Abstract
Background
Campylobacter jejuni infection represents the most frequent antecedent infection triggering the onset of the neuropathic disorders Guillain-Barré syndrome (GBS) and Miller Fisher syndrome (MFS). Although sialylated ganglioside-mimicking lipo-oligosaccharide (LOS) structures are the strongest neuropathogenic determinants in C. jejuni, they do not appear to be the only requirement for a neuropathic outcome since strains capable of their production have been isolated from patients with uncomplicated cases of enteritis. Consequently, other pathogen and/or host-related factors contribute to the onset of neurological complications. We have used comparative genomic hybridization to perform a detailed genomic comparison of strains isolated from GBS/MFS and enteritis-only patients. Our dataset, in which the gene conservation profile for 1712 genes was assayed in 102 strains, including 56 neuropathogenic isolates, represents the largest systematic search for C. jejuni factors associated with GBS/MFS to date and has allowed us to analyze the genetic background of neuropathogenic C. jejuni strains with an unprecedented level of resolution.
Results
The majority of GBS/MFS strains can be assigned to one of six major lineages, suggesting that several genetic backgrounds can result in a neuropathogenic phenotype. A statistical analysis of gene conservation rates revealed that although genes involved in the sialylation of LOS structures were significantly associated with neuropathogenic strains, still many enteritis-control strains both bear these genes and share remarkable levels of genomic similarity with their neuropathogenic counterparts. Two capsule biosynthesis genes (Cj1421c and Cj1428c) showed higher conservation rates among neuropathogenic strains compared to enteritis-control strains. Any potential involvement of these genes in neuropathogenesis must be assessed. A single gene (HS:3 Cj1135) had a higher conservation rate among enteritis-control strains. This gene encodes a glucosyltransferase that is found in some of the LOS classes that do not express ganglioside mimics.
Conclusion
Our findings corroborate that neuropathogenic factors may be transferred between unrelated strains of different genetic background. Our results would also suggest that the failure of some strains isolated from uncomplicated cases of enteritis to elicit a neuropathic clinical outcome may be due to subtle genetic differences that silence their neuropathogenic potential and/or due to host-related factors.
The microarray data has been deposited in NCBI's Gene Expression Omnibus under accession number GSE3579.
Similar content being viewed by others
Background
Campylobacter jejuni infection is a leading cause of acute bacterial gastroenteritis worldwide [1]. The widespread dissemination of C. jejuni is largely attributed to its transmission to humans by consumption of contaminated food sources frequently colonized by the bacteria. Clinical symptoms vary from mild to severe gastroenteritis to more complex and serious extraintestinal diseases, including the neuropathic disorders Guillain-Barré syndrome (GBS) or its variant Miller Fisher syndrome (MFS) [2].
GBS and MFS are acute neuropathies thought to result from a transient humoral immune response against host gangliosides in peripheral nerves [3]. GBS is the most common form of acute flaccid paralysis with incidence rates of up to 4 per 100, 000 [4]. MFS, a variant of GBS, is characterized by acute ophthalmoplegia and ataxia. The onset of these syndromes is often preceded by infectious illness and C. jejuni is the most frequent infectious agent. 30% of GBS cases and 20% of MFS cases are preceded by an infection of C. jejuni within three weeks prior to the onset of neurological symptoms [5–7]
A number of studies have sought to examine the population structure of GBS/MFS strains in the search for genetic commonalities that could account for a neuropathogenic phenotype. Initial surveys showing an over-representation of serotypes HS:19 and HS:41 among GBS strains [8–11], raised the possibility that GBS strains comprise a clonal lineage with unique virulence factors associated with GBS. For example, a high proportion of HS:19 isolates was obtained from GBS patients in countries such as Japan, despite the low prevalence of this serotype among Japanese enteritis cases, suggesting that enteritis patients infected with a strain of HS:19 serotype have an increased risk of developing GBS [8]. A similar association has been observed between Japanese MFS-related C. jejuni strains and the HS:2 serotype [10]. Subsequent studies, however, have shown substantial genetic heterogeneity in other collections of neuropathogenic strains [12–14].
Lipo-oligosaccharide (LOS) is one of the most important cell-surface structures expressed by C. jejuni, and strains associated with neuropathies express ganglioside-like LOS structures [15, 16]. The development of GBS/MFS following C. jejuni infection is thought to be related to molecular mimicry between ganglioside-like moieties on certain Campylobacter LOS classes and ganglioside epitopes on neural tissue [17], with cross-reacting anti-LOS antibodies ultimately leading to nerve damage [18]. The majority of patients with GBS subsequent to C. jejuni enteritis develop autoantibodies that react to GM1 or GD1a gangliosides [19, 20] whereas MFS patients develop anti-GQ1b antibodies [5, 21]. The LOS from GBS- and MFS-associated C. jejuni have been shown to induce anti-GM1 and anti-GQ1b antibodies in rabbits [22]. Moreover, sensitization of rabbits with GM1-like LOS of C. jejuni isolated from a GBS patient has resulted in a disease model of GBS [23].
Among the three classes of C. jejuni LOS locus (A, B, and C) initially characterized by Gilbert et al. [24], the majority of HS:19 isolates harbour a Class A LOS locus, a gene cluster implicated in the expression of ganglioside mimics [25, 26]. The A-class LOS carries the cst-II gene which, first isolated from the GBS-associated strain OH4384, encodes a bifunctional sialyltransferase capable of transferring sialic acid to either a terminal galactose residue or to a terminal sialic acid residue, resulting in linkages that lead to the production of ganglioside mimics [27]. Taken together, both cst-II and the Class A locus are currently the strongest known determinants of GBS [25, 26, 28–30] Recent studies using knockout mutants of C. jejuni and a mouse model have demonstrated the necessity of cst-II and of a related sialylation pathway gene (orf10 or neuA1, encoding a CMP-NeuAc synthetase) in the induction of anti-ganglioside antibodies [25].
Although anti-LOS cross-reactive antibodies are a major component of the development of C. jejuni induced GBS and MFS, the complete mechanism is not fully understood. Similarly, the extent to which microbial and/or host factors contribute to the development of an anti-glycolipid response and neurological symptoms remains a point of debate [31]. Cases have been described in which GBS patients with C. jejuni infections fail to display ganglioside sero-reactivity, raising the possibility that other peripheral nerve antigens are the targets in these patients [32]. Furthermore, ganglioside-like structures have been found in C. jejuni isolates from enteritis patients without GBS or MFS [33]. Recent work aiming to characterize the LOS locus of GBS/MFS-associated C. jejuni strains also suggests that there is a strong, but incomplete, correlation between neuropathogenic strains and an A or B-class LOS locus [25, 26, 29]. Although neuropathogenic strains that carry a non-A/B locus may be less prevalent, their existence raises the possibility that other factors (pathogen and/or host-related) may contribute to the onset of neurological complications. We have performed a systematic search for additional pathogen-related GBS/MFS-associated factors by performing a detailed genetic comparison of strains isolated from GBS/MFS patients, and from uncomplicated cases of enteritis (i.e. "enteritis-control strains"), by means of microarray-based Comparative Genomic Hybridization (CGH). We present here the results of this comprehensive comparative genomic survey.
Results and discussion
GBS/MFS strains are genomically heterogeneous
Previous studies have suggested the heterogeneous nature of neuropathogenic C. jejuni strains [12, 34]. We compared the CGH profiles of the 56 neuropathogenic strains in our dataset and cluster analysis confirms substantial genomic heterogeneity among the strains studied (Figure 1). However, the results also suggest the presence of several lineages distinguishable from one another based on differences among known hypervariable loci [35].
CGH profiles for the 56 neuropathogenic strains in the combined dataset. The 56 neuropathogenic strains analyzed for this study show substantially different genetic backgrounds, although most appear to belong to one of six major lineages (LIN). Three strains show unique gene conservation profiles and fail to cluster robustly with any major lineage. Branches with greater than 75% bootstrap support are shown in red. Although data is displayed including capsular genes (gray box), these genes were removed during cluster analysis to avoid biasing results. Highly divergent/Absent genes shown in red; Moderately Divergent genes are shown in blue. Legend: Hypervariable loci (L – LOS locus; F – flagellar modification locus; C – capsular locus; R/M – restriction-modification locus); Strain sets (DG: Dutch GBS; DM: Dutch MFS; JG: Japanese GBS; JM: Japanese MFS; CG: Curaçao GBS).
Clusters I and II are comprised of strains with very little genetic divergence with respect to the genome strain NCTC 11168. Cluster I includes two GBS strains (GB13 and GB14) that show among the lowest levels of divergence observed thus far with respect to the genome strain NCTC 11168. The strains in Cluster II, which include GB11 [36], are very similar to NCTC 11168, albeit with a divergence compared to that strain's C-class LOS locus. Although the bulk of the strains in clusters I and II are of the HS:2 serotype (10 of 13), three strains (GB01, GB26, and GB27) are non-HS:2. Cluster III is comprised of 4 of the 5 HS:4-complex strains in the entire dataset. Cluster IV is largely comprised of Japanese HS:2 strains harbouring significant divergences with respect to NCTC 11168 and also includes a genetically similar Dutch strain (GB25). Cluster V is comprised of neuropathogenic strains from the "Curaçao cluster", a genetically homogeneous group of enteritis and GBS strains from Curaçao that also includes the Dutch GBS/MFS isolates GB21 and MF07. Cluster VI, which at 23 strains is also the largest, includes all HS:19 strains in the dataset regardless of disease outcome or geographical source. All strains in the cluster show a high degree of genomic homogeneity with respect to one another despite the cluster containing a small number of non HS:19 strains.
Although most neuropathogenic strains appear to form part of 6 major genomic lineages, cluster analysis of the 102 strains in our survey shows that every major lineage present in the dataset includes both neuropathogenic and enteritis-only strains (Figure 2). Thus, there appears to be no lineages comprised exclusively of either neuropathogenic or enteritis-control strains. Due to the large number of HS:2 and HS:19 strains in our combined dataset, we performed all cluster analysis after removal of genes from the capsular polysaccharide locus (CPS), to remove any possible bias imparted by the expected differences at this locus. Thus, clusters predominantly composed of strains from serotypes HS:2 (clusters II and IV) and HS:19 (cluster VI) are based on genomic similarities at loci other than the CPS and are likely indicative of clonality among these strains.
CGH profile-based clustering of 102 strains included in this study. Although cluster analysis of 56 neuropathogenic strains produced 6 major lineages, inclusion of the 46 enteritis-control strains shows that these lineages are not exclusively comprised of neuropathogenic strains. Both types of strains can show substantial similarities in genomic background, which includes similarities at several hypervariable regions. The lineage (LIN) of the 56 neuropathogenic strains is shown. Highly similar enteritis-control/neuropathogenic strain pairs (boxes a through d) are shown in expanded form in Figure 3. Legend: Hypervariable loci (L – LOS locus; F – flagellar modification locus; C – capsular locus; R/M – restriction-modification locus); Strain sets (DG: Dutch GBS; DM: Dutch MFS; DE: Dutch enteritis; JG: Japanese GBS; JM: Japanese MFS; JE: Japanese enteritis; CG: Curaçao GBS; CE: Curaçao enteritis).
Statistical comparison of gene conservation rates in neuropathogenic and enteritis-control strains
In order to uncover genes associated with neuropathogenic potential, we selected a representative set of neuropathogenic and enteritis-control strains and compared the "absence rate" for each gene in the microarray in both groups of strains. Enteritis-control and neuropathogenic isolates were selected so as to represent the various lineages in our dataset and so as to minimize the effect of the unequal distribution of isolates in each lineage. After analyzing the gene content data for neuropathogenic and enteritis-control isolates, 20 genes were found to have >15% difference in absence rates. Of these, only six genes had statistically significant differences in the absence rate between both groups (Table 1). We found lower absence rates for three markers associated with A/B-class LOS loci and GBS (cgtA, neuA1, orf11) among neuropathogenic strains and although, unexpectedly, our data did not appear to show a significant association for the important GBS marker cst-II, we can attribute this to a flaw with the corresponding probe in our microarray. Of the more than 1700 genes tested in our CGH survey, only two additional genes (Cj1421c, Cj1428c or fcl) had significantly lower absence rates among neuropathogenic strains compared to the enteritis-control strains. A single gene (HS:3 Cj1135) had a significantly lower absence rate among enteritis-control strains (Table 1).
Neuropathogenic and enteritis-control strains can share remarkable levels of genetic similarity
Results from cluster analysis of the CGH data showed several instances in which a neuropathogenic strain and an enteritis-only strain clustered together with very high similarity in genomic profiles. The gene content of these strains was assessed using previously defined thresholds [37] and none of these strain pairs were found to show obvious differences in gene content in the known neuropathogenic markers associated with A/B-class LOS loci. In order to determine whether any additional differences in gene content could be correlated to differences in clinical outcome, four such strain pairs were chosen for a subsequent high-resolution comparison of CGH profiles.
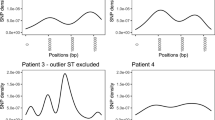
As seen in Figure 3, the gene conservation profiles of some of these strain pairs showed a remarkable degree of congruence, with CGH profiles showing Pearson Correlation coefficients greater than 0.95 on the strength of similarities at multiple hypervariable loci. Each strain pair harboured subtle differences in gene content, ranging from 5 genes for the pair of GB25 and 98652 pair to 13 genes for the pair of EC023 and GC060, however, we did not identify any common gene content differences between the neuropathogenic and enteritis-control strains among the four strain pairs we examined [see Additional file 1].
Gene conservation profiles from closely related pairs of neuropathogenic and enteritis-control strains. Strain pairs were analyzed separately to look for any potential genetic differences that could be related to differences in clinical outcome. Although each strain pair showed subtle differences in CGH profiles, none of these was common across the various strain pairs. Legend: L – LOS locus; F – flagellar modification locus; C – capsular locus; R/M – restriction-modification locus.
Conclusion
Current available evidence points to the direct involvement of genes that synthesize and transfer sialic acid to the LOS in the development of the cross-reactive anti-ganglioside antibodies thought to be the effectors in a large majority of GBS and MFS cases [25, 26]. Although the association between neuropathogenic LOS genotypes and the GBS/MFS-associated phenotype is very strong, strains that bear the requisite genes for the synthesis of ganglioside mimics have been isolated from uncomplicated cases of enteritis. Similarly, a small number of GBS/MFS-associated strains do not appear to synthesize ganglioside mimics. Thus, the incomplete penetrance of the neuropathogenic LOS genotype has raised questions regarding the possible contribution of additional factors, whether bacterial or host-related, towards the development of these neuropathies. That the host contributes to such an auto-immune response should be considered likely.
A consistent finding among various genetic surveys of C. jejuni has been that GBS strains do not appear to represent a genetic lineage distinct from enteritis-control strains [12, 14, 38–40]. Studies attempting to correlate molecular typing results to the GBS/MFS phenotype did not reveal clusters of neuropathogenic strains distinct from enteritis-control strains, and results from our study suggest that many neuropathogenic strains are genomically related to enteritis-control strains. Some of the major clusters in the dataset include strains from more than one of the three major geographical regions represented in the study. For example, some HS:19 strains from The Netherlands and Curaçao cluster with, and are genetically similar to, strains from the Japanese HS:19 cluster. Similarly, HS:2 strains from The Netherlands cluster with strains from the two Japanese HS:2 lineages. It thus appears that the genomic stability previously suggested among clonal HS:19 strains of differing geographical source [38, 41] is also valid among HS:2 strains, as has been suggested in other studies [42].
While cluster analysis is prominent in this study, the fact that neuropathogenic strains do not form a coherent cluster, and the fact that genes related to neuropathogenesis are expected to represent only a small fraction of the data, expose the shortcomings of the use of cluster analysis to define potential neuropathogenic markers. Because of this, in contrast to previous CGH-based studies [39, 40], we opted to focus our search for neuropathogenic markers on the statistical testing of each individual gene present in the array for significant over-representation or under-representation among neuropathogenic strains. Using this novel approach, we were able to obtain unambiguous statistical signal for higher conservation rates in genes associated with LOS classes A and B among neuropathogenic strains [25, 29, 43]. These results not only agree with the current hypotheses regarding the mechanism of GBS/MFS but also concur with a recent study in which GBS/MFS strains and enteritis-control strains were screened for potential neuropathogenic markers using the high resolution comparative genomic method of high-throughput Amplified Fragment Length Polymorphism (htAFLP) [44]. This study revealed 3 markers highly-associated with GBS which mapped to the LOS locus.
The strong association observed for LOS classes A and B and neuropathogenic strains in our data was found despite the extensive genomic heterogeneity in these strains. A recent study has provided direct evidence for horizontal transfer of genes, including putative neuropathogenic factors in an experimental setting [45]. Our findings and the recent description of strain GB11, a GBS-associated HS:2 strain with high genetic similarity to the genome strain NCTC 11168 but found to have an A-class LOS locus instead of a C-class locus[36] would suggest that horizontal transfer represents an important mechanism for the dissemination of neuropathogenic factors between otherwise unrelated GBS/MFS strains.
Among the known neuropathogenic genes identified in our screen cgtA and neuA1 are known to be involved in LOS biosynthesis [24]. Another gene (orf11) has been found to be associated with class A/B LOS loci [25], and it was recently shown to encode a sialic acid acetyltransferase [46]. Genes in the LOS locus needed for the synthesis of ganglioside mimics are strong GBS/MFS determinants [25, 26, 29] but neuropathogenic strains with no known sialyltransferase genes are known to exist [25]. This raises the possibility, among others, that additional bacterial factors are required to elicit neurological complications. Even though we examined every gene in our microarray for similar associations with GBS/MFS strains, after filtering the data to avoid over- or under-representation due to clonality effects, only two additional genes (Cj1421c, Cj1428c or fcl) had statistically significantly higher conservation rates among neuropathogenic strains compared to enteritis-control strains; and a single gene (HS:3 Cj1135) had a higher conservation rate among enteritis-control strains. Cj1135 is a one-domain glucosyltransferase involved in LOS biosynthesis[47], while Cj1421c and Cj1428c are involved in capsule biosynthesis although their exact functions are still undefined [48]. The potential involvement of these three genes in neuropathogenesis, if any, needs to be assessed. At the same time, the potential involvement of additional bacterially-encoded factors should not be discounted.
The microarray used to generate the bulk of the data in this study includes known GBS-associated factors and comprises greater gene diversity than that used in a previously published study of GBS strains [39]. However additional genes important to neuropathogenicity could be missing from our array. This study represents the largest systematic screen for potential neuropathogenic factors in C. jejuni but anything less than a comparative approach involving full-genome sequences is only partially complete.
One of the key findings of this study is the close genetic relationship between some neuropathogenic strains and their enteritis-associated counterparts. This has been suggested by results from various molecular typing studies, and we have been able to observe these similarities with an extremely high level of resolution. Since several scenarios could help explain the high degree of genetic similarity observed between strains with different clinical outcomes, it is important to note that highly related strains can show major differences in virulence-associated phenotypes [49]. The LOS locus also presents a unique challenge in that diversity in LOS structures can be obtained through genetic variation affecting the relevant genes [24]. Enteritis-associated strains that carry neuropathogenic genes could have their neuropathogenic potential altered or silenced through mutation, and this mechanism is likely to play a significant role in the incomplete penetrance of the putative GBS/MFS genotype.
Our data suggest that in many cases GBS/MFS-related strains might not differ in their neuropathogenic potential with respect to highly genetically related enteritis-control strains. In some cases, differences in clinical outcome are likely to be attributable to differences in host-background. GB13 and GB14, two epidemiologically related strains isolated from a family outbreak in which only one of three individuals afflicted with enteritis went on to develop neurological complications [31], serve as a reminder of how host factors are likely to play a role in the development of neuropathic clinical outcomes. At the same time we have recently shown that GB11, a GBS strain with a close genetic relationship to the genome strain NCTC 11168, appears to have acquired potential GBS factors in the form of an A-class LOS locus [36]. That study clearly underscores the valuable insight that could be gained by comparing closely related strains with differing clinical outcomes in the discovery of potential neuropathogenic factors. Future efforts should be aimed at comparative-genomic sequencing of strain pairs such as the ones described in this study in order to address whether differences in pathogen or host are responsible for differences in the clinical outcome of Campylobacteriosis cases.
Methods
Bacterial strains and Genomic DNA Isolation
One-hundred and two strains were analyzed by microarray CGH (Table 2). The "Rotterdam Dataset" is comprised of 41 strains representing GBS, MFS, and enteritis-control isolates from The Netherlands and collected between 1990 and 1999 [12, 14]. The "Curaçao Dataset" is comprised of 13 enteritis-control and GBS strains collected between 2000–2001 in the island of Curaçao (Netherlands Antilles) and is described in [50] and [51]. The "Dokkyo Dataset" is comprised of 48 strains collected from 1990–2003 in Japan and represents clinical isolates from enteritis-only, GBS, and MFS patients [52]. All bacterial cultures were initially grown on Karmali selective media (Oxoid) and subsequently on Mueller-Hinton agar plates (BACTO, Oakville, ON) for increased cell mass, (~24 hours at 37°C under microaerophilic conditions) prior to DNA isolation. Genomic DNA isolation was carried out as previously described [35].
C. jejuni NCTC 11168 Open Reading Frame DNA Microarray
Details of the microarray, including primer selection, the parameters for primer synthesis, selection of amplicons, as well as the purification and printing of DNA onto slides were previously described elsewhere [35]. A new version of the microarray became available partway through this study. This new version incorporates additional genes not present in the genome strain NCTC 11168. Additional information is available at [53].
Genomic DNA labelling
Genomic DNA was sheared into fragments ranging from 0.5 and 5 kilobases (mean size ~1.5 kilobases) using the method of Bodenteich et al. [54]. Briefly, genomic DNA was suspended in 35% glycerol and nebulized in an aerosol nebulizer (Medex, Carlsbad, CA, USA) for 45 seconds at 15 PSI. 5 μg of sheared DNA were fluorescently labelled using direct chemical coupling with the Label-IT (Mirus Corp., Madison, WI) cyanine dyes Cy3 and Cy5 as recommended by the manufacturer. Probes were purified from unincorporated dyes by sequentially passing samples through SigmaSpin (Sigma, Oakville, ON) and Qiaquick (Qiagen, Mississauga, ON) columns. Labelled DNA sample yields and dye incorporation efficiencies were calculated using the Nanodrop ND-1000 spectrophotometer (Nanodrop, Rockland, DE).
Microarray hybridizations
The hybridization profile for each strain was obtained by co-hybridizing labelled DNA from the tester strain and from the NCTC 11168 (control) strain to our microarray. Equivalent amounts (1 to 2 μg) of labelled tester and control samples with similar dye incorporation efficiencies were pooled, lyophilized, and hybridized to microarrays as previously described [35].
Microarray data acquisition and analysis
Microarrays were scanned using a Chipreader laser scanner (BioRad, Mississauga, ON) according to the manufacturer's recommendations. Spot quantification, visual inspection of potential outliers, and flagging of anomalous spots was performed using the program ArrayPro Analyzer (version 4.5; Media Cybernetics). The microarray data exported from ArrayPro was imported into the BioArray Software Environment (BASE version 1.2) [55] and is available at NCBI's Gene Expression Omnibus [56] under accession number GSE3579. Spots flagged due to poor spot morphology or low signal intensity (less than 5 × local background) were filtered out. After print-tip Loess normalization, data was used to calculate the average log ratio [log2(Signal Tester/Signal Control)] from the replicates for each gene represented on the microarray. The filtered data exported from BASE contains the average log ratio data for 1712 reporters. Log Ratio data was visualized and analyzed in TIGR's MultiExperiment Viewer (MEV version 3.0) [57]. Clustering of samples based on log ratio profile similarities was performed by the average linkage hierarchical clustering method of Eisen et al. [58], as implemented in TMEV, using Pearson correlation coefficient as a distance metric. The Support Tree method of bootstrapping implemented in TMEV was used to test the reliability of the clustering patterns (500 bootstrap re-samplings). The percentage of re-sampled trees supporting a given tree node are shown. To facilitate tree topology visualization, tree information was coded into Newick format and the trees were visualized using Treeview (version 1.6.6) [59].
Statistical testing of gene conservation rates
For statistical analysis of differential Log Ratio averages between groups of isolates we used the T-test implemented in TMEV, using a modified Bonferroni-corrected significance threshold of P < 0.05. For statistical analysis of differential gene conservation rates between groups of strains, gene conservation profiles were obtained from Log Ratio data by categorizing genes into "present", "divergent", and "absent" according to thresholds that were empirically determined previously [35, 37] To determine over- or under-representation of each gene among neuropathogenic isolates, the number of strains in which the gene was "present" and "absent"were calculated for representative groups of 32 neuropathogenic isolates and 32 enteritic isolates. P-values were then calculated for each gene on the microarray using the two-tailed Fisher's Exact test using an Microsoft Excel script developed in-house. Statistical significance of raw p-values was assessed using a threshold of P < 0.05; p-value adjustments were also performed to account for multiple testing using an in-house Microsoft Excel script that adapts the Westfall and Young permutation method to the gene conservation rate calculations described above [60]. Statistical results for the 169 genes displaying differences in conservation rates between neuropathogenic and enteritic groups are provided as Additional file 1.
Abbreviations
- GBS:
-
Guillain-Barré syndrome
- MFS:
-
Miller Fisher syndrome
- LOS:
-
Lipo-oligosaccharide
- CGH:
-
comparative-genomic hybridization.
References
Friedman CJ, Neimann J, Wegener HC, Tauxe RV: Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. Campylobacter. Edited by: Nachamkin I and Blaser M. 2000, Washington D.C., American Society for Microbiology, 121-138.
Nachamkin I, Allos BM, Ho T: Campylobacter species and Guillain-Barré syndrome. Clin Microbiol Rev. 1998, 11: 555-567.
van der Meché FG, van Doorn PA: Guillain-Barré syndrome and chronic inflammatory demyelinating polyneuropathy: immune mechanisms and update on current therapies. Ann Neurol. 1995, 37 Suppl 1: 14-31. 10.1002/ana.410370704.
Hughes RA, Rees JH: Clinical and epidemiologic features of Guillain-Barré syndrome. J Infect Dis. 1997, 176 Suppl 2: S92-S98.
Koga M, Gilbert M, Li J, Koike S, Takahashi M, Furukawa K, Hirata K, Yuki N: Antecedent infections in Fisher syndrome: a common pathogenesis of molecular mimicry. Neurology. 2005, 64: 1605-1611. 10.1212/01.WNL.0000160399.08456.7C.
Rees JH, Soudain SE, Gregson NA, Hughes RA: Campylobacter jejuni infection and Guillain-Barré syndrome. N Engl J Med. 1995, 333: 1374-9. 10.1056/NEJM199511233332102.
Jacobs BC, Rothbarth PH, van der Meche FG, Herbrink P, Schmitz PI, de Klerk MA, van Doorn PA: The spectrum of antecedent infections in Guillain-Barré syndrome: a case-control study. Neurology. 1998, 51: 1110-1115.
Kuroki S, Saida T, Nukina M, Haruta T, Yoshioka M, Kobayashi Y, Nakanishi H: Campylobacter jejuni strains from patients with Guillain-Barré syndrome belong mostly to Penner serogroup 19 and contain beta-N-acetylglucosamine residues. Ann Neurol. 1993, 33: 243-7. 10.1002/ana.410330304.
Lastovica AJ, Goddard EA, Argent AC: Guillain-Barré syndrome in South Africa associated with Campylobacter jejuni O:41 strains. J Infect Dis. 1997, 176 Suppl 2: 139-43.
Yuki N, Takahashi M, Tagawa Y, Kashiwase K, Tadokoro K, Saito K: Association of Campylobacter jejuni serotype with antiganglioside antibody in Guillain-Barré syndrome and Fisher's syndrome. Ann Neurol. 1997, 42: 28-33. 10.1002/ana.410420107.
Allos BM, Lippy FT, Carlsen A, Washburn RG, Blaser MJ: Campylobacter jejuni strains from patients with Guillain-Barré syndrome. Emerg Infect Dis. 1998, 4: 263-268.
Endtz HP, Ang CW, van den BN, Duim B, Rigter A, Price LJ, Woodward DL, Rodgers FG, Johnson WM, Wagenaar JA, Jacobs BC, Verbrugh HA, van BA: Molecular characterization of Campylobacter jejuni from patients with Guillain-Barré and Miller Fisher syndromes. J Clin Microbiol. 2000, 38: 2297-2301.
Engberg J, Nachamkin I, Fussing V, McKhann GM, Griffin JW, Piffaretti JC, Nielsen EM, Gerner-Smidt P: Absence of clonality of Campylobacter jejuni in serotypes other than HS:19 associated with Guillain-Barré syndrome and gastroenteritis. J Infect Dis. 2001, 184: 215-220. 10.1086/322010.
Dingle KE, van den BN, Colles FM, Price LJ, Woodward DL, Rodgers FG, Endtz HP, van BA, Maiden MC: Sequence typing confirms that Campylobacter jejuni strains associated with Guillain-Barré and Miller-Fisher syndromes are of diverse genetic lineage, serotype, and flagella type. J Clin Microbiol. 2001, 39: 3346-3349. 10.1128/JCM.39.9.3346-3349.2001.
Yuki N, Taki T, Inagaki F, Kasama T, Takahashi M, Saito K, Handa S, Miyatake T: A bacterium lipopolysaccharide that elicits Guillain-Barré syndrome has a GM1 ganglioside-like structure. J Exp Med. 1993, 178: 1771-1775. 10.1084/jem.178.5.1771.
Yuki N, Taki T, Takahashi M, Saito K, Yoshino H, Tai T, Handa S, Miyatake T: Molecular mimicry between GQ1b ganglioside and lipopolysaccharides of Campylobacter jejuni isolated from patients with Fisher's syndrome. Ann Neurol. 1994, 36: 791-3. 10.1002/ana.410360517.
Yuki N: Carbohydrate mimicry: a new paradigm of autoimmune diseases. Curr Opin Immunol. 2005, 17: 577-582. 10.1016/j.coi.2005.09.004.
Hafer-Macko C, Hsieh ST, Li CY, Ho TW, Sheikh K, Cornblath DR, McKhann GM, Asbury AK, Griffin JW: Acute motor axonal neuropathy: an antibody-mediated attack on axolemma. Ann Neurol. 1996, 40: 635-644. 10.1002/ana.410400414.
Ho TW, Willison HJ, Nachamkin I, Li CY, Veitch J, Ung H, Wang GR, Liu RC, Cornblath DR, Asbury AK, Griffin JW, McKhann GM: Anti-GD1a antibody is associated with axonal but not demyelinating forms of Guillain-Barré syndrome. Ann Neurol. 1999, 45: 168-173. 10.1002/1531-8249(199902)45:2<168::AID-ANA6>3.0.CO;2-6.
Ogawara K, Kuwabara S, Mori M, Hattori T, Koga M, Yuki N: Axonal Guillain-Barré syndrome: relation to anti-ganglioside antibodies and Campylobacter jejuni infection in Japan. Ann Neurol. 2000, 48: 624-631. 10.1002/1531-8249(200010)48:4<624::AID-ANA9>3.0.CO;2-O.
Jacobs BC, Endtz H, van der Meche FG, Hazenberg MP, Achtereekte HA, van Doorn PA: Serum anti-GQ1b IgG antibodies recognize surface epitopes on Campylobacter jejuni from patients with Miller Fisher syndrome. Ann Neurol. 1995, 37: 260-264. 10.1002/ana.410370218.
Ang CW, de Klerk MA, Endtz HP, Jacobs BC, Laman JD, van der Meche FG, van Doorn PA: Guillain-Barré syndrome- and Miller Fisher syndrome-associated Campylobacter jejuni lipopolysaccharides induce anti-GM1 and anti-GQ1b Antibodies in rabbits. Infect Immun. 2001, 69: 2462-9. 10.1128/IAI.69.4.2462-2469.2001.
Yuki N, Susuki K, Koga M, Nishimoto Y, Odaka M, Hirata K, Taguchi K, Miyatake T, Furukawa K, Kobata T, Yamada M: Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barré syndrome. Proc Natl Acad Sci U S A. 2004, 101: 11404-11409. 10.1073/pnas.0402391101.
Gilbert M, Karwaski MF, Bernatchez S, Young NM, Taboada E, Michniewicz J, Cunningham AM, Wakarchuk WW: The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J Biol Chem. 2002, 277: 327-337. 10.1074/jbc.M108452200.
Godschalk PC, Heikema AP, Gilbert M, Komagamine T, Ang CW, Glerum J, Brochu D, Li J, Yuki N, Jacobs BC, van BA, Endtz HP: The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barré syndrome. J Clin Invest. 2004, 114: 1659-1665. 10.1172/JCI200415707.
Koga M, Gilbert M, Takahashi M, Li J, Koike S, Hirata K, Yuki N: Comprehensive analysis of bacterial risk factors for the development of Guillain-Barré syndrome after Campylobacter jejuni enteritis. J Infect Dis. 2006, 193: 547-555. 10.1086/499969.
Gilbert M, Brisson JR, Karwaski MF, Michniewicz J, Cunningham AM, Wu Y, Young NM, Wakarchuk WW: Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-mhz 1H and 13C NMR analysis. J Biol Chem. 2000, 275: 3896-3906. 10.1074/jbc.275.6.3896.
van Belkum A, van den Braak N, Godschalk P, Ang W, Jacobs B, Gilbert M, Wakarchuk W, Verbrugh H, Endtz H: A Campylobacter jejuni gene associated with immune-mediated neuropathy. Nat Med. 2001, 7: 752-753. 10.1038/89831.
Parker CT, Horn ST, Gilbert M, Miller WG, Woodward DL, Mandrell RE: Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J Clin Microbiol. 2005, 43: 2771-2781. 10.1128/JCM.43.6.2771-2781.2005.
Koga M, Takahashi M, Masuda M, Hirata K, Yuki N: Campylobacter gene polymorphism as a determinant of clinical features of Guillain-Barré syndrome. Neurology. 2005, 65: 1376-1381. 10.1212/01.wnl.0000176914.70893.14.
Ang CW, van Doorn PA, Endtz HP, Merkies IS, Jacobs BC, de Klerk MA, van Koningsveld R, van der Meche FG: A case of Guillain-Barré syndrome following a family outbreak of Campylobacter jejuni enteritis. J Neuroimmunol. 2000, 111: 229-33. 10.1016/S0165-5728(00)00369-6.
Jacobs BC, van Doorn PA, Schmitz PI, Tio-Gillen AP, Herbrink P, Visser LH, Hooijkass H, van der Meche FG: Campylobacter jejuni infections and anti-GM1 antibodies in Guillain-Barré syndrome. Ann Neurol. 1996, 40: 181-187. 10.1002/ana.410400209.
Aspinall GO, Fujimoto S, McDonald AG, Pang H, Kurjanczyk LA, Penner JL: Lipopolysaccharides from Campylobacter jejuni associated with Guillain-Barré syndrome patients mimic human gangliosides in structure. Infect Immun. 1994, 62: 2122-2125.
Duim B, Ang CW, van BA, Rigter A, van Leeuwen NW, Endtz HP, Wagenaar JA: Amplified fragment length polymorphism analysis of Campylobacter jejuni strains isolated from chickens and from patients with gastroenteritis or Guillain-Barré or Miller Fisher syndrome. Appl Environ Microbiol. 2000, 66: 3917-3923. 10.1128/AEM.66.9.3917-3923.2000.
Taboada EN, Acedillo RR, Carrillo CD, Findlay WA, Medeiros DT, Mykytczuk OL, Roberts MJ, Valencia CA, Farber JM, Nash JH: Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J Clin Microbiol. 2004, 42: 4566-4576. 10.1128/JCM.42.10.4566-4576.2004.
Gilbert M, Godschalk PC, Karwaski MF, Ang CW, van BA, Li J, Wakarchuk WW, Endtz HP: Evidence for acquisition of the lipooligosaccharide biosynthesis locus in Campylobacter jejuni GB11, a strain isolated from a patient with Guillain-Barré syndrome, by horizontal exchange. Infect Immun. 2004, 72: 1162-1165. 10.1128/IAI.72.2.1162-1165.2004.
Taboada EN, Acedillo RR, Luebbert CC, Findlay WA, Nash JH: A new approach for the analysis of bacterial microarray-based Comparative Genomic Hybridization: insights from an empirical study. BMC Genomics. 2005, 6: 78-10.1186/1471-2164-6-78.
Fujimoto S, Allos BM, Misawa N, Patton CM, Blaser MJ: Restriction fragment length polymorphism analysis and random amplified polymorphic DNA analysis of Campylobacter jejuni strains isolated from patients with Guillain-Barré syndrome. J Infect Dis. 1997, 176: 1105-1108.
Leonard EE, Tompkins LS, Falkow S, Nachamkin I: Comparison of Campylobacter jejuni isolates implicated in Guillain-Barré syndrome and strains that cause enteritis by a DNA microarray. Infect Immun. 2004, 72: 1199-203. 10.1128/IAI.72.2.1199-1203.2004.
Champion OL, Gaunt MW, Gundogdu O, Elmi A, Witney AA, Hinds J, Dorrell N, Wren BW: Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc Natl Acad Sci U S A. 2005, 102: 16043-16048. 10.1073/pnas.0503252102.
Nachamkin I, Engberg J, Gutacker M, Meinersman RJ, Li CY, Arzate P, Teeple E, Fussing V, Ho TW, Asbury AK, Griffin JW, McKhann GM, Piffaretti JC: Molecular population genetic analysis of Campylobacter jejuni HS:19 associated with Guillain-Barré syndrome and gastroenteritis. J Infect Dis. 2001, 184: 221-226. 10.1086/322008.
Siemer BL, Harrington CS, Nielsen EM, Borck B, Nielsen NL, Engberg J, On SL: Genetic relatedness among Campylobacter jejuni serotyped isolates of diverse origin as determined by numerical analysis of amplified fragment length polymorphism (AFLP) profiles. J Appl Microbiol. 2004, 96: 795-802. 10.1111/j.1365-2672.2004.02205.x.
Nachamkin I, Liu J, Li M, Ung H, Moran AP, Prendergast MM, Sheikh K: Campylobacter jejuni from patients with Guillain-Barré syndrome preferentially expresses a GD(1a)-like epitope. Infect Immun. 2002, 70: 5299-5303. 10.1128/IAI.70.9.5299-5303.2002.
Godschalk P, Bergman M, Gorkink R, Simons G, van den Braak N, Lastovica A, Endtz H, Verbrugh H, van Belkum A: Identification of DNA sequence variation in Campylobacter jejuni strains associated with the Guillain-Barre syndrome by high-throughput AFLP analysis. BMC Microbiology. 2006, 6: 32-10.1186/1471-2180-6-32.
Phongsisay V, Perera VN, Fry BN: Exchange of lipooligosaccharide synthesis genes creates potential Guillain-Barre syndrome-inducible strains of Campylobacter jejuni. Infect Immun. 2006, 74: 1368-1372. 10.1128/IAI.74.2.1368-1372.2006.
Houliston RS, Endtz HP, Yuki N, Li J, Jarrell HC, Koga M, van BA, Karwaski MF, Wakarchuk WW, Gilbert M: Identification of a sialate O-acetyltransferase from Campylobacter jejuni: demonstration of direct transfer to the C-9 position of terminal alpha-2, 8-linked sialic acid. J Biol Chem. 2006, 281: 11480-11486. 10.1074/jbc.M512183200.
Gilbert M, Godschalk PCR, Parker CT, Endtz HP, Wakarchuk WW: Genetic bases for the variation in the lipooligosaccharide outer core of Campylobacter jejuni and possible association of glycosyltransferase genes with post-infectious neuropathies. Campylobacter: Molecular and Cellular Biology. Edited by: J.M.Ketley and M.E.Konkel . 2005, Norwich, UK, Horizon Bioscience, 219-248.
Karlyshev AV, Champion OL, Churcher C, Brisson JR, Jarrell HC, Gilbert M, Brochu D, St MF, Li J, Wakarchuk WW, Goodhead I, Sanders M, Stevens K, White B, Parkhill J, Wren BW, Szymanski CM: Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol Microbiol. 2005, 55: 90-103. 10.1111/j.1365-2958.2004.04374.x.
Carrillo CD, Taboada E, Nash JHE, Lanthier P, Kelly J, Lau PC, Verhulp R, Mykytczuk O, Sy J, Findlay WA, Amoako K, Gomis S, Willson P, Austin JW, Potter A, Babiuk L, Allan B, Szymanski CM: Genome-wide Expression Analyses of Campylobacter jejuni NCTC11168 Reveals Coordinate Regulation of Motility and Virulence by flhA. J Biol Chem. 2004, 279: 20327-20338. 10.1074/jbc.M401134200.
Duim B, Godschalk PC, van den BN, Dingle KE, Dijkstra JR, Leyde E, van der PJ, Colles FM, Endtz HP, Wagenaar JA, Maiden MC, van BA: Molecular evidence for dissemination of unique Campylobacter jejuni clones in Curacao, Netherlands Antilles. J Clin Microbiol. 2003, 41: 5593-5597. 10.1128/JCM.41.12.5593-5597.2003.
Endtz HP, van West H, Godschalk PC, de HL, Halabi Y, van den BN, Kesztyus BI, Leyde E, Ott A, Verkooyen R, Price LJ, Woodward DL, Rodgers FG, Ang CW, van KR, van BA, Gerstenbluth I: Risk factors associated with Campylobacter jejuni infections in Curacao, Netherlands Antilles. J Clin Microbiol. 2003, 41: 5588-5592. 10.1128/JCM.41.12.5588-5592.2003.
Takahashi M, Koga M, Yokoyama K, Yuki N: Epidemiology of Campylobacter jejuni isolated from patients with Guillain-Barré and Fisher syndromes in Japan. J Clin Microbiol. 2005, 43: 335-9. 10.1128/JCM.43.1.335-339.2005.
Campychip Description. [http://ibs-isb.nrc-cnrc.gc.ca/glycobiology/campychips_e.html]
Bodenteich AS, Chissoe Y, Wang F, Roe BA: Shotgun cloning as the strategy of choice to generate templates for high throughput dideoxynucleotide sequencing. Automated DNA sequencing and analysis techniques. Edited by: Adams MD, Fields C and Venter C. 1994, London, UK: Academic Press, 42-50.
Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C: BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 2002, 3: SOFTWARE0003-10.1186/gb-2002-3-8-software0003.
National Center for Biotechnology Information's Gene Expression Omnibus. 2006, [http://www.ncbi.nlm.nih.gov/projects/geo/]
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J: TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003, 34: 374-8.
Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998, 95: 14863-14868. 10.1073/pnas.95.25.14863.
Page RD: TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996, 12: 357-8.
Westfall PH, Young SS: Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment. 1993, New York, John Wiley and Sons
Acknowledgements
Funding for this work has been provided through the National Research Council's Genomics and Health Initiative, Phase II and The Human Frontier Science Program (RGP 38/2003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declares that there are no competing interests.
Authors' contributions
ENT designed M-CGH experiments, carried out downstream data analysis, and drafted the manuscript. RRA performed hybridizations, performed preliminary data analysis and assisted with downstream data analysis. MK characterized the Japanese strains. PCRG characterized the Dutch and Curaçao strains. JHEN, MG, and AvB conceived of the study, and participated in its design and coordination and helped to draft the manuscript. HPE, and NY participated in the conception and supervised the design of the study. All authors submitted comments on drafts and read and approved the final manuscript.
Electronic supplementary material
12864_2006_1072_MOESM1_ESM.xls
Additional file 1: Statistical assessment of genes displaying differential conservation rates among enteritic and neuropathogenic isolates. The table represents the statistical assessment of the conservation rates among the genes that have been observed to be differentially conserved enteritic versus neuropathogenic isolates. (XLS 55 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Taboada, E.N., van Belkum, A., Yuki, N. et al. Comparative genomic analysis of Campylobacter jejuni associated with Guillain-Barré and Miller Fisher syndromes: neuropathogenic and enteritis-associated isolates can share high levels of genomic similarity. BMC Genomics 8, 359 (2007). https://doi.org/10.1186/1471-2164-8-359
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-8-359