Abstract
Background
The phylum Apicomplexa is an early-branching eukaryotic lineage that contains a number of important human and animal pathogens. Their complex life cycles and unique cytoskeletal features distinguish them from other model eukaryotes. Apicomplexans rely on actin-based motility for cell invasion, yet the regulation of this system remains largely unknown. Consequently, we focused our efforts on identifying actin-related proteins in the recently completed genomes of Toxoplasma gondii, Plasmodium spp., Cryptosporidium spp., and Theileria spp.
Results
Comparative genomic and phylogenetic studies of apicomplexan genomes reveals that most contain only a single conventional actin and yet they each have 8–10 additional actin-related proteins. Among these are a highly conserved Arp1 protein (likely part of a conserved dynactin complex), and Arp4 and Arp6 homologues (subunits of the chromatin-remodeling machinery). In contrast, apicomplexans lack canonical Arp2 or Arp3 proteins, suggesting they lost the Arp2/3 actin polymerization complex on their evolutionary path towards intracellular parasitism. Seven of these actin-like proteins (ALPs) are novel to apicomplexans. They show no phylogenetic associations to the known Arp groups and likely serve functions specific to this important group of intracellular parasites.
Conclusion
The large diversity of actin-like proteins in apicomplexans suggests that the actin protein family has diverged to fulfill various roles in the unique biology of intracellular parasites. Conserved Arps likely participate in vesicular transport and gene expression, while apicomplexan-specific ALPs may control unique biological traits such as actin-based gliding motility.
Similar content being viewed by others
Background
The phylum Apicomplexa contains several protozoan pathogens that cause severe disease in mammals, including humans. Members such as Plasmodium falciparum, and P. vivax, which cause severe human malaria, and Theileria parva and T. annulata, which are responsible for economic losses in cattle in Africa, result in profound medical, social, and economic effects [1, 2]. Others such as Toxoplasma gondii, Cryptosporidium parvum and C. hominis are primarily health threats in HIV+/AIDS and immunosuppressed populations [3].
Apicomplexans are primarily obligate intracellular parasites that rely on actin-based motility for cell invasion [4]. Invasion occurs by active parasite motility that is coupled to timed secretion of proteins from specialized apical secretory organelles, which are a hallmark feature of this phylum [5, 6]. The apical secretory organelles (called micronemes, rhoptries, and dense granules) release their contents in a highly regulated fashion upon host cell interactions [7]. Microneme proteins provide adhesion to the host cells and supply the traction needed for invasion. Rhoptry and dense granule proteins function in the establishment and maintenance of a protective, intracellular niche called the parasitophorous vacuole (reviewed in [8]). Understanding how motility and invasion are regulated is crucial to elucidating the pathobiology of these organisms, yet we know relatively little about how these functions are controlled at the cellular level.
Apicomplexans are characterized by a unique cytoskeleton that is distinct from that of other eukaryotes [9]. At their apical end is a specialized microtubule-organizing center called the polar ring complex, which coordinates a series of singlet microtubules called the subpellicular microtubules [10, 11]. The remarkable stability of these microtubules provides a defined shape and polarity to the cells that is necessary for motility and invasion [12]. The subpellicular microtubules encompass the apical secretory organelles and may play a role in trafficking to the apical end of the cell. Apicomplexans also regulate their actin cytoskeleton differently, maintaining a large pool of soluble actin, both globular and in short, unstable filaments [13–15]. During motility, actin filaments must rapidly assemble to support gliding and then turnover rapidly to prevent unwanted movement. Actin regulation is thus crucial to the control of motility. In other eukaryotes, a large family of actin-related proteins helps control many cytoskeletal functions including vesicle transport and actin-based motility.
Actin-related proteins (Arps) are conserved across all eukaryotes and some prokaryotes. Although all members share a common actin-fold and an overall sequence similarity to actin [16–18], individual Arps carry out a variety of biochemical and structural roles in the cell [19]. These include roles in cell division [20], translocation of cargo along microtubules via dynein [21, 22], actin polymerization [23], and transcriptional regulation via chromatin/ heterochromatin remodeling [24–26]. Currently, more than 11 classes of Arps have been reported from a broad range of eukaryotes including plants, animals, fungi, and protozoans (i.e. Dictyostelium, Acanthamoeba, and Tetrahymena). In each case, the Arp groups link the separate kingdoms both by protein similarity and common biochemical functions. Despite their apparent conservation among the majority of eukaryotes, no Arps have been previously described in the Apicomplexa.
Complete genome sequences have recently been provided for a variety of apicomplexan parasites. A cursory examination of these genomes reveals multiple actins and actin related proteins; however, these have been inconsistently identified and annotated. The complex biology of these parasites led us to examine actin-related proteins in this phylum relative to other eukaryotes using a combination of phylogenetic and reciprocal BLAST analyses. Our findings reveal a complexity of actin-related proteins not previously appreciated and define both conserved and unique members of this protein family within the Apicomplexa.
Results and discussion
Phylogenetic comparisons of actin-like proteins in apicomplexans
We searched the recently completed genomes of Toxoplasma gondii, Plasmodium spp., Cryptosporidium spp., and Theileria spp. for actin-related proteins using conventional actins and conserved Arp proteins from organisms spanning several phyla including mammals, plants, flies, worms, yeast, and protozoa [see Additional File 1]. BLAST analysis identified over 60 candidate actin-related proteins in total among the apicomplexan genomes examined in this study (Table 1). Reciprocal BLASTP searches using each of these apicomplexan actin-like proteins against the NCBI CDD database revealed that the majority of them contain a conserved actin domain (pfam00022) (Table 1). However, at present individual actin-related protein groups have not been defined by distinct domains or motifs common to members of only one group. Consequently, we sought to establish relationships between the apicomplexan actin-like proteins and conventional Arps using sequence alignment and phylogenetic analyses. Candidate actin-related proteins were aligned with a broader spectrum of Arps from a variety of eukaryotic taxa and bacterial actin-like proteins using CLUSTALX [27]. The relative divergence of actin-like proteins was determined by Neighbor-Joining distance analysis using the phylogenetic analysis program PAUP*4.01b [28]. The resulting bootstrapped phylogram is shown in Fig. 1. Parsimony analysis revealed a similar branching pattern for the major Arp groups, but was less able to resolve deep branching groups (i.e. Arp4 and various apicomplexan specific ALPs), likely due to the divergence of these sequences (Fig. 2). We have focused primarily on the relationships supported by distance analysis, since this methodology is more appropriate for highly divergent sequences.
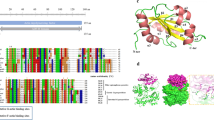
Phylogenetic comparisons of actin and actin-related proteins in apicomplexans and model organisms. In addition to conventional actins, apicomplexans contain conserved Arp1, Arp4, and Arp6 proteins; however, they do not encode Arp2 or Arp3 orthologues. Many apicomplexan proteins do not group with any of the known Arp clades. These have been divided further into proteins that are highly conserved among all the apicomplexans (i.e. ALP1, ALP2, ALP3) and those that are organism-specific (i.e. ALP5, ALP7, ALP8). Phylogenetic analysis was performed using PAUP*4.0b10 and the BioNeighbor-Joining algorithm (BioNJ) to determine the divergence distances among taxa. Consensus trees were bootstrapped for 1000 replicates and drawn according to the 50% majority-rule. Conventional actin was defined as the out-group. Subgroups of Arps and ALPs have been highlighted to define the boundaries between groups. Taxa are as follows: At = Arabidopsis thaliana, Bs = Bacillus subtilis, Ce = Caenorhabditis elegans, Cp = Cryptosporidium parvum, Dd = Dictyostelium discodium, Dm = Drosophila melanogaster, Hs = Homo sapiens, Pf = Plasmodium falciparum, Sc = Saccharomyces cerevisiae, Tg = Toxoplasma gondii, Tp = Theileria parva, Tt = Tetrahymena thermophila. Bootstrap values ≥90% are represented by the black nodes ●, values ≥ 75% are represented by the gray nodes  , and values ≥50% are denoted by the white nodes ○.
, and values ≥50% are denoted by the white nodes ○.
Parsimony analysis of actin and actin-related proteins from apicomplexans and other taxa. Phylogenetic analysis performed using parsimony resulted in groupings that mirrored distance analysis. All major classes of Arp and ALP groups are maintained except for the Arp4 group, which differs slightly from the BioNJ consensus tree in other eukaryotic taxa as well as in the apicomplexans. Relationships were calculated in PAUP*4.01b using the heuristic algorithm and verified by bootstrapping (>100 replicates). Consensus trees were drawn according to the bootstrap 50% majority-rule. Conventional actins were defined as the out-group. Subgroups of Arps and ALPs have been highlighted to define the boundaries between groups. Taxa are defined in Figure 1. Bootstrap values ≥90% are represented by the black nodes ●, values ≥ 75% are represented by the gray nodes  , and values ≥50% are denoted by the white nodes ○.
, and values ≥50% are denoted by the white nodes ○.
Our analysis reveals that the apicomplexans all encode a single conventional actin (with the exception of Plasmodium which has two conventional actins), and the remaining proteins form a total of 10 distinct actin-related protein groups (Fig. 1). Three of these groups were shown to belong to well-characterized Arps including Arp1, Arp4, and Arp6 (Fig. 1). In contrast, we discovered that several other apicomplexan actin-like proteins (ALPs) were unique to this phylum, as they did not group with any of the conventional Arps (i.e. ALP1 and ALP2)(Fig.1); therefore, we have used the designation actin-like protein (ALP) to differentiate the apicomplexan-specific protein groups. A comparison of actin-like proteins within the Apicomplexa is summarized in Table 1. The remaining ALPs were specific to a subset of apicomplexans such as Toxoplasma (i.e. ALPs 8, and 9), Cryptosporidium (i.e. ALP7) and Plasmodium (i.e. ALP5). Several of these groups also contain paralogues, for example ALP5a and ALP5b in Plasmodium (Fig.1). While some ALPs appear as deep branches (i.e. TpALP4b, PfALP2b, CpALP7a and CpALP7b, TgALP2a, TgALP3) they were grouped and hence named in part based on BLAST results (Table 1) and phylogenetic analysis of apicomplexan ALPs compared in the absence of other organisms (data not shown). Our findings suggest that some actin-like proteins play roles that are conserved across all eukaryotes while other members of this group have diverged to fulfill specific roles within the Apicomplexa.
The two key features most prominent about the apicomplexan actin-like proteins are their strong conservation of the Arp1 protein (a major component of the dynactin complex) and their remarkable lack of both Arp2 and Arp3 homologues (subunits of the Arp2/3 actin polymerization complex) (Fig. 1). The presence of a highly conserved Arp1 and the absence of Arp2 and Arp3 orthologues have important biochemical implications for these parasites regarding vesicular trafficking and actin regulation, respectively. Arp1 is an essential component of the dynactin complex involved in vesicular trafficking [29, 30] while Arp2/3 forms a multi-subunit complex that is the primary means of regulating actin polymerization in eukaryotic cells [31, 32]. For these reasons, we conducted a more comprehensive study of the protein components that constitute these complexes.
The dynactin complex
Dynactin is a microtubule-associated complex that is critical for tethering cellular cargo to the cytoplasmic motor protein dynein [30]. Cytoplasmic dynein consists of heavy, light, intermediate, and intermediate light chains in addition to several regulatory subunits [33]. We searched the P. falciparum and C. parvum genomes to identify components of this complex using text word searches. Convincing orthologues for all of the subunits were found in both parasites as shown by reciprocal BLASTP and the presence of conserved pfam domains (Table 2). These hits were then used to identify orthologues in other apicomplexan genomes by BLASTP as verified by both significant BLAST E-values and the presence of conserved pfam domains (Table 2). Somewhat surprisingly, a complete complex was not readily identified in Theileria, with the exception of subunits for heavy and light chains (Table 2).
Based on the presence of a conserved dynein complex in a majority of apicomplexans, we thought it reasonable to search for evidence of a dynactin complex. The dynactin complex consists of several protein subunits that are grouped into two domains: the Arp1 rod and a flexible arm region. The protein subunits of the Arp1 rod are more highly conserved between eukaryotes than the remaining dynactin subunits [30]; therefore, we focused our efforts on defining homologues to these proteins in the apicomplexans. The subunits comprising the Arp1 rod include Arp1, Arp11, capping protein (CapZ), p62, p25, p27, and actin (see [30] for a complete review of the dynactin complex).
We used database searches to identify the dynactin subunits within apicomplexans. Sequences from mammals, flies, worms, and protozoa were compared against the NBCI nr database and the respective genomic databases of Toxoplasma, Plasmodium, and Cryptosporidium (Table 3) [see Additional file 1]. Arp1 was readily identified in Plasmodium, Cryptosporidium, and Toxoplasma, although it is apparently absent in Theileria (Table 1). Highly conserved orthologues of the p25, p27, and p62 subunits were found in Toxoplasma, Plasmodium, and Cryptosporidium as shown by both significant BLASTP E-values and the presence of conserved pfam domains (Table 3).
The Arp1 rod contains a short filament of Arp1 subunits [34] that is capped at both ends. The (+) or barbed end is terminated by capping protein [35] and the (-) or pointed end by the actin-related protein Arp11 [36]. Toxoplasma and Plasmodium both contain β subunits of capping protein, and the α subunit in Plasmodium showed a significant BLASTP E-value and conserved pfam motif (Table 3). The α subunit reported for Toxoplasma is highly divergent (NCBI AAU93918) and does not have significant matches, although BLASTP searches turn up a number of α subunit orthologues (Table 3). Additionally only the β subunit was identified in Cryptosporidium (Table 3). Capping protein always exists as an α/β dimer [37] and it is possible the α subunit is divergent in Toxoplasma and Cryptosporidium and hence difficult to recognize at present. Our phylogenetic analysis of the Arps did not show strong affinities between any of the ALP proteins and the Arp11 group (Fig. 1). However, we have included TgALP3, PfALP3, and CpALP3 as possible Arp11 orthologues based on their sequence similarity to the Arp11 proteins in BLASTP searches (Table 3).
Dynamitin is a component of the flexible arm region of the dynactin complex [30]. We identified proteins with recognizable dynamitin domains in Toxoplasma and Plasmodium, but not Cryptosporidium (Table 3). The remaining subunits of the dynactin complex were not detected by BLAST or protein domain searches in these organisms. However since these other subunits are less well conserved, failure to detect them by BLAST is not surprising.
The identification of apicomplexan orthologues to all the subunits of the Arp1 rod, and the presence of dynamitin in Toxoplasma and Plasmodium, provides strongly supportive evidence that a functional complex exists in these parasites. Theileria appears to be an exception to this pattern as neither Arp1 or the other subunits were recognized. The conserved complex in parasites likely carries out duties analogous to the dynactin in other eukaryotes. One possible role for this complex would be the directed delivery of secretory protein vesicles as has been described in other systems [29]. Secretory protein trafficking occurs via an ER-Golgi mediated pathway [38] and dynactin could provide the transportation by which cargo vesicles reach their specialized secretory organelles at the apical pole. Apical secretion is an important component of cellular invasion and maintenance of this polarization is thus vital to the survival of the parasite.
The Arp2/3 actin polymerization complex
The Arp2/3 complex consists of 7 subunits that regulate actin polymerization at the leading edge in motile cells [23], as well as providing a propulsive force to move endosomes throughout the cytoplasm [39, 40]. Arp2/3 is a major nucleator of actin polymerization in most eukaryotic cells; however, our phylogenetic analyses of the apicomplexan actin-related proteins did not show homologues to either Arp2 or Arp3 (Fig. 1). Notably, Arp2 and Arp3 homologues have been previously annotated in both the Plasmodium and Cryptosporidium genome databases (PfArp3: CAD51790, PfArp2: CAD49164, CpArp3: EAK88375, and CpArp2, EAK88162). These proteins correspond to our annotations PfALP5a, PfALP5b, CpALP7a, and CpALP7b, respectively. Phylogenetic comparisons do not support these previously proposed annotations, but rather indicate that these actin-like proteins are part of other ALP groups (Fig. 1).
A recent analysis of the actin family from model organisms was utilized to derive predictive models for grouping Arp groups in a variety of taxa [41]. Importantly, this analysis also found Arp1, Arp4, and Arp6 homologues among the Apicomplexa (Plasmodium and Cryptosporidium genomes) but failed to identify orthologues of Arp2, Arp3 or other Arp groups [41]. Collectively, these findings indicate apicomplexans do not encode a conserved Arp2/3 complex.
We also searched for the other 5 subunits of the Arp2/3 complex that are known as actin-related protein complex 1 (ARPC1)/p41, ARPC2/p34, ARPC3/p21, ARPC4/p20, and ARPC5/p16. A separately recognized domain is only described for ARPC4/p20 (pfam05856), perhaps reflecting the divergence of the remaining subunits across the many taxa where they are readily identified by BLAST. We conducted genome-wide BLAST searches of apicomplexans as described above using ARPC proteins from mammals, flies, yeast, plants, and protozoa (see Table 4) [see Additional file 1]. No proteins with similarity to subunits ARPC2, 3, and 5 were found in any of the four apicomplexan genomes.
Potential orthologues to the ARPC1/p41 were found in Plasmodium and Cryptosporidium (Table 3): both of these proteins contain WD40 repeats, which are a distinguishing feature of the ARPC1/p41 proteins in other eukaryotes [42]. This analysis was supported by BLAST and also by protein domain searches using Prosite, which identified WD40 repeat domains in both proteins (Pfam PF00400, SMART domain SM00320). WD40 repeats mediate protein-protein interactions and are involved in regulating numerous biological functions in addition to their role in actin nucleation [43, 44]. Since ARPC1/p41 is not necessary for the overall cohesiveness of the Arp2/3 subunits [45], we can hypothesize this protein may serve an alternative function outside of the Arp2/3 complex in Plasmodium and Cryptosporidium.
Surprisingly, Cryptosporidium encodes a conserved ARPC4/p20 subunit as shown by BLAST analysis and by Prosite domain similarity (pfam PF05856) (Table 4). In other eukaryotes, ARPC4 forms a stable heterodimeric complex with ARPC2/p34 that comprises the structural core of the Arp2/3 complex [45]. In the absence of ARPC4/p20, Arp2/3 complexes are not formed [45], underscoring its importance to the protein scaffold. It is therefore unusual that Cryptosporidium would retain a close orthologue to one subunit and completely lack the other (Table 4). Additionally, the ARPC2/ARPC4 heterodimer binds actin filaments and is thought to be necessary for branching of daughter filaments from existing mother filaments [45]. Actin in Toxoplasma does not appear to be branched [46], thus it is unclear why Cryptosporidium maintains an ARPC4 homologue (Table 4).
The presence of remnant ARPC1/p41 homologues in Plasmodium and Cryptosporidium, and ARPC4/p20 in Cryptosporidium indicates that the complex may have been functional at one time in these parasites; however, they either have since lost the complex completely or the subunits have diverged to the extent that they are no longer recognizable. Support for this hypothesis comes from other alveolates, such as the closely-related but deeper branching ciliate lineages [47]. The ciliate Tetrahymena thermophilia encodes a canonical Arp2/3 complex with easily recognizable Arp2 (AAN73249), Arp3 (AAN73250), ARPC2 (4.m00362), ARPC3 (43.m00326) and ARPC4 (152.m00065) subunits. Loss of a functional complex in the apicomplexans may have resulted from their highly specialized, intracellular lifestyles. Deciphering how apicomplexans control actinfilament turnover is thus an intriguing and unanswered question. We postulate that evolution of alternative proteins, such as the ALP1 proteins (Fig. 1), could enable parasites to regulate actin polymerization in a more streamlined mechanism, yet maintain the overall function of the complex.
The ALP1 group of apicomplexan-specific proteins is phylogenetically similar to both the Arp2 and Arp3. ALP1 in Toxoplasma is the second closest paralogue to conventional actin with which it shares 37% identity and 57% similarity (Table 1). Moreover, TgALP1 is 49% identical to PfALP1 and 45% identical to CpALP1, indicating the ALP1 proteins are highly conserved within this phylum (Table 1). These phylogenetic properties, in conjunction with the lack of any obvious Arp2 or Arp3 homologues, lead us to hypothesize that the ALP1 proteins may play a corresponding or complementary role to these two proteins in the apicomplexans.
Arps and chromatin remodeling
In addition to their cytoskeletal roles, actin and Arps function in the nucleus as components of chromatin-modifying and chromatin-remodeling protein complexes [48]. These Arps include Arp4, Arp5, Arp6 and Arp8 [26, 49–51]. Arps 7 and 9 are yeast-specific and do not have homologues in other eukaryotes [48]. Our studies show the apicomplexans encode conventional Arp4 and Arp6 orthologues (Fig. 1).
Chromatin-modifying and -remodeling machinery are involved in DNA replication, DNA repair mechanisms and transcriptional regulation [52]. Arp4 is present in several complexes including the NuA4 histone acetyltransferase and several members of the ATP-dependent SWI2-SNF2 family of chromatin-remodeling complexes [51, 53]. In yeast, Arp6 is also a member of SWR1, a subgroup of the SWI2/SNF2 chromatin-remodeling complexes [50]. Other roles for Arp6 include transcriptional deactivation via heterochromatin-remodeling in Drosophila and vertebrates [24].
Changes in gene expression are important means of regulating function and such changes have been shown to play a role in parasite stage-differentiation [54–56]. The Plasmodium and Cryptosporidium genomes appear to lack many common transcription factors leading to the hypothesis that these parasites rely heavily on chromatin-remodeling for transcriptional control [57, 58]. This is supported by the fact that apicomplexans appear to contain several components of the SWI2/SNF2 ATPase chromatin-remodeling machinery [57, 58]. Recently, Saksouk et. al. showed the first direct correlation of histone acetylation and methylation to stage-specific gene expression in Toxoplasma [59], supporting the importance of chromatin modification and remodeling in these parasites. The presence of conserved Arp4 and Arp6 orthologues suggests that actin-related proteins participate in chromatin remodeling in apicomplexans similar to other eukaryotes.
Conclusion
Comprehensive analysis of the genome content of these parasites combined with phylogenetic groupings has allowed us to propose potential functions for many of these Arp/ALP groups. Our findings indicate that apicomplexans encode a variety of actin-like proteins (ALPs) that likely participate in actin-based motility, vesicle transport, and transcriptional regulation through chromatin remodeling. Delineating their respective functions will ultimately enrich our understanding of these parasites, and also the evolution of the actin family as a whole.
Methods
Assembly of actin-like proteins from apicomplexans and other alveolates
Comprehensive BLAST searches were performed against the T. gondii genome database (ToxoDB Release v3.0) using 27 protein sequences from Arp1 through Arp4 that represented major taxa including mammals, plants, flies, worms, yeast, and protozoa. Actin-like proteins were identified in Plasmodium spp. (PlasmoDB Release v4.3) and C. parvum (NCBI nr database and CryptoDB Release v3.0) by combination of tBLASTn and BLASTP searches using the above conserved Arps or Toxoplasma candidate actin-like sequences. BLAST searches were restricted to only high quality "hits" (e-value of ≤ .0001). In the case where only nucleotide data was available, the matching nucleotide sequence was translated using the GENESCAN webserver [60] using Arabidopsis thaliana to predict exon-intron structures. In these cases, the resulting amino acid sequence predictions were used in all subsequent analyses. Once identified, candidate sequences were entered into a "reverse" BLAST search of the NCBI database [61] to determine if there was a reciprocal best match to the protein used to identify it.
Protein candidates from Tetrahymena thermophila were obtained via BLASTP searches of the NCBI nr database comparing Arps from model organisms and by searching the Tetrahymena genome database [62]. Searches of the Tetrahymena genome database were done using tBLASTn and restricted to TIGR predicted proteins.
Preliminary sequence data was obtained from The Institute for Genomic Research website [63], ToxoDB [64], PlasmoDB [65], CryptoDB [66].
A complete list of all taxa and accession/contig numbers used in these studies is provided [see Additional file 1].
CLUSTALX alignments
The above candidate actin-like protein sequences were compared with a larger repertoire of Arp proteins from a variety of eukaryotes [67] and bacterial actin-like proteins retrieved from NCBI. All sequences were entered into the alignment program CLUSTALX [27] using pairwise parameters set as: gap opening penalty = 15.0, gap extension penalty = 0.10; and multiple alignment parameters set as: gap opening penalty = 15.0, gap extension penalty = 0.30, delay divergent sequences (%) = 25. All other parameters were set to the default settings. Clustal alignments used in this analysis are posted at [68].
Phylogenetic analysis
CLUSTAL alignments were entered into the phylogenetic analysis program PAUP4.0b10 for Macintosh [28]. Only regions of the alignments with conservation across all taxa were included in the analyses. The optimality criterion was set to distance (mean character difference, minimal evolution, negative branches = 0) and 1000 bootstrap replicates were performed using the BioNeighbor-Joining (BioNJ) algorithm. Alternatively, a full heuristic algorithm was used for parsimony analysis, supported by bootstrapping for > 100 replicates. Consensus trees were drawn according to the Bootstrap 50% majority-rule and conventional actins were defined as the out-group.
Dynactin and Arp2/3 complex subunits
Highly conserved subunits of both the dynactin and Arp2/3 complexes were retrieved from NCBI nr for model organisms [see Additional file 1]. These proteins were used in BLASTP searches of the Toxoplasma [64], Plasmodium [65], Cryptosporidium [61, 66], Theileria [61], and Tetrahymena [61, 62] databases for candidate orthologues, as described above. Candidate proteins were used in a "reverse" BLAST of the NCBI database [61] to determine their relatedness to the proteins used to identify them.
Abbreviations
- ALP:
-
Actin-like protein
- Arp:
-
actin-related protein
- ARPC:
-
actin related protein complex
- capZ:
-
capping protein
- CDD conserved domain database:
-
NCBI, National Center for Biotechnology Information, pfam, protein family database.
References
Nahlen BL, Korenromp EL, Miller JM, Shibuya K: Malaria risk: Estimating clinical episodes of malaria. Nature. 2005, 437: E3-10.1038/nature04178.
Billiouw M, Vercruysse J, Marcotty T, Speybroeck N, Chaka G, Berkvens D: Theileria parva epidemics: a case study in eastern Zambia. Vet Parasitol. 2002, 107: 51-63. 10.1016/S0304-4017(02)00089-4.
Kaplan JE, Jones JL, Dykewicz CA: Protists as opportunistic pathogens: public health impact in the 1990s and beyond. J Eukaryot Microbiol. 2000, 47: 15-20. 10.1111/j.1550-7408.2000.tb00004.x.
Sibley LD: Invasion strategies of intracellular parasites. Science. 2004, 304: 248-253. 10.1126/science.1094717.
Dubremetz JF, Schwartzman JD: Subcellular organelles of Toxoplasma gondii and host cell invasion. Res Immunol. 1993, 144: 31-33. 10.1016/S0923-2494(05)80093-8.
Dubremetz JF: Host cell invasion by Toxoplasma gondii. Trends Microbiol. 1998, 6:
Carruthers VB, Sibley LD: Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997, 73: 114-123.
Blackman MJ, Bannister LH: Apical organelles of Apicomplexa: biology and isolation by subcellular fractionation. Mol Biochem Parasitol. 2001, 117: 11-25. 10.1016/S0166-6851(01)00328-0.
Morrissette NS, Sibley LD: Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev. 2002, 66: 21-38. 10.1128/MMBR.66.1.21-38.2002.
Morrissette NS, Murray JM, Roos DS: Subpellicular microtubules associate with an intramembranous particle lattice in the protozoan parasite Toxoplasma gondii. J Cell Sci. 1997, 110: 35-42.
Russell DG, Burns RG: The polar ring of coccidian sporozoites: a unique microtubule-organizing centre. J Cell Sci. 1984, 65: 193-207.
Bannister LH, Mitchell GH: The role of the cytoskeleton in Plasmodium falciparum merozoite biology: an electron-microscopic view. Ann Trop Med Parasitol. 1995, 89: 105-111.
Dobrowolski JM, Niesman IR, Sibley LD: Actin in the parasite Toxoplasma gondii is encoded by a single copy gene, ACT1 and exists primarily in a globular form. Cell Motil Cytoskel. 1997, 37: 253-262. 10.1002/(SICI)1097-0169(1997)37:3<253::AID-CM7>3.0.CO;2-7.
Poupel O, Tardieux I: Toxoplasma gondii motility and host cell invasiveness are drastically impaired by jasplakinolide, a cyclic peptide stabilizing F-actin. Microbes Infect. 1999, 1: 653-662. 10.1016/S1286-4579(99)80066-5.
Schmitz S, Grainger M, Howell SA, Calder LJ, Gaeb M, Pinder JC, Holder AA, Veigel C: Malaria parasite actin filaments are very short. J Molec Biol. 2005, 349: 113-125. 10.1016/j.jmb.2005.03.056.
Poch O, Winsor B: Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast. 1997, 13: 1053-1058. 10.1002/(SICI)1097-0061(19970915)13:11<1053::AID-YEA164>3.0.CO;2-4.
Frankel S, Mooseker MS: The actin-related proteins. Curr Opin Cell Biol. 1996, 8: 30-37. 10.1016/S0955-0674(96)80045-7.
Schroer TA, Fyrberg E, Cooper JA, Waterston RH, Helfman D, Pollard TD, Meyer DI: Actin-related protein nomenclature and classification. J Cell Biol. 1994, 127: 1777-1778. 10.1083/jcb.127.6.1777.
Schafer DA, Schroer TA: Actin-related proteins. Annu Rev Cell Dev Biol. 1999, 15: 341-363. 10.1146/annurev.cellbio.15.1.341.
Karki S, Holzbaur EL: Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 1999, 11: 45-53. 10.1016/S0955-0674(99)80006-4.
Clark SW, Meyer DI: Centractin is an actin homologue associated with the centrosome. Nature (Lond). 1992, 359: 246-250. 10.1038/359246a0.
Schroer TA, Sheetz MP: Two activators of microtubule-based vesicle transport. J Cell Biol. 1991, 115: 1309-1318. 10.1083/jcb.115.5.1309.
Mullins RD, Stafford WF, Pollard TD: Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997, 136: 331-343. 10.1083/jcb.136.2.331.
Kato M, Sasaki M, Mizuno S, Harata M: Novel actin-related proteins in vertebrates: similarities of structure and expression pattern to Arp6 localized on Drosophila heterochromatin. Gene. 2001, 268: 133-140. 10.1016/S0378-1119(01)00420-6.
Shen X, Ranallo R, Choi E, Wu C: Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003, 12: 147-155. 10.1016/S1097-2765(03)00264-8.
Szerlong H, Saha A, Cairns BR: The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBOJ. 2003, 22: 3175-3187. 10.1093/emboj/cdg296.
Higgins DG, Thompson JD, Gibson TJ: Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996, 266: 382-402.
Swofford DL: Phylogenetic Analysis Using Parsimony (* and other methods). 2002, Sunderland: Sinauer Associates
Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW: Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991, 115: 1639-1650. 10.1083/jcb.115.6.1639.
Schroer TA: Dynactin. Annu Rev Cell Dev Biol. 2004, 20: 759-779. 10.1146/annurev.cellbio.20.012103.094623.
Pollard TD, Beltzner CC: Structure and function of the Arp2/3 complex. Curr Opin Struct Biol. 2002, 12: 768-774. 10.1016/S0959-440X(02)00396-2.
Machesky LM, Gould KL: The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999, 11: 117-121. 10.1016/S0955-0674(99)80014-3.
Vale RD: The molecular motor toolbox for intracellular transport. Cell. 2003, 112: 467-480. 10.1016/S0092-8674(03)00111-9.
Bingham JB, Schroer TA: Self-regulated polymerization of the actin-related protein Arp1. Curr Biol. 1999, 9: 223-226. 10.1016/S0960-9822(99)80095-5.
Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA: Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994, 126: 403-412. 10.1083/jcb.126.2.403.
Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA: Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J Cell Biol. 1999, 147: 307-320. 10.1083/jcb.147.2.307.
Wear MA, Yamashita A, Kim K, Maeda Y, Cooper JA: How capping protein binds the barbed end of the actin filament. Curr Biol. 2003, 13: 1531-1537. 10.1016/S0960-9822(03)00559-1.
Joiner KA, Roos DS: Secretory traffic in the eukaryotic parasite Toxoplasma gondii: less is more. J Cell Biol. 2002, 157: 557-563. 10.1083/jcb.200112144.
Taunton J, Rowning BA, Coughlin ML, Wu M, Moon RT, Mitchison TJ, Larabell CA: Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J Cell Biol. 2000, 148: 519-530. 10.1083/jcb.148.3.519.
Fehrenbacher K, Huckaba T, Yang HC, Boldogh I, Pon L: Actin comet tails, endosomes and endosymbionts. J Exp Biol. 2003, 206: 1977-1984. 10.1242/jeb.00240.
Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B: Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 2005, 16: 5736-5748. 10.1091/mbc.E05-06-0508.
Welch MD, Iwamatsu A, Mitchison TJ: Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997, 385: 265-269. 10.1038/385265a0.
Li D, Roberts R: WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001, 58: 2085-2097.
Pan F, Egile C, Lipkin T, Li R: ARPCl/Arc40 mediates the interaction of the actin-related protein 2 and 3 complex with Wiskott-Aldrich syndrome protein family activators. J Biol Chem. 2004, 279: 54629-54636. 10.1074/jbc.M402357200.
Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD: Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Molec Cell. 2001, 8: 1041-1052. 10.1016/S1097-2765(01)00393-8.
Wetzel DM, Håkansson S, Hu K, Roos DS, Sibley LD: Actin filament polymerization regulates gliding motility by apicomplexan parasites. Mol Biol Cell. 2003, 14: 396-406. 10.1091/mbc.E02-08-0458.
Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF: A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000, 290: 972-977. 10.1126/science.290.5493.972.
Blessing CA, Ugrinova GT, Goodson HV: Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004, 14: 435-442. 10.1016/j.tcb.2004.07.009.
Doyon Y, Cote J: The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004, 14: 147-154. 10.1016/j.gde.2004.02.009.
Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C: ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2003, 303: 343-348. 10.1126/science.1090701.
Shen X, Mizuguchi G, Hamiche A, Wu C: A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000, 406: 541-544. 10.1038/35020123.
Ehrenhofer-Murray AE: Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004, 271: 2335-2349. 10.1111/j.1432-1033.2004.04162.x.
Olave IA, Reek-Peterson SL, Crabtree GR: Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002, 71: 755-781. 10.1146/annurev.biochem.71.110601.135507.
Pologe LG: Aberrant transcription and the failure of Plasmodium falciparum to differentiate into gametocytes. Mol Biochem Parasitol. 1994, 68: 35-43. 10.1016/0166-6851(94)00142-1.
Spielmann T, Beck HP: Analysis of stage-specific transcription in Plasmodium falciparum reveals a set of genes exclusively transcribed in ring stage parasites. Mol Biochem Parasitol. 2000, 111: 453-458. 10.1016/S0166-6851(00)00333-9.
Singh U, Brewer JL, Boothroyd JC: Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol Microbiol. 2002, 44: 721-733. 10.1046/j.1365-2958.2002.02903.x.
Templeton TJ, Iyer LM, Anantharaman V, Enomoto S, Abrahante JE, Subramanian GM, Hoffman SL, Abrahamsen MS, Aravind L: Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004, 14: 1686-1695. 10.1101/gr.2615304.
Meissner M, Soldati D: The transcription machinery and the molecular toolbox to control gene expression in Toxoplasma gondii and other protozoan parasites. Microbes Infect. 2005, 7: 1376-1384. 10.1016/j.micinf.2005.04.019.
Saksouk N, Bhatti MM, Keiffer S, Smith AT, Musset K, Garin JF, Sullivan WJ, Cesbron-Delauw MF, Hakimi MA: Histone modifying complexes regulate gene expression pertinent to the differentiation of protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005, 25: 10301-10314. 10.1128/MCB.25.23.10301-10314.2005.
GENSCAN Webserver at MIT. [http://genes.mit.edu/GENSCAN.html]
National Center for Biotechnology Information. [http://www.ncbi.nlm.nih.gov/]
Tetrahymena genome database. [http://www.ciliate.org/]
The Institute for Genome Research. [http://www.tigr.org]
Toxoplasma genome database. [http://ToxoDB.org]
Plasmodium genome database. [http://www.plasmodb.org/]
Cryptosporidium genome database. [http://www.cryptodb.org/]
Goodson HV, Hawse WF: Molecular evolution of the actin family. J Cell Sci. 2002, 115: 2619-2622.
Arp superfamily alignments. [http://www.sibleylab.wustl.edu/Publications.htm]
Acknowledgements
We thank Feng Chen, David Roos (U. Pennsylvania) for help with homology searches, Scott Handley (Washington University) for advice in phylogenetic analyses; Holly Goodson (Notre Dame), Matt Welch (UC Berkeley) and Trina Schroer (Johns Hopkins) for stimulating discussions; and Bill Sullivan (Indiana Univ.) and his colleagues for kindly providing data prior to publication.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
JLG and LDS devised the overall strategy for these studies. JLG performed all database analyses, sequence alignments, and phylogenetic comparisons. JLG authored the text of this manuscript and LDS provided comments and revisions to the final version of this text.
Electronic supplementary material
12864_2005_380_MOESM1_ESM.pdf
Additional File 1: Taxa. Listing of the taxa and accession numbers for protein alignments and BLAST analyses used in the present study. (PDF 51 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Gordon, J.L., Sibley, L.D. Comparative genome analysis reveals a conserved family of actin-like proteins in apicomplexan parasites. BMC Genomics 6, 179 (2005). https://doi.org/10.1186/1471-2164-6-179
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-6-179