is at least 3 times smaller than the next smallest archaeal genome (see Table 1), we favour the latter possibility (Fig. 2b) [35, 46, 58].
Interpretive diagrams showing the suggested evolutionary stages where genes for some of the signature proteins that are specific for the Crenarchaeota and Euryarchaeota as well as some of the Crenarchaeota subgroups, likely originated. The top diagram (A) indicates the evolutionary interpretation of the signature proteins in the absence of any other information, whereas that below (B) indicates our interpretation of this data taking into consideration other relevant information discussed in the text. The branching pattern shown here is unrooted and the proteins that are shared by all archaea were introduced in a common ancestor of all archaea. The dotted line for N. equitans in (B) indicates that its placement within Euryarchaeota lineage is uncertain. The abbreviations T and AF in these figures as well as others refer to tables and Additional files.
Of the proteins that are uniquely present in all archaea, PAB0063 corresponds to tRNA nucleotidyltransferase (CCA-adding enzyme), which builds and repairs the 3' end of tRNA [59]. Functionally similar enzymes are also present in bacteria and eukaryotes (assigned as Class II), but their sequences share very little homology with the archaeal CCA-adding enzyme (Class I), which explains why no homologs were detected in any bacteria or eukaryotes in blast searches. The main mechanistic difference between class I and class II enzymes is that the tRNA substrate is required to fully define the nucleotide binding site in class I enzyme, whereas class II has a preformed nucleotide binding site that recognizes CTP and ATP in the absence of tRNA [60]. Another protein PAB0316 is assigned as archaeal type DNA primase, which also has its synonymous counterparts in bacterial and eukaryotic species, but shows very little homology to them [61, 62]. In the same way, protein PAB1633 is annotated as a PilT family ATPase, which showed very little similarity to bacterial ATPases involved in type IV pili biogenesis [54]. Further studies of this protein could provide insights into novel aspects of the archaeal flagellar system. A number of other proteins viz. PAB1716, PAB0018a, PAB0075, PAB0475 and PAB2104, have also been assigned putative functions based on sequence analysis, but their exact roles in archaeal cells remains to be determined. Interestingly, for protein PAB0075, two gene copies with acceptable E-values are also present in the genomes of Dehalococcoides ethenogenes 195, Dehalococcoides sp. CBDB1 and Dehalococcoides sp. BAV1, which belong to Chloroflexi [2]. Because no homologue of PAB0075 is present in other bacteria, it is likely that this protein was transferred from archaea to the common ancestor of Dehalococcoides followed by a gene duplication event.
Table 2(b) lists 20 additional proteins, which are specific to archaea but missing in a small number of species. Because these proteins are present in most Euryarchaeota as well as Crenarchaeota species, but not detected in Bacteria or Eukaryotes except one LGT case (PAB2342, see note in Table 2), we consider them also to be distinctive characteristics of most Archaea. Of these proteins, 11 proteins (viz. PAB0654, PAB0950, PAB1135, PAB1906, PAB7388, PAB0547, PAB0552, PAB0623, PAB1272, PAB1429 and PAB1721) are mainly missing in the 4 Thermoplasmata species. Thermoplasmata are thermoacidophilic archaea which lack cell envelope [19, 55, 63](see Table 1). Some studies have suggested that high temperature and very low intracellular pH exert selective pressure favouring smaller genomes [19]. Thus, it is possible that genes for these proteins were selectively lost in the Thermoplasmata lineage. Most of these proteins are of unknown function. However, 8 of them have been assigned putative functions with the title of "archaeal type"'. For example, PAB0301 is archaeal sugar kinase, PAB0950 is archaeal transcription factor E α-subunit, PAB1387 is archaeal flagella accessory protein, PAB7094 is archaeal chromatin protein, and PAB0552 is archaeal type Holliday junction resolvase. These proteins do not show detectable sequence similarity to their counterparts in Bacteria or Eukaryotes, and some studies indicate that they also differ in terms of their structure, function or interaction with other cell components [64, 65].
(b) Proteins that are specific for Crenarchaeota
As mentioned in the introduction, the Archaea are divided into 2 main groups, Crenarchaeota and Euryarchaeota, based on 16S rRNA trees as well many other gene trees and characteristics. The Crenarchaeota are also indicated to differ from Euryarchaeota in terms of their ribosome structure [30, 31]. In comparison to Euryarchaeota, which contain physiologically and metabolically diverse groups of organisms, the Crenarchaeota were thought to be a pure collection of extreme thermophiles and most members metabolize sulfur. However, recent studies indicate that Crenarchaeota are much more diverse in their physiology and ecology than was previously believed [28, 66]. Many species living in the cold ocean also belong to this group based on their branching pattern in 16S rRNA trees, although most of them have not been cultivated [67]. Currently, this phylum is comprised of one single class Thermoprotei containing three orders: Thermoproteales, Desulfurococcales and Sulfolobales. Fortunately, every order has a completely sequenced representative (see Table 1)[50, 51, 68, 69], which provide a platform to explore the characteristics that are unique to crenarchaeal species. Comparative genomic surveys have revealed some molecular features that are shared by crenarchaea but not euryarchaea, such as the lack of histones, absence of the FtsZ-MinCDE system and distinctive rRNA operon organization [69]. Lake et al. have also identified distinctive differences in ribosome structure and an insert in elongation factor EF-G and EF-Tu, which can be used to distinguish Crenarchaeota from Euryarchaeota [6, 30, 70]. However, these features are not unique characteristics of the Crenarchaeota.
Blast searches on each ORF from the genomes of A. pernix and S. acidocaldarius DSM 639 [49, 50] have identified 11 proteins which are shared by all five crenarchaeal species, but whose homologs are not found in other archaea, or any bacteria or eukaryotes with only 3 exceptions (see Table 3(a)). The genes for these proteins likely evolved in a common ancestor of the Crenarchaeota and they provide potential molecular markers for species from this phylum. Additionally, 22 proteins that are listed in Table 3(b) are only found in A. pernix and three Sulfolobus genomes. These proteins suggest that Aeropyrum and Sulfolobus may have shared a common ancestor exclusive of Pyrobaculum. However, we have also come across 9 proteins that are shared by Aeropyrum and Pyrobaculum (Table 3(c)) and 14 proteins that are exclusively present in the 3 Sulfolobus species and Pyrobaculum (see Table 3(d)). Hence, based upon the species distributions of these proteins, the relationships among the Aeropyrum, Sulfolobales and Pyrobaculum are not entirely clear (Fig. 2a). In phylogenetic trees Thermoproteales (i.e. Pyrobaculum) branches consistently earlier than Desulfurococcales (i.e. Aeropyrum) and Sulfolobales (Fig. 1) [32, 44]. This observation in conjunction with the fact that Aeropyrum and Sulfolobus share larger numbers of proteins in common with each other suggests that these two groups likely shared a common ancestor exclusive of Pyrobaculum (Fig. 2b). The proteins that are only found in Aeropyrum and Pyrobaculum, or in Sulfolobus and Pyrobaculum, most likely evolved in a common ancestor of the crenarchaea, but were subsequently lost in either the Sulfolobales or A. pernix lineages.
In addition to these proteins that are uniquely present in either all sequenced Crenarchaeota genomes or different groups of Crenarchaeota species, these analyses have also identified 264 proteins that are unique for the Sulfolobales species (see Additional file 1). Of these, 184 proteins are present in all 3 sequenced Sulfolobus genomes, whereas the remaining 80 are present in at least two of the three Sulfolobus genomes. In this work, since blast analyses were not carried out on all three Sulfolobus genomes, it is likely that the numbers of genes or proteins that are uniquely shared by only two Sulfolobus genomes is much higher than indicated here. Chen et al. [50] have previously analyzed the genome of S. acidocaldarius DSM 639 and indicated the presence of 107 genes that were specific for Crenarchaeota and 866 genes that were specific to Sulfolobus genus. However, in the present work, relatively few genes that are uniquely shared by various Crenarchaeota species were identified. This difference could be due to more stringent criteria that we have employed for identification of proteins that are specific to different groups. The genome of Thermofilum pendens Hrk 5, which belongs to Thermoproteales, has also been partially sequenced and information for large numbers of genes/proteins from this species is available in the NCBI database. By carrying out blast searches on each ORF from P. aerophilum genome [51], we have identified 42 proteins that are only found in the above 2 Thermoproteales species (see Additional file 2). The numbers of proteins shared by these two species will likely increase once complete genome of T. pendens becomes available. Many of these proteins are expected to provide markers for the Thermoproteales order.
(c) Proteins that are specific for Euryarchaeota
The Euryarchaeota, which comprise a majority of the cultured and sequenced archaea, is a morphologically, metabolically and physiologically diverse collection of species as evidenced by the presence in this group of various methanogens, extreme halophiles, cell wall-less archaea and sulfate reducing microbes [2, 13]. No unique biochemical or molecular characteristic that is commonly shared by all of the different lineages is known. The present study has identified 20 proteins that are only found in Euryarchaeota species with 3 exceptions (see Table 4). In this Table, the first 7 proteins (Table 4(a)) are present in most euryarchaeota species. Of these proteins, PAB0082 and PAB2404 were found in all sequenced euryarchaeota species. PAB2404 was also present in N. equitans, supporting its placement within the Euryarchaeota [35, 46]. The protein PAB0082 is annotated as archaeosine tRNA-ribosyltransferase (ArcTGT), which catalyzes the exchange of guanine with a free 7-cyano-7-deazaguanine (preQ0) base, as the first step in the biosynthesis of an archaea-specific modified base, archaeosine (7-formamidino-7-deazaguanosine) [71]. It should be mentioned that there is another protein PAB0740 in the same genome, which is also annotated and experimentally confirmed as ArcTGT [72]. The latter belongs to a family of proteins that are highly conserved in all archaea species (including Crenarchaeota) and some bacteria. It seems that PAB0082 might be involved in RNA modification since it possesses a PUA domain (named after pseudouridine synthase and archaeosine transglycosylase), but its function is likely different from PAB0740. The protein PAB2404, which is annotated as DNA polymerase II large subunit, is highly conserved within Euryarchaeota, but is not found anywhere else except in Nanoarchaeum. This enzyme is the major DNA replicase in Euryarchaeota and also a distinctive molecular marker for this group [73, 74]. The genes for the above proteins likely evolved in a common ancestor of Euryarchaeota (Fig. 2) and they provide molecular markers for this diverse group of organisms.
Another 13 proteins listed in Table 4(b) are found in almost all euryarchaeota, but they are missing in Thermoplasmata. Their distribution suggests that either Thermoplasmata is a deep branching lineage within Euryarchaeota or that the genes for these proteins have been selectively lost from Thermoplasmata [55]. Of these proteins, PAB0188 is also present in N. equitans supporting its placement with Euryarchaeota. Five other proteins from the first two columns in Table 4 (viz. MMP0243, Ta0062, VNG1263c, MMP1287, and VNG2408c) are also not found in the 4 Thermococci species. These results can again be explained by either selective loss of these genes from these particular groups or deeper branching of these lineages within the Euryarchaeota species. On the basis of proteins listed in Table 4, although one can infer that Thermoplasmata and Thermococci are deeper branching lineages within Euryarchaeota in comparison to methanogens, their relative branching order cannot be resolved.
(d) Proteins that are specific for different main groups within Euryarchaeota
Proteins specific for methanogenic archaea and their various subgroups
Currently, the methanogens form the largest group within the Euryarchaeota. They are distinguished from all other prokaryotes by their ability to obtain all or most of their energy via the reduction of CO2 to methane or by the process of methanogenesis. In the Bergey's manual [75], the methanogenes are divided into 5 distinct orders (viz. Methanobacteriales, Methanococcales, Methanomicrobiales, Methanosarcinales and Methanopyrales). Some studies have suggested that these organisms possess a set of unique enzymes which are responsible for methanogenesis, such as coenzyme M, Factor 420 and methanopterin [76]. However, no systematic study has been carried out thus far to identify proteins that are uniquely present in different methanogens. Our blast searches of proteins from different methanogens have led to identification of 31 proteins, which are uniquely found in various methanogenic archaea. Twenty of these 31 proteins are present in all sequenced methanogens, while 11 proteins are missing only in M. stadtmanae, which is a human intestinal inhabitant (see notes in Table 5). This archaeon generate methane by reduction of methanol with H2 and lacks many proteins present in the genomes of other methanogens [77, 78]. Thus, it is highly likely that the 11 proteins missing in M. stadtmanae were selectively lost from this species. Therefore, it is very likely that the genes for these 31 proteins that are commonly shared by virtually all methanogens (Table 5(a)) evolved in a common ancestor of all methanogens.
These analyses have also identified 10 proteins that are uniquely shared by various methanogens as well as A. fulgidus (see Table 5(b)). The genes for these proteins likely evolved in a common ancestor of A. fulgidus and various methanogenic archaea and they point to a close relationship between these two groups of organisms (Fig. 3). Ten additional proteins are present in A. fulgidus as well as various Methanosarcinales and M. hungatei (Methanomicrobiales) (Table 5(c)). It is likely that the genes for these proteins also evolved in a common ancestor of A. fulgidus and various methanogenic archaea, but they were selectively lost in other methanogens. Of the proteins that are commonly shared by A. fulgidus and various methanogenic archaea, MMP0607 is reported to be a novel repressor of nif and glnA genes, which are involved in nitrogen assimilation [79]. Interestingly, 2 homologs of this protein are also found in 3 Dehalococcoides species, but nowhere else, which are very likely due to LGT. Protein MMP0984 is the ε-subunit of carbon-monoxide dehydrogenase complex, which is made up of five subunits in different methanogens [80]. The epsilon subunits are required for the reversible oxidation of CO to CO2 [81]. All of the other components could be found in a few bacterial species, while the ε-subunit is restricted to methanogenic archaea and A. fulgidus [82, 83]. Protein MMP1499 is identified as a transcriptional regulator with a Helix-turn-helix (HTH) motif, but its exact role has not been reported.
An interpretive diagram showing the evolutionary stages where genes for different proteins that are specific for methanogenic archaea likely originated. The 10 proteins that are uniquely shared by A. fulgidus and various methanogenic archaea indicate that this lineage is the closest ancestor of all methanogens.
Among the genes that are uniquely shared by various methanogenic archaea (or these archaea plus A. fulgidus), two large gene clusters responsible for methanogenesis are found. The proteins MMP1346, MMP1560–MMP1564 and MMP1566–MMP1567 (Table 5) are parts of an eight-component complex, coenzyme M methyltransferase (Mtr), which catalyzes an energy-conserving, sodium-ion-translocating step in methanogenesis from H2 and CO2 [84]. M. maripaludis contains all of the known Mtr subunits, but the gene coding for MtrF is fused into the N-terminal region of MtrA [53]. All other methanogenic archaeal genomes contain complete set of mtr genes. It is of interest to note that for the protein MMP1567 (MtrH), homologues with low E-values are also found in two Desulfitobacterium hafniense strains as well as in three Rhizobiales species (Aminobacter lissarensis, Methylobacterium chloromethanicum, and Hyphomicrobium chloromethanicum; α-proteobacteria) (see note in Table 5). These three rhizobiae species can use methyl halides as a sole source of carbon and energy, and all of them possess a set of cmu genes which are essential for methyl chloride degradation [85]. In particular, the CmuB protein which is homologous to MMP1567 transfers a methyl group to methylcobalamin:H4 folate (H4F), which is analogous to the reverse of the reaction catalyzed by MtrH in archaea [86]. In view of the sequence and functional similarity between MtrH and CmuB proteins, it is likely that the mtrH gene was laterally transferred from a methanogenic archaeon to the common ancestor of the above three rhizobiae species to serve the new functional role. The function of the laterally transferred mtrH related gene in D. hafniense is not known at present.
The proteins MMP1555–MMP1559 in Table 5 form another gene cluster, encoding the subunits of Methyl-coenzyme M reductase (MCR). This complex catalyzes the final reaction of the energy conserving pathway in which methylcoenzyme M and coenzyme B are converted to methane and the heterodisulfide CoM-S-S-CoB [87, 88]. Except for these proteins, the other proteins listed in Table 5 are of putative or unknown functions. It is likely that these proteins are involved in some aspects of methanogenesis or other unknown pathways unique to methanogenic archaea. These proteins provide molecular markers for methanogens, which can be used for identification of new archaeal species capable of methane production.
The blast searches of the M. maripaludis [53] and M. kandleri [56] genomes have identified 10 proteins that are uniquely shared by all of the following species belonging to the orders Methanobacteriales (M. thermoautotrophicus), Methanococcales (M. jannaschii, M. maripaludis) and Methanopyrales (M. kandleri) (Table 6(b)). Of these, only 2 proteins are present in M. stadtmanae, which is also a Methanobacteriales that has lost most of its genes due to its adaptation to the human intestine [78]. The genes for these 10 proteins likely evolved in a common ancestor of the above groups of methanogens (Fig. 3), which corresponds to the cluster of methanogenic archaea referred to as "Class I methanogens" [13]. Interestingly, these studies have also identified 10 proteins that are uniquely shared by these methanogenic orders and M. hungatei (see Table 6(a)), which branches distantly in phylogenetic trees [13]. The unique presence of these proteins in these methanogens suggests that species from these groups shared a common ancestor exclusive of other methanogenic archaea (Fig. 3).
Fifteen additional proteins discovered in this work (Table 6(c)) are uniquely present in M. kandleri and various Methanobacteriales indicating that these two groups are more closely related to each other than the Methanococcales (Fig. 3). We have also come across 7 proteins that are uniquely shared by Methanococcales and Methanobacteriales (Table 6(d)), and 4 proteins that are only present in Methanococcales and Methanopyrales (Table 6(e)). The most likely explanation to account for the species distributions of these latter proteins is that their genes also originated in a common ancestor of the above three groups of methanogens, but were selectively lost in either the Methanobacteriales or Methanopyrales lineages. These analyses have also identified 14 additional proteins that are uniquely present in all 5 Methanosarcinales species (Table 6(f)), as well as 7 proteins that are only found in various Methanosarcinales and M. hungatei (Table 6(g)). Lastly, these studies have also identified 55 proteins that are uniquely present in M. maripaludis and M. jannaschii (Methanococcales, see Additional file 3(a)) and 68 proteins that are only present in M. burtonii and 3 Methanosarcina species, all belonging to the Methanosarcinaceae family (see Additional file 3(b)) (Fig. 3) indicating that they are likely distinctive characteristics of species from these groups.
Of the proteins that are uniquely found in Methanococcales, Methanobacteriales, Methanopyrales and Methanomicrobiales, 12 proteins viz. MMP1448–MMP1454, MMP1456, MMP1458–MMP1460 and MMP1467 are from a big gene cluster eha, which encodes the multisubunit membrane-bound [Ni-Fe] hydrogenase [89]. Two of these proteins, MMP1456 and MMP1458, are only found in Methanococcales (Table 6(e)). The whole eha operon is composed of 20 ORFs in the genome of M. thermoautotrophicus and of these only these 12 proteins are restricted to these methanogens while the other subunits have counterparts in bacteria. The precise roles of these 12 proteins, which are predicted to be integral membrane proteins in the hydrogenase complex, have not been determined [89]. Among the other proteins that are specific for these groups of methanogens, MMP0127 and MMP1716 are Hmd homologs, which catalyze the reversible dehydrogenation of N5, N10-methylenetetrahydromethanopterin [90]. In the proteins that are specific for the Methanococcales (see Additional file 3(a)), one large gene cluster (MMP0233–MMP0240) is found, but no information is available concerning its possible function. Except for these proteins, all other proteins that are specific for these methanogenic archaea are of unknown or putative function.
Proteins that are specific for Thermococci
Thermococci are obligately thermophilic, strictly anaerobic cocci, which are able to convert elemental sulfur to hydrogen sulfide. Thus, they are so called "extremely thermophilic sulfur metabolizer", which comprise one of the main functional groups within Euryarchaeota. According to the Bergey's Manual [75], the class Thermococci contains only one family, Thermococcaceae, consisting of 2 genera: Thermococcus and Pyrococcus. Currently, 4 species from this family have been completely sequenced (Pyrococcus abyssi, P. horikoshii, P. furiosus and Thermococcus kodakarensis; see Table 1) [52, 91–93]. The blast searches on each protein from P. abyssi have identified 141 proteins that are shared by all 4 of these species (see Additional file 4(a)). All of these proteins show high degree of conservation within Thermococci and they do not have homologs in any other prokaryotes or eukaryotes except one possible LGT event (PAB1493, see note in Additional file 4). The genes for these proteins have likely evolved in a common ancestor of the Thermococci (Fig. 3). Of these proteins, PAB1510 is annotated as TBP-interacting protein (TIP), which forms complex with TBP (TATA-binding protein) to regulate transcription [94]. It is known that the archaeal transcription machinery is strikingly similar to that in eukaryotes [23], but no TBP-binding component was found in archaeal species until the discovery of the TIP in T. kodakaraensis [95, 96]. Most other Themococci-specific proteins are of unknown function, although in a few cases limited similarity to domains in known protein families have been noted. A number of proteins (viz. PAB0643–PAB0644.1n; PAB1821–PAB1826) are clustered together in the P. abyssi genome, and it is possible that they may form functional units and are involved in related functions.
Cohen et al. [52] have reported a large number of proteins which are restricted to the Pyrococcus genus. However, a number of proteins from their list are also found in T. kodakarensis KOD1 [93], whose genome was not available when their work was published. Some proteins are not specific for either Pyrococcus or Thermococci according to our criteria and some of them are only found in one species – P. abyssi. Our analysis of the P. abyssi GE5 genome has also identified 43 proteins that are unique to the Pyrococcus genus (see Additional file 4(b)). Again, almost all of these proteins are of unknown function except PAB2241, which is annotated as RNase P, but this annotation seems arbitrary as it does not show significant sequence similarity to known RNases. The proteins that are uniquely found in the 3 Pyrococcus genomes likely evolved in a common ancestor of this genus (Fig. 4).
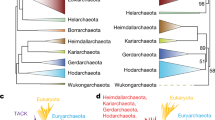
A summary diagram showing the branching order of different groups within archaea based upon species distribution patterns of various archaeal-specific proteins. The arrows mark the suggested evolutionary stages where proteins that are uniquely shared by the indicated groups were introduced. The details of these proteins can be found in the indicated tables (T) or Additional files (AF). The branching pattern shown here is unrooted. The dotted line for N. equitans indicates that its placement within Euryarchaeota is uncertain. The dotted line extending from the proteins found in all archaea indicates that one cannot use this to root the archaeal tree.
Proteins that are specific for Halobacteria
Extreme halophiles constitute another major class within Euryarchaeota. They require 5–10 times the salinity of seawater (ca. 3–5 M NaCl) for optimal growth [17, 97]. In order to grow in such high salinity environments, they have developed a set of physiological adaptation, such as: high internal concentration of potassium chloride, acidic proteome with low pI value, high GC content with GC bias in the wobble position, unique chloride pumps to maintain osmotic balance, etc. [17, 98, 99]. Among archaea, halobacteria also have the unique ability to use solar energy to generate a proton gradient to synthesize ATP. So far, the Class Halobacteria harbors one family with 15 genera and 4 species have been completely sequenced, including Halobacterium sp. NRC-1, Haloarcula marismortui, H. walsbyi and Natronomonas pharaonis [54, 98, 100, 101]. By performing blast searches on each protein in the Halobacterium sp. NRC-1 genome, we have identified 127 proteins, which are only present in all 4 Halobacteria species with only 3 exceptions (see Additional file 5).
Of the proteins listed in this Table, VNG0016H, VNG1096H, VNG2414H and VNG2563H are annotated as DNA-binding proteins or regulators because of the presence of HTH domain, but their exact functions have not been reported. VNG0667G is an ATP-binding protein of ABC transporter family. Several other proteins, such as VNG2089H and VNG2628H, have also been assigned possible functions based on weak similarity to known conserved domains in the CDD database [102], but their exact functions remain to be determined. Because of their high degree of conservation and uniqueness to halobacteria, the genes for these proteins likely evolved in a common ancestor of Halobacteria (Fig. 4) and they are presumably involved in unique physiological functions related to their adaptation to the hypersaline environment. Because of their specificity for Halobacteria, these proteins provide useful biomarkers for this group of species.
In addition to these proteins that are specific to all sequenced halobacterial species, we have also identified a large number of proteins either uniquely shared by 3 halobacterial species or only found in 2 halobacterial species (see Additional files 6 and 7). Surprisingly, these proteins are present in different combinations of halobacterial species. The four-halobacterial species are from 4 different genera within the Halobacteriales order and their relationships are unclear at present. The largest numbers of these proteins (i.e. 56) are uniquely shared by the Haloarcula, Haloquadratum and Natronomonas species, followed by 49 proteins that are restricted to Haloquadratum and Haloarcula. These results suggest that of these three species, Haloquadratum and Haloarcula are more closely related to each other and that Halobacterium might be the deepest branching of the four available halobacterial species (Fig. 4). However, the genome size of these halobacterial species varies and some of these protein sequences are present on plasmids found in these species, which makes it difficult to reliably infer their relationships solely based on the number of shared proteins. Among the proteins that are specific for halobacteria, only few have been assigned possible functions. Protein VNG2178H is annotated as PhiH1-like repressor and VNG0584H is assigned as a Rieske Fe-S protein. Two additional proteins VNG1720H and VNG2562H have been annotated as iron-binding proteins because of their similarity with FhuD and TroA_a domains, respectively [102]. All of the other proteins are of unknown function.
Proteins that are specific for Thermoplasmata
The Thermoplasmata group is comprised of cell wall-less archaea, which resemble the bacterial Mycoplasma species [63]. Generally, they are thermoacidophilic, aerobic or facultative anaerobic, and are able to reduce sulfur to H2S under anaerobic conditions [19, 55]. To date, this class include three families-Thermoplasmaceae, Picrophilaceae, and Ferroplasmaceae, each represented by one genus [103, 104]. Three complete genomes from this class (T. acidophilum, T. volcanium and P. torridus) are available at present (see Table 1) [19, 55, 63] and Ferroplasma acidarmanus Fer1 genome is draft assembled and sequence information for this is also available in the NCBI database. Our analyses have uncovered 77 proteins that are uniquely present in all four species belonging to this class (see Additional file 8(a)) (Fig. 4). Most of these proteins are present in all four available genomes, but a few are missing in one or two species, which is probably due to gene loss. Besides, we have also identified 33 proteins, which are shared only by the two Thermoplasma species (see Additional file 8(b)) and 17 proteins unique to P. torridus and F. acidarmanus (see Additional file 8(c)). The latter proteins indicate that species from Picrophilaceae and Ferroplasmaceae families are more closely related to each other (Fig. 4). All of these proteins are of unknown or predicted functions.
Proteins restricted to several archaeal lineages or showing sporadic distribution
In addition to the above proteins that are restricted to specific lineages of archaea, we have also identified 63 proteins, which are shared by several archaeal groups (see Table 7). The distribution pattern of these proteins could provide useful insights concerning the phylogenetic relationship between different groups. However, their distribution patterns could also be explained by gene losses in specific lineages or LGT between particular groups. Table 7 shows many proteins that are uniquely shared by various methanogenic archaea, Archaeoglobus and Thermococci. The first 5 proteins in Table 7(a) (PAB0076, PAB0138, PAB0965, PAB1927 and PAB1994) are present in all of the Thermococci and most of the methanogens. Four of these proteins are also present in A. fulgidus. The next 13 proteins in this Table are also uniquely found in most of the Thermococci as well as a number of methanogens and also in many cases in A. fulgidus. In addition, 6 proteins listed in Table 7(b) are only found in various Thermococci and A. fulgidus. These results suggest a closer relationship between the methanogenic archaea, A. fulgidus and Thermococci within the Euryarchaeota lineage. In conjunction with our earlier inference that A. fulgidus forms an outgroup of the methanogenic archaea, these results suggest that the above three groups are related in the following manner: Thermococci → A. fulgidus → Methanogens.
Although the relationship suggested above is the most likely explanation for the observed results, we have also come across three proteins (VNG1263c, MMP11287 and VNG2408c) that are uniquely present in various Halobacteria, A. fulgidus and different methanogens. To account for their species distribution, one has to postulate that their genes have been selectively lost from the Thermococci. In addition, 9 proteins are only found in various Halobacteria as well as Methanosarcinales and Methanomicrobiales (Table 7(c)). Their distribution requires again either selective gene losses from other lineages or LGT from Halobacteria to these methanogens.
Our analyses have also uncovered 30 proteins that are uniquely shared by species from Thermoplasmata and Sulfolobus (see Table 7(d)). Among these proteins, 7 are present in all Thermoplasmata and Sulfolobus species for which sequence information is available, while the remainder are missing in 1 or more species. It has been reported that there has been much lateral gene transfer between T. acidophilum and S. solfataricus, both of which inhabit the same environment [55]. However, the shared presence of these proteins in these two groups could also result from a unique shared ancestry of these thermo-acidophilic archaea.
Another 43 Archaea-specific proteins are sporadically present in different archaeal species (see Additional file 9). A number of proteins in this group are present in a limited number (between 3 to 6) of archaeal species belonging to different groups. There are 2 possible explanations that can account for their sporadic distribution: First, it is possible that some of these genes are the remnants of sequences that also originated in an ancestral lineage of Archaea but they have been selectively lost in many species because they are not required for growth. Second, the sporadic presence of these genes in a number of archaeal species can also be explained if some of these genes originally evolved in a particular group or species of archaea and then transferred to other archaea by LGT [105]. However, in view of the observed specificity of these genes/proteins for archaea, the LGTs in these cases need to be selective and limited to within archaea.
Conclusion
Comparative analyses of sequenced archaeal genomes presented here have led to identification of large numbers of proteins that are distinctive characteristics of either all archaea or its different main groups. Based upon these proteins, all of the main groups within Archaea (e.g. Crenarchaeota, Euryarchaeota, Halobacteria, Thermococci, Thermoplasmata, Methanogens) and their subgroups can now be clearly distinguished in molecular terms. The species distribution of these signature proteins strongly suggests that their genes have evolved or originated at various stages in the evolution of archaea, but once evolved, they are indicated to be generally stably retained in various descendents of these lineages with minimal gene loss or LGTs. Based upon the species distributions of these proteins, the evolutionary stages where the genes for these proteins have likely evolved are shown in Fig. 4. The evolutionary relationships among archaea have thus far been mainly inferred on the basis of their branching in phylogenetic trees based on 16S rRNA and certain protein sequences [2, 7, 13, 23–25]. The results of our analyses although they support many inferences reached based on phylogenetic trees (viz. identification of all of the main clades in phylogenetic trees in molecular terms) (Fig. 1) [2, 7, 13, 23–25], they also differ from them in important regards. In particular, our results shed important light on certain phylogenetic relationships that were very puzzling or were not resolved based on earlier studies. Some of these novel inferences are discussed below.
In phylogenetic trees based on 16S rRNA and various proteins sequences, the methanogenic archaea form at least two distinct clusters (see Fig. 1) [13, 29, 34, 56, 106]. In addition, in many of these trees, M. kandleri branches distinctly from all other methanogenic archaea [13, 34, 48]. The methanogenic archaea in these trees are interspersed by other groups of non-methanogenic archaea such as Halobacteriales, Archaeoglobus, Thermoplasmatales and Thermococcales (see Fig. 1) [13, 34, 48]. This has led to important questions concerning the origin of methanogenesis i.e. whether it evolved only once and its absence in the intervening lineages [13, 29, 35, 76]. To account for these results, it has been suggested that methanogenesis evolved once in a common ancestor of the above groups, i.e. different methanogenic archaea, Halobacteriales, Archaeoglobus, Thermoplasmatales and also possibly Thermococcales, comprising virtually all euryarchaeota, but that the various genes involved in this process were subsequently lost from different groups except the methanogens [13, 29, 56]. This scenario, in essence, proposes that the common ancestor of different physiologically and metabolically distinct groups within euryarchaeota was a methanogen and this capability was independently lost in all other lineages.
In contrast to this proposal, our phylogenomics analyses have identified 31 proteins that are uniquely present in virtually all methanogens, as well as many proteins that are specifically shared by different subgroups of methanogens. Of these proteins only about 1/3 are indicated to be directly involved in methanogenesis and the cellular functions of others are presently not known. The unique presence of such large numbers of proteins by nearly all methanogens, but none of the above groups of archaea, strongly indicates that the genes for these proteins evolved in a common ancestor of various methanogens. These results strongly suggest that all methanogenic archaea form a mononphyletic lineage exclusive of all other groups of archaea (Fig. 4). Importantly, these studies have also identified 10 proteins that are uniquely shared by all methanogens as well as by A. fulgidus. In contrast, we have not come across any protein that various methanogenic archaea uniquely share with any of the Halobacterales or Thermoplasmatales. These observations are highly significant because they strongly suggest that Archaeoglobus and all of the methanogens shared a common ancestor exclusive of all other archaea. In other words, the ancestral lineage that led to the origin of methanogenesis very likely evolved from the Archaeoglobus lineage (Fig. 4). It is also significant that of the proteins that are uniquely shared by Archaeoglobus and methanogens, several form part of complexes that are important for nitrogen assimilation and methanogenesis. These results support the view that these characteristics have their origin within the Archaeoglobus lineage.
The present work also provides clarification regarding the phylogenetic position of M. kandleri. In phylogenetic trees based on 16S rRNA or different protein sequences, the branching of this species is highly variable [13, 34, 47, 48] and it often forms the deepest branch within the Euryarchaeota. In the present work, we have identified 31 proteins that are uniquely shared by all methanogens including M. kandleri, as well as 10 proteins that M. kandleri specifically shares with various Methanobacteriales and Methanococcales, and 15 additional proteins that are only found in M. kandleri and the two Methanobacteriales species (M. thermoautotrophicus and M. stadtmanae). These observations reliably place M. kandleri with other methanogenic archaea with the Methanobacteriales as its closest relatives (Fig. 4). Our results also suggest a closer relationship of the Thermococcales to the Archaeoglobus and methanogenic archaea, although this relationship is not as strongly supported as between Archaeoglobus and Methanogens.
The observed differences in the evolutionary relationships among methanogens based upon phylogenomics analyses versus those by traditional phylogenetic methods can in principle be accounted for by three explanations. First, it is possible that the branching patterns of various clades in phylogenetic trees are misleading and they have been affected by factors such as long branch attraction effect [107, 108]. Second, the polyphyletic branching of methanogens can also be explained (as indicated earlier) if the genes uniquely shared by all methanogens evolved in an early branching lineage such as M. kandleri, but subsequently they were either completely or partially lost from various non-methanogenic (viz. Halobacteriales, Thermoplasmatales and Archaeoglobus) groups that lie in between the two methanogenic clusters (Fig. 1). Third, lateral transfer of these genes from one methanogenic archaea to all others can also explain these results. Of these possibilities, we favour the first explanation, as the last two require extensive gene loss or LGT from (or into) multiple independent lineages.
The present work also supports the placement of N. equitans within the Euryarchaeota lineage. N. equitans has a very small genome (only 0.49 Mb), which is at least 3 times smaller than any other archaeal genome. Due to its very small size, there are only 6 genes that N. equitans uniquely shares with all other archaea. However, our analysis indicates that whereas N. equitans shares a few genes (PAB2404 and PAB 0188) with most of the Euryarchaeota, it does not share any gene uniquely with most of the Crenarchaeota species, indicating its closer affinity for the former lineage. Although our analysis of the N. equitans genome has not revealed any strong signals indicating its specific affinity for any of the Euryarchaeota groups, the shared presence of some proteins by N. equitans and Thermococci (and in some cases also A. fulgidus and methanogens) suggest that it may be related to the Thermococci. However, because of the extensive gene losses that have occurred in this genome, we are not able to draw any reliable inference in this regard. Therefore, although we have depicted N. equitans as a deep branching lineage within Euryarchaeota (Fig. 4), based upon our analysis, its placement within Euryarchaeota is not resolved.
The present work also suggests that Thermoplasmatales might be a deeper branching lineage within Euryarchaeota in comparison to the Thermococcales, Halobacteriales, Archaoglobous and Methanogens. This inference is suggested by the observation that a number of proteins that are uniquely present in almost all other Euryarcheota species are missing in the Thermoplasmatales. Although the absence of these proteins in the Thermoplasmatales can be explained by specific gene loss, the possibility that the genes for at least some of these proteins have evolved after the branching of Thermoplasmatales deserves serious consideration. The deeper branching of the Thermoplasmatales within the Euryarchaeota will place it closer to the Crenarchaeota. Such a placement could prove helpful in understanding why so many genes (i.e. 30) are uniquely shared by various Thermoplasmatales and the Sulfolobales.
For the archaeal-specific proteins identified in the present work, sequence information at present is available from only a limited number of archaeal species. Hence, it is important to obtain information for these genes/proteins from other archaeal species to confirm whether these proteins are distinctive characteristics of the specified groups or a subgroup of such species. These proteins in addition to their utility for phylogenetic and taxonomic studies also provide valuable means for understanding archaeal biology [35, 38]. The cellular functions of most of these proteins are not known and further studies in this regard should prove very helpful in the discovery of novel biochemical and physiological characteristics that are unique to either all or different groups of archaea [38]. Lastly, the primary sequences of many of these genes/proteins are also highly conserved and they provide novel means for identification of different groups of archaea in various environmental settings by means of PCR amplification and other molecular biological and immunological methods.
Methods
Identification of Archaea-specific proteins
To identify proteins which are specific for Archaea or its various subgroups, all proteins in the genomes of A. pernix K1 (APE), S. acidocaldarius DSM 639 (Saci), P. aerophilum str. IM2 (PAE), P. abyssi GE5 (PAB), M. maripaludis S2 (MMP), M. kandleri AV19 (MK), M. burtonii DSM 6242 (Mbu), Halobacterium sp. NRC-1 (VNG), H. walsbyi DSM 16790 (HQ), T. acidophilum DSM 1728 (Ta) and P. torridus DSM 9790 (PTO), were analyzed. Protein-protein blast searches were carried out on each individual protein using the default parameters, without the low complexity filter, to identify different proteins where all significant hits were from archaea [109]. The results of blast searches were inspected for sudden increase in Expected values (E-values) from the last archaeal species in the search to the first non-archaeal organism. The proteins that were of interest generally involved a large increase in E-values from the last archaeal hit to the first hit from any other organism. Further, the E values of these latter hits were expected to be in a range higher than 10-4, which indicates a weak level of similarity that could occur by chance. However, higher E-values are sometimes acceptable for smaller proteins as the magnitude of the E-value depends upon the length of the query sequence.
All promising proteins identified by the above criteria were further analyzed using the position-specific iterated (PSI) blast program. In the present work, a protein was considered to be archaeal-specific if all hits producing significant alignments were from the indicated groups of archaeal species. However, we have also retained a few proteins where 1 or 2 isolated species from other groups (e.g. bacteria or eukaryotes) also had acceptable E-values. We consider these proteins to be also archaea-specific and their presence in isolated unrelated species is very likely due to lateral gene transfer. For all archaea-specific proteins described here, the protein ID, accession number and their possible functions (also COG or CDD number [102, 110]) are presented in Tables 2, 3, 4, 5, 6, 7, 8 and Additional files 1, 2, 3, 4, 5, 6, 7, 8, 9. All proteins indicated in various tables are specific for the Archaea based on these criteria unless otherwise mentioned.
Phylogenetic analyses
Phylogenetic analyses was carried out on a concatenated sequence alignment of 31 universally distributed proteins [45]. The information regarding these proteins is provided in the Additional file 10. For each of these proteins sequences from all 29 archaeal species were downloaded and multiple sequence alignments were created using ClustalX 1.83 program. A concatenated sequence alignment for all 31 proteins was imported into Gblocks 0.91b [111] to remove poorly aligned region. The resulting final alignment of 6,252 amino acid sites was used for phylogenetic analyses. A NJ tree based on this dataset was constructed by TREECON 1.3b program with Kimura two-parameter model distance [112]; Maximum-Likelihood tree were computed under a WAG+F model plus a gamma distribution with four categories by TREE-PUZZLE 5.2 [113, 114]; Maximum-Parsimony tree were obtained by Mega 3.1 package [115]. All of the trees were bootstrapped 100 times.
References
Woese CR, Kandler O, Wheelis ML: Towards A Natural System of Organisms - Proposal for the Domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990, 87: 4576-4579. 10.1073/pnas.87.12.4576.
Ludwig W, Klenk HP: Overview:A phylogenetic backbone and taxonomic framework for prokaryotic systamatics. Bergey's Manual of Systematic Bacteriology. Edited by: D.R. B and R.W. C. 2001, Berlin, Springer-Verlag, 49-65.
Pace NR: A molecular view of microbial diversity and the biosphere. Science. 1997, 276: 734-740. 10.1126/science.276.5313.734.
Skophammer RG, Herbold CW, Rivera MC, Servin JA, Lake JA: Evidence that the Root of the Tree of Life is not within the Archaea. Mol Biol Evol. 2006
Karlin S, Brocchieri L, Trent J, Blaisdell BE, Mrazek J: Heterogeneity of genome and proteome content in bacteria, archaea, and eukaryotes. Theor Popul Biol. 2002, 61: 367-390. 10.1006/tpbi.2002.1606.
Gupta RS: Protein Phylogenies and Signature Sequences: A Reappraisal of Evolutionary Relationships Among Archaebacteria, Eubacteria, and Eukaryotes. Microbiol Mol Biol Rev. 1998, 62: 1435-1491.
Olsen GJ, Woese CR, Overbeek R: The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994, 176: 1-6.
Gupta RS: What are archaebacteria: Life's third domain or monoderm prokaryotes related to Gram-positive bacteria? A new proposal for the classification of prokaryotic organisms. Mol Microbiol. 1998, 29: 695-708. 10.1046/j.1365-2958.1998.00978.x.
Mayr E: Two empires or three?. Proc Natl Acad Sci USA. 1998, 95: 9720-9723. 10.1073/pnas.95.17.9720.
Gupta RS: The Natural Evolutionary Relationships Among Prokaryotes. Crit Rev Microbiol. 2000, 26: 111-131. 10.1080/10408410091154219.
Cavalier-Smith T: The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol. 2002, 52: 7-76.
Woese CR: The universal ancestor. Proc Natl Acad Sci USA. 1998, 95: 6854-6859. 10.1073/pnas.95.12.6854.
Gribaldo S, Brochier-Armanet C: The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci. 2006, 361: 1007-1022. 10.1098/rstb.2006.1841.
Gupta RS: Molecular Sequences and the Early History of Life. Microbial Phylogeny and Evolution: Concepts and Controversies. Edited by: Sapp J. 2005, New York, Oxford University Press, 160-183.
Woese CR: Bacterial Evolution. Microbiol Rev. 1987, 51: 221-266.
Barns SM, Fundyga RE, Jeffries MW, Pace NR: Remarkable Archaeal Diversity Detected in A Yellowstone-National-Park Hot-Spring Environment. Proc Natl Acad Sci U S A. 1994, 91: 1609-1613. 10.1073/pnas.91.5.1609.
Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S: Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001, 11: 1641-1650. 10.1101/gr.190201.
Gonzalez JM, Sheckells D, Viebahn M, Krupatkina D, Borges KM, Robb FT: Thermococcus waiotapuensis sp. nov., an extremely thermophilic archaeon isolated from a freshwater hot spring. Arch Microbiol. 1999, 172: 95-101. 10.1007/s002030050745.
Futterer O, Angelov A, Liesegang H, Gottschalk G, Schleper C, Schepers B, Dock C, Antranikian G, Liebl W: Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc Natl Acad Sci U S A. 2004, 101: 9091-9096. 10.1073/pnas.0401356101.
Schleper C, Jurgens G, Jonuscheit M: Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005, 3: 479-488. 10.1038/nrmicro1159.
Jones WJ, Nagle DP, Whitman WB: Methanogens and the Diversity of Archaebacteria. Microbiol Rev. 1987, 51: 135-177.
Lange M, Ahring BK: A comprehensive study into the molecular methodology and molecular biology of methanogenic Archaea. FEMS Microbiol Lett. 2001, 25: 553-571.
Olsen GJ, Woese CR: Archaeal genomics: An overview. Cell. 1997, 89: 991-994. 10.1016/S0092-8674(00)80284-6.
Brown JR, Doolittle WF: Archaea and the prokaryote-to-eukaryote transition. Microbiol Rev. 1997, 61: 456-502.
Brendel V, Brocchieri L, Sandler SJ, Clark AJ, Karlin S: Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J Mol Evol. 1997, 44: 528-541. 10.1007/PL00006177.
Walsh DA, Doolittle WF: The real 'domains' of life. Curr Biol. 2005, 15: R237-R240. 10.1016/j.cub.2005.03.034.
Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV: Comparative genomics of the archaea (Euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999, 9: 608-628.
Burggraf S, Huber H, Stetter KO: Reclassification of the crenarchaeal orders and families in accordance with 16S rRNA sequence data. Int J Syst Bacteriol. 1997, 47: 657-660.
Bapteste E, Brochier C, Boucher Y: Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea. 2005, 1: 353-363.
Lake JA, Henderson E, Oakes M, Clark MW: Eocytes - A New Ribosome Structure Indicates A Kingdom with A Close Relationship to Eukaryotes. Proc Natl Acad Sci U S A. 1984, 81: 3786-3790. 10.1073/pnas.81.12.3786.
Lake JA: Evolving Ribosome Structure - Domains in Archaebacteria, Eubacteria, Eocytes and Eukaryotes. Annu Rev Biochem. 1985, 54: 507-530. 10.1146/annurev.bi.54.070185.002451.
Brochier C, Forterre P, Gribaldo S: An emerging phylogenetic core of Archaea: phylogenies of transcription and translation machineries converge following addition of new genome sequences. BMC Evol Biol. 2005, 5:
Woese CR, Olsen GJ: Archaebacterial Phylogeny - Perspectives on the Urkingdoms. Syst Appl Microbiol. 1986, 7: 161-177.
Brochier C, Forterre P, Gribaldo S: Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biol. 2004, 5:
Makarova KS, Koonin EV: Evolutionary and functional genomics of the Archaea. Curr Opin Microbiol. 2005, 8: 586-594. 10.1016/j.mib.2005.08.003.
Graham DE, Overbeek R, Olsen GJ, Woese CR: An archaeal genomic signature. Proc Natl Acad Sci U S A. 2000, 97: 3304-3308. 10.1073/pnas.050564797.
Karlin S: Global dinucleotide signatures and analysis of genomic heterogeneity. Curr Opin Microbiol. 1998, 1: 598-610. 10.1016/S1369-5274(98)80095-7.
Galperin MY, Koonin EV: 'Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004, 32: 5452-5463. 10.1093/nar/gkh885.
Griffiths E, Ventresca MS, Gupta RS: BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species. BMC Genomics. 2006, 7: 14-10.1186/1471-2164-7-14.
Kainth P, Gupta RS: Signature Proteins that are Distinctive of Alpha Proteobacteria . BMC Genomics. 2005, 6: 94-10.1186/1471-2164-6-94.
Gao B, Paramanathan R, Gupta RS: Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie van Leeuwenhoek. 2006, 90: 69-91. 10.1007/s10482-006-9061-2.
Gupta RS, Griffiths E: Chlamydiae-specific proteins and indels: novel tools for studies. Trends Microbiol. 2006
Gupta RS: Molecular signatures (unique proteins and conserved Indels) that are specific for the epsilon proteobacteria (Campylobacterales). BMC Genomics. 2006, 7: 167-
Matte-Tailliez O, Brochier C, Forterre P, Philippe H: Archaeal phylogeny based on ribosomal proteins. Mol Biol Evol. 2002, 19: 631-639.
Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P: Toward automatic reconstruction of a highly resolved tree of life. Science. 2006, 311: 1283-1287. 10.1126/science.1123061.
Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P: Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales?. Genome Biol. 2005, 6:
Burggraf S, Stetter KO, Rouviere P, Woese CR: Methanopyrus-Kandleri - An Archael Methanogen Unrelated to All Other Known Methanogens. Syst Appl Microbiol. 1991, 14: 346-351.
Rivera MC, Lake JA: The phylogeny of Methanopyrus kandleri. Int J Syst Bacteriol. 1996, 46: 348-351.
Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y: Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999, 6: 83-101. 10.1093/dnares/6.2.83.
Chen LM, Brugger K, Skovgaard M, Redder P, She QX, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk HP, Garrett RA: The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol. 2005, 187: 4992-4999. 10.1128/JB.187.14.4992-4999.2005.
Fitz-Gibbon ST, Ladner H, Kim UJ, Stetter KO, Simon MI, Miller JH: Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc Natl Acad Sci U S A. 2002, 99: 984-989. 10.1073/pnas.241636498.
Cohen GN, Barbe V, Flament D, Galperin M, Heilig R, Lecompte O, Poch O, Prieur D, Querellou J, Ripp R, Thierry JC, Van der Oost J, Weissenbach J, Zivanovic Y, Forterre P: An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol Microbiol. 2003, 47: 1495-1512. 10.1046/j.1365-2958.2003.03381.x.
Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, de Macario EC, Dodsworth JA, Gillett W, Graham DE, Hackett M, Haydock AK, Kang A, Land ML, Levy R, Lie TJ, Major TA, Moore BC, Porat I, Palmeiri A, Rouse G, Saenphimmachak C, Soll D, Van Dien S, Wang T, Whitman WB, Xia Q, Zhang Y, Larimer FW, Olson MV, Leigh JA: Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol. 2004, 186: 6956-6969. 10.1128/JB.186.20.6956-6969.2004.
Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ, Hough DW, Maddocks DG, Jablonski PE, Krebs MP, Angevine CM, Dale H, Isenbarger TA, Peck RF, Pohlschroder M, Spudich JL, Jung KH, Alam M, Freitas T, Hou SB, Daniels CJ, Dennis PP, Omer AD, Ebhardt H, Lowe TM, Liang R, Riley M, Hood L, DasSarma S: Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A. 2000, 97: 12176-12181. 10.1073/pnas.190337797.
Ruepp A, Graml W, Santos-Martinez ML, Koretle KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W: The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000, 407: 508-513. 10.1038/35035069.
Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, Shakhova VV, Belova GI, Aravind L, Natale DA, Rogozin IB, Tatusov RL, Wolf YI, Stetter KO, Malykh AG, Koonin EV, Kozyavkin SA: The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc Natl Acad Sci U S A. 2002, 99: 4644-4649. 10.1073/pnas.032671499.
Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, Beeson KY, Bibbs L, Bolanos R, Keller M, Kretz K, Lin XY, Mathur E, Ni JW, Podar M, Richardson T, Sutton GG, Simon M, Soll D, Stetter KO, Short JM, Noordewier M: The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci U S A. 2003, 100: 12984-12988. 10.1073/pnas.1735403100.
Huber H, Hohn MJ, Stetter KO, Rachel R: The phylum Nanoarchaeota: Present knowledge and future perspectives of a unique form of life. Res Microbiol. 2003, 154: 165-171. 10.1016/S0923-2508(03)00035-4.
Cho HD, Verlinde CL, Weiner AM: Archaeal CCA-adding enzymes - Central role of a highly conserved beta-turn motif in RNA polymerization without translocation. J Biol Chem. 2005, 280: 9555-9566. 10.1074/jbc.M412594200.
Xiong Y, Li F, Wang JM, Weiner AM, Steitz TA: Crvstal structures of an archaeal class ICCA-adding enzyme and its nucleotide complexes. Mol Cell. 2003, 12: 1165-1172. 10.1016/S1097-2765(03)00440-4.
Iyer LM, Koonin EV, Leipe DD, Aravind L: Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005, 33: 3875-3896. 10.1093/nar/gki702.
Ito N, Nureki O, Shirouzu M, Yokoyama S, Hanaoka F: Crystallization and preliminary X-ray analysis of a DNA primase from hyperthermophilic archaeon Pyrococcus horikoshii. J Biochem (Tokyo). 2001, 130: 727-730.
Kawashima T, Amano N, Koike H, Makino S, Higuchi S, Kawashima-Ohya Y, Watanabe K, Yamazaki M, Kanehori K, Kawamoto T, Nunoshiba T, Yamamoto Y, Aramaki H, Makino K, Suzuki M: Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc Natl Acad Sci U S A. 2000, 97: 14257-14262. 10.1073/pnas.97.26.14257.
Daugherty M, Vonstein V, Overbeek R, Osterman A: Archaeal shikimate kinase, a new member of the GHMP-kinase family. J Bacteriol. 2001, 183: 292-300. 10.1128/JB.183.1.292-300.2001.
Aravind L, Makarova KS, Koonin EV: Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000, 28: 3417-3432. 10.1093/nar/28.18.3417.
Dworkin M, al : The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 2001, Springer-Verlag, [http://link.springer-ny.com/link/service/books/10125/].3
Knittel K, Losekann T, Boetius A, Kort R, Amann R: Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol. 2005, 71: 467-479. 10.1128/AEM.71.1.467-479.2005.
Kawarabayasi Y, Hino Y, Horikawa H, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, Fukui S, Nagai Y, Nishijima K, Otsuka R, Nakazawa H, Takamiya M, Kato Y, Yoshizawa T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Masuda S, Yanagii M, Nishimura M, Yamagishi A, Oshima T, Kikuchi H: Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 2001, 8: 123-140. 10.1093/dnares/8.4.123.
She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CCY, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PMK, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J: The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001, 98: 7835-7840. 10.1073/pnas.141222098.
Rivera MC, Lake JA: Evidence That Eukaryotes and Eocyte Prokaryotes Are Immediate Relatives. Science. 1992, 257: 74-76. 10.1126/science.1621096.
Ishitani R, Nureki O, Fukai S, Kijimoto T, Nameki N, Watanabe M, Kondo H, Sekine M, Okada N, Nishimura S, Yokoyama S: Crystal structure of archaeosine tRNA-guanine transglycosylase. J Mol Biol. 2002, 318: 665-677. 10.1016/S0022-2836(02)00090-6.
Aravind L, Koonin EV: Novel predicted RNA-binding domains associated with the translation machinery. J Mol Evol. 1999, 48: 291-302. 10.1007/PL00006472.
Shen Y, Tang XF, Matsui E, Matsui I: Subunit interaction and regulation of activity through terminal domains of the family D DNA polymerase from Pyrococcus horikoshii. Biochem Soc Trans. 2004, 32: 245-249. 10.1042/BST0320245.
Cann IKO, Ishino Y: Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics. 1999, 152: 1249-1267.
Garrity GM, Holt JG: The road map to the manual. Bergey's Manual of Systematic Bacteriology. Edited by: Boone DR and Castenholz RW. 2001, Berlin, Springer-Verlag, 119-166. 2nd
Reeve JN, Nolling J, Morgan RM, Smith DR: Methanogenesis: Genes, genomes, and who's on first?. J Bacteriol. 1997, 179: 5975-5986.
Vandewijngaard WMH, Creemers J, Vogels GD, Vanderdrift C: Methanogenic Pathways in Methanosphaera-Stadtmanae. FEMS Microbiol Lett. 1991, 80: 207-212. 10.1016/0378-1097(91)90596-3.
Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK: The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H-2 for methane formation and ATP synthesis. J Bacteriol. 2006, 188: 642-658. 10.1128/JB.188.2.642-658.2006.
Lie TJ, Leigh JA: A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol Microbiol. 2003, 47: 235-246. 10.1046/j.1365-2958.2003.03293.x.
Murakami E, Ragsdale SW: Evidence for intersubunit communication during acetyl-CoA cleavage by the multienzyme CO dehydrogenase/acetyl-CoA synthase complex from Methanosarcina thermophila - Evidence that the beta subunit catalyzes C-C and C-S bond cleavage. J Biol Chem. 2000, 275: 4699-4707. 10.1074/jbc.275.7.4699.
Lu WP, Jablonski PE, Rasche M, Ferry JG, Ragsdale SW: Characterization of the Metal Centers of the Ni/Fe-S Component of the Carbon-Monoxide Dehydrogenase Enzyme Complex from Methanosarcina-Thermophila. J Biol Chem. 1994, 269: 9736-9742.
Lindahl PA, Chang B: The evolution of acetyl-CoA synthase. Orig Life Evol Biosph. 2001, 31: 403-434. 10.1023/A:1011809430237.
Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, Richardson DL, Kerlavage AR, Graham DE, Kyrpides NC, Fleischmann RD, Quackenbush J, Lee NH, Sutton GG, Gill S, Kirkness EF, Dougherty BA, McKenney K, Adams MD, Loftus B, Peterson S, Reich CI, Mcneil LK, Badger JH, Glodek A, Zhou LX, Overbeek R, Gocayne JD, Weidman JF, McDonald L, Utterback T, Cotton MD, Spriggs T, Artiach P, Kaine BP, Sykes SM, Sadow PW, DAndrea KP, Bowman C, Fujii C, Garland SA, Mason TM, Olsen GJ, Fraser CM, Smith HO, Woese CR, Venter JC: The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997, 390: 364-&. 10.1038/37052.
Harms U, Weiss DS, Gartner P, Linder D, Thauer RK: The energy conserving N5-methyltetrahydromethanopterin:coenzyme M methyltransferase complex from Methanobacterium thermoautotrophicum is composed of eight different subunits. Eur J Biochem. 1995, 228: 640-648. 10.1111/j.1432-1033.1995.0640m.x.
Warner KL, Larkin MJ, Harper DB, Murrell JC, McDonald IR: Analysis of genes involved in methyl halide degradation in Aminobacter lissarensis CC495. FEMS Microbiol Lett. 2005, 251: 45-51. 10.1016/j.femsle.2005.07.021.
McAnulla C, Woodall CA, McDonald IR, Studer A, Vuilleumier S, Leisinger T, Murrell JC: Chloromethane utilization gene cluster from Hyphomicrobium chloromethanicum strain CM2(T) and development of functional gene probes to detect halomethane-degrading bacteria. Appl Environ Microbiol. 2001, 67: 307-316. 10.1128/AEM.67.1.307-316.2001.
Grabarse W, Mahlert F, Duin EC, Goubeaud M, Shima S, Thauer RK, Lamzin V, Ermler U: On the mechanism of biological methane formation: Structural evidence for conformational changes in methyl-coenzyme M reductase upon substrate binding. J Mol Biol. 2001, 309: 315-330. 10.1006/jmbi.2001.4647.
Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK: Crystal structure of methyl coenzyme M reductase: The key enzyme of biological methane formation. Science. 1997, 278: 1457-1462. 10.1126/science.278.5342.1457.
Tersteegen A, Hedderich R: Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases - Transcription of the operons and sequence analysis of the deduced proteins. Eur J Biochem. 1999, 264: 930-943. 10.1046/j.1432-1327.1999.00692.x.
Hartmann GC, Klein AR, Linder M, Thauer RK: Purification, properties and primary structure of H-2-forming N-5,N(10)methylenetetrahydromethanopterin dehydrogenase from Methanococcus thermolithotrophicus. Arch Microbiol. 1996, 165: 187-193.
Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S: Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998, 5: 55-76. 10.1093/dnares/5.2.55.
Robb FT, Maeder DL, Brown JR, DiRuggiero J, Stump MD, Yeh RK, Weiss RB, Dunn DM: Genomic sequence of hyperthermophile, Pyrococcus furiosus: Implications for physiology and enzymology. Methods Enzymol. 2001, 330: 134-157.
Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T: Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005, 15: 352-363. 10.1101/gr.3003105.
Yamamoto T, Matsuda T, Sakamoto N, Matsumura H, Inoue T, Morikawa M, Kanaya S, Kai Y: Acta Crystallogr D Biol Crystallogr. Acta Crystallogr D Biol Crystallogr. 2003, 59: 372-374. 10.1107/S090744490202142X.
Qureshi SA, Bell SD, Jackson SP: Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997, 16: 2927-2936. 10.1093/emboj/16.10.2927.
Matsuda T, Morikawa M, Haruki M, Higashibata H, Imanaka T, Kanaya S: Isolation of TBP-interacting protein (TIP) from a hyperthermophilic archaeon that inhibits the binding of TBP to TATA-DNA. FEBS Lett. 1999, 457: 38-42. 10.1016/S0014-5793(99)01005-4.
Muller V, Oren A: Metabolism of chloride in halophilic prokaryotes. Extremophiles. 2003, 7: 261-266. 10.1007/s00792-003-0332-9.
Falb M, Pfeiffer F, Palm P, Rodewald K, Hickmann V, Tittor J, Oesterhelt D: Living with two extremes: Conclusions from the genome sequence of Natronomonas pharaonis. Genome Res. 2005, 15: 1336-1343. 10.1101/gr.3952905.
DasSarma S: Genome sequence of an extremely halophilic archaeon. Microbial Genomes. Edited by: Fraser CM, Read TD and Nelson KE. 2004, totowa, new jersey, humana press, 383-399.
Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu YL, Gan RR, Hung PL, Date SV, Marcotte E, Hood L, Ng WV: Genome sequence of Haloarcula marismortui: A halophilic archaeon from the Dead Sea. Genome Res. 2004, 14: 2221-2234. 10.1101/gr.2700304.
Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, Pfeiffer F, Oesterhelt D: The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity. BMC Genomics. 2006, 7: 169-10.1186/1471-2164-7-169.
Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWweese-Scott C, Geer LY, Gwadz M, He SQ, Hurwitz DI, Jackson JD, Ke ZX, Lanczycki CJ, Liebert CA, Liu CL, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang DC, Bryant SH: CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005, 33: D192-D196. 10.1093/nar/gki069.
Golyshina OV, Pivovarova TA, Karavaiko GI, Kondrat'eva TF, Moore ERB, Abraham WR, Lunsdorf H, Timmis KN, Yakimov MM, Golyshin PN: Ferroplasma acidiphilum gen. nov., sp nov., an acidophilic, autotrophic, ferrous-iron-oxidizing, cell-wall-lacking, mesophilic member of the Ferroplasmaceae fam. nov., comprising a distinct lineage of the Archaea. Int J Syst Evol Microbiol. 2000, 50: 997-1006.
Garrity GM, Bell JA, liburn TG: Taxonomic outline of the prokaryotes. bergey's manual of systematic bacteriology. 2004, 2
Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau MER, Nesbo CL, Case RJ, Doolittle WF: Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet. 2003, 37: 283-328. 10.1146/annurev.genet.37.050503.084247.
Boone DR: Bergey's Manual of systematic bacteriology. 2001, Springer, 1: 2nd
Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F: Heterotachy and long-branch attraction in phylogenetics. BMC Evol Biol. 2005, 5:
Felsenstein J: Cases in which parsimony and compatibility methods will be positively misleading. Systematic Zoology. 1978, 27: 401-410. 10.2307/2412923.
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Tatusov RL, Galperin MY, Natale DA, Koonin EV: The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28: 33-36. 10.1093/nar/28.1.33.
Castresana J: Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000, 17: 540-552.
van de Peer Y, De Wachter R: TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994, 10: 569-570.
Schmidt HA, Strimmer K, Vingron M, von Haeseler A: TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002, 18: 502-504. 10.1093/bioinformatics/18.3.502.
Whelan S, Goldman N: A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001, 18: 691-699.
Kumar S, Tamura K, Nei M: MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004, 5: 150-163. 10.1093/bib/5.2.150.
Acknowledgements
We thank Inas Radhi, Amy Mok, and Gayathri Vaidyanathan for assistance in carrying out blast searches on some of the archaeal genomes. We also thank Venus Wong for computer support that facilitated the blastp analyses. This work was supported by a research grant from the National Science and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
BG carried out blast searches on different proteins and was responsible for the initial evaluation of the results. RSG conceived and directed this study and was responsible for the final evaluation of the results. BG prepared a rough draft of the manuscript under RSG's directions, which was revised and modified by RSG. All authors have read and approved the final manuscript.
Electronic supplementary material
12864_2006_799_MOESM1_ESM.pdf
Additional file 1: Proteins specific to Sulfolobales. These proteins are indicated to be specific for the either all three sequenced Sulfolobus species (S. solfataricus, S. acidocaldarius and S. tokodaii) or two of these based on Blastp and PSI-Blast searches. The proteins only found in a single Sulfolobus species are not listed here. (PDF 11 KB)
12864_2006_799_MOESM2_ESM.pdf
Additional file 2: Proteins specific to Pyrobaculum and Thermofilum. For the proteins listed in this table, significant hits in Blastp and PSI-Blast searches are only observed for Pyrobaculum and Thermofilum. Because the genome of Thermofilum pendens is only partially sequenced, additional proteins of this kind may be found. (PDF 8 KB)
12864_2006_799_MOESM3_ESM.pdf
Additional file 3: Proteins specific for particular groups of methanogens. For the proteins listed in this table, significant hits in Blastp and PSI-Blast searches are only observed for (a) Methanococcales and (b) Methanosarcinaceae. (PDF 10 KB)
12864_2006_799_MOESM4_ESM.pdf
Additional file 4: Proteins specific for Thermococci. The proteins listed in this table are specific for either various (a) Thermococci (i.e. Thermococcus and Pyrococcus) or (b) various Pyrococci. (PDF 10 KB)
12864_2006_799_MOESM5_ESM.pdf
Additional file 5: Proteins specific for various Halobacteria. The proteins listed in this Table are specifically found in various sequenced halobacteria species as determined by Blastp and PSI-Blast searches. (PDF 10 KB)
12864_2006_799_MOESM6_ESM.pdf
Additional file 6: Proteins specific for various Halobacteria. These proteins are also specific for Halobacteria. However, unlike the proteins listed in Additional file 5, they are present in only three of the four sequenced halobacterial genomes. (PDF 9 KB)
12864_2006_799_MOESM7_ESM.pdf
Additional file 7: Proteins specific for various Halobacteria. These proteins are also specific for Halobacteria but they are found in only two of the four sequenced halobacterial genomes. (PDF 9 KB)
12864_2006_799_MOESM8_ESM.pdf
Additional file 8: Proteins specific for Thermoplasmata. These proteins are specific for either (a) all sequenced Thermoplasmata species, or (b) specific for only Thermoplasma, or (c) specific for Picrophilus and Ferroplasma. (PDF 10 KB)
12864_2006_799_MOESM9_ESM.pdf
Additional file 9: Archaeal-specific proteins with sporadic distribution. These proteins are specific for Archaea but they show sporadic distribution in different groups. (PDF 8 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Gao, B., Gupta, R.S. Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics 8, 86 (2007). https://doi.org/10.1186/1471-2164-8-86
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-8-86