Abstract
Background
Genes specifically expressed in the oocyte play key roles in oogenesis, ovarian folliculogenesis, fertilization and/or early embryonic development. In an attempt to identify novel oocyte-specific genes in the mouse, we have used an in silico subtraction methodology, and we have focused our attention on genes that are organized in genomic clusters.
Results
In the present work, five clusters have been studied: a cluster of thirteen genes characterized by an F-box domain localized on chromosome 9, a cluster of six genes related to T-cell leukaemia/lymphoma protein 1 (Tcl1) on chromosome 12, a cluster composed of a SPErm-associated glutamate (E)-Rich (Speer) protein expressed in the oocyte in the vicinity of four unknown genes specifically expressed in the testis on chromosome 14, a cluster composed of the oocyte secreted protein-1 (Oosp-1) gene and two Oosp-related genes on chromosome 19, all three being characterized by a partial N-terminal zona pellucida-like domain, and another small cluster of two genes on chromosome 19 as well, composed of a TWIK-Related spinal cord K+ channel encoding-gene, and an unknown gene predicted in silico to be testis-specific. The specificity of expression was confirmed by RT-PCR and in situ hybridization for eight and five of them, respectively. Finally, we showed by comparing all of the isolated and clustered oocyte-specific genes identified so far in the mouse genome, that the oocyte-specific clusters are significantly closer to telomeres than isolated oocyte-specific genes are.
Conclusion
We have studied five clusters of genes specifically expressed in female, some of them being also expressed in male germ-cells. Moreover, contrarily to non-clustered oocyte-specific genes, those that are organized in clusters tend to map near chromosome ends, suggesting that this specific near-telomere position of oocyte-clusters in rodents could constitute an evolutionary advantage. Understanding the biological benefits of such an organization as well as the mechanisms leading to a specific oocyte expression in these clusters now requires further investigation.
Similar content being viewed by others
Background
Mammalian oocyte is the only known cell able to activate the zygotic genome after fertilization. Its cytoplasm is also able to reprogram the nucleus of a differentiated cell in cloning experiments. Therefore, it is likely that several genes specifically expressed in the oocytes are responsible for this ability to program diploid genomes. It is the case for the so-called maternal genes such as Maternal Antigen That Embryo Required (MATER), Zygotic Arrest 1 (Zar1), Stella and nucleoplasmin 2 (Npm2) that are all required for normal embryonic development beyond the 1- or 2-cell stage [1, 2]. Moreover, new observations have shown that loss of function mutations in the two oocyte-specific genes GDF-9 and BMP-15 are responsible for severe alterations of ovarian folliculogenesis, these alterations being different depending on the hetero- or homozygous state of the individuals in the ovine species [3, 4]. So genes specifically expressed in the oocyte seem to play key roles in oogenesis, ovarian folliculogenesis, fertilization and/or early embryonic development. In an attempt to identify novel oocyte-specific genes, several groups have used both mRNA differential display [5] and in silico subtraction approaches [6]. By using the latter, we have recently identified six genes of the oogenesin family [7], these genes being present on chromosome 4 in a cluster of almost 1 Mb composed of twelve oogenesin paralogous genes. We have also identified six genes presenting similarities with NALP5/MATER [8] which are specifically expressed in the mouse oocyte, three being clusterized on chromosome 7. We have experimentally validated the specificity of expression for three oogenesin and five NALP genes [7, 8]. The fact that these two groups of genes are localized in two clusters indicates that they likely originate from duplications of an ancestral gene.
In the present work, we have used the same in silico approach to identify new oocyte-specific genes that are also localized in clusters in the mouse genome. In particular, we have identified three new loci in the mouse genome containing several genes specifically expressed in the oocyte, and two loci containing genes specifically expressed in male and female germ cells, which may thus be called "germ cell loci". We have experimentally verified the specificity of expression for eight of them by RT-PCR and five of them by in situ hybridization. Moreover, we have compared the map position of all the "oocyte-clusters" identified so far with that of the other "isolated" oocyte-specific genes. We could show that oocyte-specific clustered genes are significantly closer to telomeres than oocyte-specific isolated genes. Our results emphasize a new vision of gene regulation, where large-scale genome organization likely plays an important function.
Results
In silico identification of oocyte-specific genes
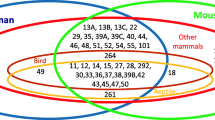
By using the Digital Differential Display (DDD) software, and as previously described [7, 8], we found in silico a list of approximately 60 genes for which ESTs were exclusively detected in mouse egg libraries. After localization on the mouse genome [9], we noticed that several of these genes were neighbored by genes for which ESTs were exclusively present in egg and/or testis libraries as well. Finally, the number of oocyte-specific genes approached one hundred. Amongst these genes were found most of the well-known oocyte specific genes such as GDF-9, Mater or Zar1, validating the methodology, as well as the 12 oogenesins and the 6 NALP/Mater-like genes [7, 8]. In the present work, we have focused our attention towards five other clusters of genes (Fig. 1).
Localization of germ-cell-specific cluster in mouse chromosome 9, 12, 14 and 19 [9]. The presence of mRNAs or ESTs of these different genes in male and female germ cells was assessed by experimental (RT-PCR, Southern blot and in situ hybridization) and/or in silico methodologies as described in Methods. O stands for Ovary/Egg; T stands for Testis.
The first cluster is localized on chromosome 9 and is composed of thirteen genes corresponding to the GenBank accession numbers AK054339, AK087710, AK087709, AK087808, XM_284540, XM_486447, XM_486263, XM_488195, XM_356253, AK054298, AK087669, XM_356191 and AK007274. All of these genes encode proteins that contain an N-terminal F-box domain as well as WD40 repeats in the C-terminal region. As the gene corresponding to the GenBank accession number AK087709 is named "F-box only protein 12" (FBXO12), we have renamed the twelve other genes FBXO12A, FBXO12B, FBXO12C, FBXO12D, FBXO12E, FBXO12F, FBXO12G, FBXO12H, FBXO12I, FBXO12J, FBXO12K and FBXO12L (Table 1 & Fig. 1).
The second cluster contains the so-called Tcl1 (T-cell leukemia/lymphoma protein 1) and Tcl1b genes, and maps on chromosome 12. This cluster contains six genes related to Tcl1 gene and is predicted to be oocyte-specific in silico (Fig. 1 & Table 1). The protein encoded by this gene shares similarities with the protein encoded by the gene MTCP1, also involved in chromosomal translocations in T-cell proliferative disease. Its structure consists of an eight-stranded β-barrel with a particular topology, with the N- and C-terminal halfs being linked by a pseudo dyad [10].
We have identified another cluster on chromosome 14, which was composed of five genes. One of them encoded for a protein that belongs to the Speer (SPErm-associated glutamate (E)-Rich protein) family, characterized by a very high proportion of alpha-helical secondary structure. We have thus renamed this gene SpeerA (Genbank identification number XM_138939). The corresponding Speer proteins are weakly similar to proteins that contain interacting domain with cytoskeletal as well as nuclear matrix proteins such as actin and dynein. In silico, ESTs of SpeerA are present in egg libraries but also in round spermatid libraries (Table 1). The ESTs corresponding to the four nearest neighboring genes, corresponding to the GenBank identification numbers AK076711, AK005833, AK016429 and AK019699, have been exclusively detected in silico in testis libraries (Fig. 1 & Table 1). The gene corresponding to the AK016429 accession number encoded for a protein that presented no clear similarity with any known protein. The three other genes likely encoded for untranslated RNAs, as no long ORF could be detected in their sequence.
Two other clusters map to chromosome 19. The first one is composed of three genes (Genbank identification numbers XM_129237, AF420487, and XM_129191). The corresponding three protein sequences share about 47% of similarity and can be aligned with the 150 first amino acids of domains of the Zona-Pellucida family (Fig. 2). Secondary structure predictions indicate mainly a β-fold for this domain, in which the conserved Cys78 and Cys98 might form a disulfide bond, according to experimental data obtained on the corresponding Cys61 and Cys81 of ZP3 [11]. The C-terminal ends of Oosp proteins (not represented) are highly variable and cannot be aligned with the much more large C-terminal part of ZP domains. The gene corresponding to the GenBank accession number AF420487 is the well-known "oocyte secreted protein 1" (Oosp1) [12]. So we have renamed the two other genes Oosp2 and Oosp3 (Table 1 & Fig 1).
Alignment of Oosp protein sequence with the N-terminal part of the ZP domain (150 amino acids) of ZP3. Identities and similarities are shown with black boxes and black frames, respectively. An hypothetical disulfide bond is suspected between Cys78 and Cys98 according to experimental data obtained on the corresponding Cys61 and Cys81 of ZP3 [11].
The second cluster on chromosome 19 is composed of an oocyte specific gene, corresponding to the XM_285304 GenBank identification number (Fig. 1), the protein of which is similar to a TWIK-Related spinal cord K+ (Trik) channel. The other gene, corresponding to the AK015359 number, was predicted to be testis-specific in silico (Table 1), and encoded for an unknown protein (Fig. 1).
Expression analysis
By analysis of RT-PCR products on BET-stained gels, FBXO12B, FBXO12D, Tcl1, Tcl1b3, Tcl1b4, SpeerA and Oosp3 appeared to be exclusively expressed in the ovary, whereas the gene corresponding to the AK076711 GenBank identification number appeared to be exclusively expressed in the testis (data not shown). By contrast, actin was readily amplified in all tissues. To increase the sensitivity of detection of these transcripts, Southern blot experiments were performed on RT-PCR products, confirming these results, except for Tcl1 and SpeerA genes for which a slight expression was also detected in the testis (Fig. 3).
In situ hybridization confirmed that within the ovary, SpeerA, FBXO12B, FBXO12D, Tcl1 and Oosp3 were exclusively expressed in oocytes (Fig. 4). Transcripts were detected in primary follicles, as well as in early and large antral follicles (Fig. 4), except for FBXO12D and Tcl1, for which transcripts were only slightly if at all detected in large antral follicles (Fig. 4).
Metagenome analysis
Altogether, the chromosome localization of 92 oocyte-specific genes was known, amongst the ~100 in silico-predicted oocyte-specific genes recovered. Amongst these 92 genes, 48 were organized in 7 "oocyte-clusters" (Table 2). We did not consider the two "germ-cell clusters" on chromosomes 14 and 19 in this analysis, but we included the oogenesin and Nalp9 clusters previously described by our group [7, 8], as well as the Obox cluster described by Rajkovic et al. (2002). The other 44 oocyte-genes were considered as "isolated" in the mouse genome. For these latter genes, the average distance expressed in relative distance from the chromosome end ranged from 2.5% to 49.8% (average = 24 ± 16%). By contrast, the 48 oocyte-specific genes organized in clusters mapped between 5.6% and 20.6% from the closest chromosome end (average = 12.6 ± 5.3%). A Student t test was performed to compare the two categories, considering only one position for each cluster, and resulted in a value of 0.035. Similar analysis was performed taking into account the number of genes before and after the locus/cluster under scrutiny. This analysis gave rise to similar results. Therefore, oocyte-specific genes that are organized in clusters map significantly nearest chromosome ends than those that are "isolated".
Discussion
The present work and two previous identifications of the oogenesin genes [7] and of the mater family genes [8] demonstrate that the in silico subtraction is an efficient and reliable methodology to identify new oocyte-specific genes. By localizing these genes on the mouse genome, we have discovered other "germ-cell" specific genes, organized in clusters. RT-PCR as well as in situ hybridization experiments confirmed their specificity of expression in the ovary and/or in the testis. As for GDF-9, Mater, Zar1 or Stella, these genes may play important roles in folliculogenesis, fertilization and/or early embryonic development.
Amongst the "oocyte-specific clusters" presented here, the Tcl1/1B has previously been described. Indeed, in a previous work on human TCL1 and TCL1B genes, the six murine Tcl1 and Tcl1b1-5 orthologous genes were already shown to be expressed in oocytes and two-cell embryos but not in other adult tissues except in lymphoid cell lines, where overexpression of Tcl1 in these cells leads to leukemia or lymphoma [13]. Tcl1 was also shown to be expressed in testis, as previously described [14, 15], its overexpression inducing the formation of testicular seminomas [16]. Interestingly, Tcl1-/- female mice are sterile, as early embryos are unable to undergo normal cell cleavage up to 8-cell stage, the majority of embryos being even unable to proceed beyond the 4- to 8-cell stage [16]. Therefore, like Mater and Zar1, Tcl1 can be considered as a maternal effect gene as well. It is possible that part of the effects of this gene family is to promote cell survival and/or proliferation by activating the Akt kinase as previously observed [17, 18].
The F-box proteins, encoded by the thirteen genes localized on chromosome 9 cluster, might specifically interact with other proteins in the oocyte. Indeed, the F-box motif is present in numerous proteins known to serve as a link between a target protein and an ubiquitin-conjugating enzyme. In particular, it has been shown that the Early Mitotic Inhibitor (Emi)-1, which contains an F-box domain, is able to regulate mitosis progress by inhibiting premature anaphase promoting complex/cyclosome (APC) activation [19], the APC complex being composed of an ubiquitin ligase coupled with the SCF (Skp1-Cullin-F-box protein) complex [20]. So it is possible that the oocyte-specific F-Box proteins would participate in the stabilization of the meiotic process at the prophase I stage.
Concerning the cluster on chromosome 14, in situ hybridization showed a high level of expression of SpeerA gene in the mouse oocyte, corroborating previous results from Matzuk's group [6]. By analyzing the RT-PCR results, we found that this gene was also expressed in the testis, likely in male germ cells. The Speer family has recently been described as a new family of 14 genes. Interestingly, SPEER-1, SPEER-2, and SPEER-4D were recently shown to be expressed specifically in the mouse testis [21]. These genes have open reading frames of approximately 200-220 amino acids, and encode for proteins that exhibit a high proportion of alpha-helical secondary structure, comprising approximately 15% glutamate residues. According to their possible interaction with cytoskeletal proteins such as actin or dynein, it has been hypothesized that the SPEER may be nuclear matrix proteins involved in the reorganization of the spermatocyte/spermatid nucleus [21]. SpeerA, discovered in our study (XM_138939), shares about 34% amino acid sequence similarity with SPEER-4A gene, which could suggest that it plays a similar role in the oocyte. Of note, in contrast to most of the oocyte clusters, the four testis-specific genes neighboring the SpeerA gene were not structurally related to each other.
One of the clusters on chromosome 19 contains the well-known oocyte-specific gene Oosp-1 [12], and two structurally similar genes, these three genes clearly sharing a ZP-like domain. This suggests that these genes encode for structural proteins involved in the formation of certain components of the zona pellucida. A structural analysis of the sequences of these proteins is underway in our laboratory to further investigate the structural specificity of these proteins. The biological roles of the two genes present in the other cluster on chromosome 19, as well as the role of the testis-specific genes present on the chromosome 14 cluster are unknown.
Overall, for all of these genes, further experiments as well as structural analysis are now necessary to investigate their biological roles in oocyte and follicle maturation. Investigating such a role will require functional studies such as identification of partners and/or targeted invalidation.
As we previously discussed, the targeted invalidation of MATER gene leads to female infertility due to a blockage of early embryonic development [22]. This suggests that the oocyte-specific MATER-like genes, i.e. the Nalp9A-F genes that we have identified [8] are not able to compensate the absence of the MATER/Nalp5 product in Mater-/- mice. Similarly, invalidation of Tcl1 gene in the mouse was not clearly compensated by the five oocyte-specific Tcl1B genes. Overall, this suggests that despite an apparent redundancy in the expression of these structurally similar genes, every member of the oocyte Nalp and Tcl1 family of paralogous genes seem to have very specific biological roles in female reproduction. Invalidation of each of these genes is now necessary to study their specific biological role. Moreover, as expected, most oocyte-specific genes organized in clusters are paralogous genes, originating presumably from a common ancestral gene by successive duplication events. It is the case for the oogenesin, Nalp9, Fbxo12, Tcl1 as well as the Osp1-3 genes. It is clearly not the case for the clusters localized on 14A3 and 19D3 (Fig. 1). Nevertheless, such an organization of "oocyte- or germ-cell clusters" suggests the presence of specific genomic regulatory regions in the vicinity of these genes.
As we depicted in figure 1, the SpeerA gene is localized on a cluster on chromosome 14, in the vicinity of four unknown testis-specific genes. On chromosome 19, the oocyte-specific gene related to TWIK-Related spinal cord K+ channel is localized near the gene corresponding to AK015359 GenBank number, which is predicted in silico to be testis-specific as well. Interestingly, we have previously shown that in the oogenesin cluster on chromosome 4, the oogenesin-4 gene is also slightly expressed in the testis. Of note, Pramel1 gene, which is localized near oogenesin4, has also been described as a male germ-cell specific genes [7, 23]. Similarly, in the oocyte-specific Nalp cluster on chromosome 7, we have shown that Nalp9D is also clearly expressed in the testis, presumably in germ cells. So the notion of oocyte-specific cluster would not be systematically restricted to female germ cells, and would be extended in some cases to "germ-cells cluster" as suggested in this work.
One striking observation of our work is the difference in chromosome localization between oocyte-specific genes organized in clusters and oocyte-genes that are not. If genes were located at random on the chromosome and that the distance to the closest end is measured, then the expected range would be 0% to 50%, and one would expect to reach an average distance of 25% if enough genes are taken into consideration. In our study, isolated genes were very close to this value (24%) whereas clusters were located in average at half of this distance (12%). Gene clusters presumably originate from local gene duplications [24], which are now recognized as efficient motors of diversification in metazoans. Of note, the location of gene families near telomeres has a deep sense in terms of rapid evolution. Indeed, subtelomeric sequences are recognized as the most rapidly evolving regions in the genome, maybe due to the high level of recombination present in these regions [25]. On the other hand, clusters of genes are the entry gates for accumulation of mutations, without the risk of noxious consequences for the organism, and therefore enable the development of new functions, as clearly illustrated by the variants of the human hemoglobin beta chain, which adapts the foetus to successive oxygen environments during development [26, 27]. For mammals with a high reproductive potential, such as mice and other rodents, the possibility of rapid evolution for genes involved in ovarian function may be a great advantage. Therefore, one can hypothesize that the specific near telomere position of oocyte-specific gene clusters corresponds to an evolutionary advantage.
Interestingly, the genes that we have described here are strictly silenced in non-ovarian tissues, and one may hypothesize that the specific near telomere position of oocyte clusters would contribute towards such a silencing. In particular, it has been shown that telomeric and pericentric regions of chromosomes are mainly composed of heterochromatin in most eukaryotic genomes. Moreover, euchromatic genes relocated by chromosomal rearrangement or by transposition in the neighbourhood of heterochromatin can be silenced in drosophila [28]. In yeast, a similar phenomenon of reversible gene silencing near telomere has been called Telomere Position Effect (TPE) [29, 30]. The presence of TPE has also been observed in human cells [31]. Actually, a very specific nuclear topology would explain such a phenomenon. Indeed in yeast, the 32 telomeres cluster at the nuclear periphery in 8 to 10 groups, organized in compartments rich in histone-binding silencing factors [32]. Testing if the telomeres would be organized in very specific compartments in oocyte and in other tissues requires further investigations.
Conclusion
We have studied five clusters of genes specifically expressed in female, some of them being also expressed in male germ-cells. Moreover, contrarily to non-clustered oocyte-specific genes, those that are organized in clusters tend to map near chromosome ends, suggesting that this specific near-telomere position of oocyte-clusters could result in an evolutionary advantage. Understanding the biological roles as well as the mechanisms underlying such a specific expression now requires further investigation.
Methods
In silico identification of oocyte-specific genes
As previously described [7, 8] three cDNA libraries derived from mouse unfertilized eggs (dbEST library ID.10029), 2-cell egg (dbEST library ID.5391) and in vitro fertilized eggs (dbEST library ID.2589) were submitted to digital differential display analysis [33] to identify oocyte-specific ESTs that are not found in several non normalized cDNA libraries from different adult somatic non-tumoral tissues (brain, kidney, stomach, liver, lung, spleen, muscle, heart, skin, bone marrow, adipose tissue, adrenal gland). The physical localization of identified genes on the mouse chromosomes was retrieved from http://www.ncbi.nlm.nih.gov/mapview[9]. The presence of specific domains within the corresponding proteins was determined on http://smart.embl-heidelberg.de/[34] as well by a Blast search of homologous proteins [35]
RT-PCR analyses
Total RNA was extracted from whole adult tissues (ovary, liver, kidney, testis, heart, brain, muscle, adrenal gland and spleen) using RNAble reagent according to the manufacturer's procedure (Eurobio, Les Ulis, France). Reverse transcription was performed for 1 hour at 42°C in a total volume of 20 μl with 2 μg total RNA per sample following standard procedure. Five μl of the cDNA product was amplified by PCR using primers described in Table 3. RT-PCR products were also analyzed by Southern blot. Briefly, the RT-PCR products were fractionated on 1% agarose gel, transferred to Hybond-N+ membrane (Amersham-Pharmacia) and hybridized with the corresponding cDNA fragment labeled by random priming (1 × 106 cpm/ml) as described previously [36].
PCR products were purified from the agarose using the gel extraction kit QIAEX II (Qiagen, Hilden, Germany) and inserted into pGEM-T vector (Promega, Madison, WI, USA). Identity of the selected clones was checked by sequencing.
In situ hybridization
Frozen ovaries from 3 female mice in di-estrus, 3 female mice in proestrus and 3 female mice in estrus were serially sectioned (10 μm) with a cryostat to perform in situ hybridization experiments using 35S-labeled cRNAs probes as previously described [37]. The specificity of the hybridization signals was assessed by comparing the hybridization of the cRNA antisense with the corresponding sense probes. Histological determination of follicular size and degree of atresia was performed on adjacent sections stained with Feulgen [37].
Metagenome analysis
For the metagenome analysis, we have taken account for all the oocyte-specific genes identified so far by the in silico methodology (data not shown), including well known genes (Mater, Zar1), as well as the 12 genes of the oogenesin family [7], and the 6 genes similar to NALP5/MATER [8] that we have previously identified. A total of 92 genes were analyzed, 44 of which are isolated on the chromosomes while 48 were organized in 7 clusters (Table 2). To study the relative distribution of these two subgroups of genes, their relative distance to the closest chromosome end was calculated, and a global Student t-test was calculated to compare the two subgroups. To avoid any bias due to the fact that genes in clusters often comes from duplication of ancestral genes, bias which would artificially increase their coefficient in such a test, we have considered only one position for each cluster. Moreover, although genes are not strictly homogenously distributed along the chromosomes, the clusterized and the isolated oocytes genes considered for the analysis do cover all the mouse chromosomes, thereby limiting the risk of a bias.
References
Dean J: Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J Reprod Immunol. 2002, 53: 171-180. 10.1016/S0165-0378(01)00100-0.
Varani S, Matzuk MM: Phenotypic effects of knockout of oocyte-specific genes. Ernst Schering Res Found Workshop. 2002, 63-79.
Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM: Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996, 383: 531-535. 10.1038/383531a0.
Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, Beattie AE, Davis GH, Ritvos O: Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet. 2000, 25: 279-283. 10.1038/77033.
Zeng F, Schultz RM: Gene expression in mouse oocytes and preimplantation embryos: use of suppression subtractive hybridization to identify oocyte- and embryo-specific genes. Biol Reprod. 2003, 68: 31-39. 10.1095/biolreprod.102.007674.
Rajkovic A, Yan MSC, Klysik M, Matzuk M: Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil Steril. 2001, 76: 550-554. 10.1016/S0015-0282(01)01966-5.
Dade S, Callebaut I, Mermillod P, Monget P: Identification of a new expanding family of genes characterized by atypical LRR domains. Localization of a cluster preferentially expressed in oocyte. FEBS Lett. 2003, 555: 533-538. 10.1016/S0014-5793(03)01341-3.
Dade S, Callebaut I, Paillisson A, Bontoux M, Dalbies-Tran R, Monget P: In silico identification and structural features of six new genes similar to MATER specifically expressed in the oocyte. Biochem Biophys Res Commun. 2004, 324: 547-553. 10.1016/j.bbrc.2004.09.086.
Mus musculus genome view. [http://www.ncbi.nlm.nih.gov/mapview]
Hoh F, Yang YS, Guignard L, Padilla A, Stern MH, Lhoste JM, van Tilbeurgh H: Crystal structure of p14TCL1, an oncogene product involved in T-cell prolymphocytic leukemia, reveals a novel beta-barrel topology. Structure. 1998, 6: 147-155. 10.1016/S0969-2126(98)00017-3.
Boja ES, Hoodbhoy T, Fales HM, Dean J: Structural characterization of native mouse zona pellucida proteins using mass spectrometry. J Biol Chem. 2003, 278: 34189-34202. 10.1074/jbc.M304026200.
Yan C, Pendola FL, Jacob R, Lau AL, Eppig JJ, Matzuk MM: Oosp1 encodes a novel mouse oocyte-secreted protein. Genesis. 2001, 31: 105-110. 10.1002/gene.10010.
Virgilio L, Narducci MG, Isobe M, Billips LG, Cooper MD, Croce CM, Russo G: Identification of the TCL1 gene involved in T-cell malignancies. Proc Natl Acad Sci U S A. 1994, 91: 12530-12534.
Hallas C, Pekarsky Y, Itoyama T, Varnum J, Bichi R, Rothstein JL, Croce CM: Genomic analysis of human and mouse TCL1 loci reveals a complex of tightly clustered genes. Proc Natl Acad Sci U S A. 1999, 96: 14418-14423. 10.1073/pnas.96.25.14418.
Narducci MG, Virgilio L, Engiles JB, Buchberg AM, Billips L, Facchiano A, Croce CM, Russo G, Rothstein JL: The murine Tcl1 oncogene: embryonic and lymphoid cell expression. Oncogene. 1997, 15: 919-926. 10.1038/sj.onc.1201246.
Narducci MG, Fiorenza MT, Kang SM, Bevilacqua A, Di Giacomo M, Remotti D, Picchio MC, Fidanza V, Cooper MD, Croce CM, Mangia F, Russo G: TCL1 participates in early embryonic development and is overexpressed in human seminomas. Proc Natl Acad Sci U S A. 2002, 99: 11712-11717. 10.1073/pnas.182412399.
Pekarsky Y, Koval A, Hallas C, Bichi R, Tresini M, Malstrom S, Russo G, Tsichlis P, Croce CM: Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci U S A. 2000, 97: 3028-3033. 10.1073/pnas.040557697.
Laine J, Kunstle G, Obata T, Sha M, Noguchi M: The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell. 2000, 6: 395-407. 10.1016/S1097-2765(00)00039-3.
Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK: Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001, 105: 645-655. 10.1016/S0092-8674(01)00361-0.
Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD: The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000, 10: 429-439. 10.1016/S0962-8924(00)01834-1.
Spiess AN, Walther N, Muller N, Balvers M, Hansis C, Ivell R: SPEER--a new family of testis-specific genes from the mouse. Biol Reprod. 2003, 68: 2044-2054. 10.1095/biolreprod.102.011593.
Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM: Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000, 26: 267-268. 10.1038/81547.
Wang PJ, McCarrey JR, Yang F, Page DC: An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001, 27: 422-426. 10.1038/86927.
Ohno S: Gene duplication and the uniqueness of vertebrate genomes circa 1970-1999. Semin Cell Dev Biol. 1999, 10: 517-522. 10.1006/scdb.1999.0332.
Der-Sarkissian H, Vergnaud G, Borde YM, Thomas G, Londono-Vallejo JA: Segmental polymorphisms in the proterminal regions of a subset of human chromosomes. Genome Res. 2002, 12: 1673-1678. 10.1101/gr.322802.
Coates ML: Hemoglobin function in the vertebrates: an evolutionary model. J Mol Evol. 1975, 6: 285-307.
Proudfoot NJ, Shander MH, Manley JL, Gefter ML, Maniatis T: Structure and in vitro transcription of human globin genes. Science. 1980, 209: 1329-1336.
Weiler KS, Wakimoto BT: Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995, 29: 577-605. 10.1146/annurev.ge.29.120195.003045.
Gottschling DE, Aparicio OM, Billington BL, Zakian VA: Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990, 63: 751-762. 10.1016/0092-8674(90)90141-Z.
Kyrion G, Liu K, Liu C, Lustig AJ: RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993, 7: 1146-1159.
Baur JA, Zou Y, Shay JW, Wright WE: Telomere position effect in human cells. Science. 2001, 292: 2075-2077. 10.1126/science.1062329.
Gotta M, Gasser SM: Nuclear organization and transcriptional silencing in yeast. Experientia. 1996, 52: 1136-1147.
Digital Differential Display. [http://www.ncbi.nlm.nih.gov/UniGene/info_ddd.html]
Web-based tool SMART (Simple Molecular Architecture Research Too). [http://smart.embl-heidelberg.de/]
Sequence comparison tool BLAST (Basic Local Alignment Search Tool). [http://www.ncbi.nlm.nih.gov/BLAST/]
Gay E, Seurin D, Babajko S, Doublier S, Cazillis M, Binoux M: Liver-specific expression of human insulin-like growth factor binding protein-1 in transgenic mice: repercussions on reproduction, ante- and perinatal mortality and postnatal growth. Endocrinology. 1997, 138: 2937-2947. 10.1210/en.138.7.2937.
Besnard N, Pisselet C, Monniaux D, Locatelli A, Benne F, Gasser F, Hatey F, Monget P: Expression of messenger ribonucleic acids of insulin-like growth factor binding protein-2, -4, and -5 in the ovine ovary: localization and changes during growth and atresia of antral follicles. Biol Reprod. 1996, 55: 1356-1367.
Mashimo T, Glaser P, Lucas M, Simon-Chazottes D, Ceccaldi PE, Montagutelli X, Despres P, Guenet JL: Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2',5'-oligoadenylate synthetases. Genomics. 2003, 82: 537-552. 10.1016/S0888-7543(03)00176-9.
Rajkovic A, Yan C, Yan W, Klysik M, Matzuk MM: Obox, a family of homeobox genes preferentially expressed in germ cells. Genomics. 2002, 79: 711-717. 10.1006/geno.2002.6759.
Acknowledgements
We wish thank Claude Cahier and his team as well as Eric JeanPierre for expert animal care. We wish also to acknowledge Joëlle Dupont for advices and Thierry Delpuech for bacterial technical assistance. We thank Pascal Mermillod, Svetlana Uzbekova, and Sophie Pennetier for helpful discussion, as well as Alice Pierre for her appreciated help.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
AP and SD performed most of the in silico investigation, as well as the molecular biology and the in situ hybridization experiments. IC contributed to the structural analysis of the Oosp-cluster genes. MB participated to the in situ hybridization experiments and DV performed most of the metagenome analysis of the oocyte-genes organized in clusters. PM was assisted by RD-T to coordinate this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Paillisson, A., Dadé, S., Callebaut, I. et al. Identification, characterization and metagenome analysis of oocyte-specific genes organized in clusters in the mouse genome. BMC Genomics 6, 76 (2005). https://doi.org/10.1186/1471-2164-6-76
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-6-76