Abstract
Background
A yeast strain lacking the two genes SSA1 and SSA2, which encode cytosolic molecular chaperones, acquires thermotolerance as well as the mild heat-shocked wild-type yeast strain. We investigated the genomic response at the level of mRNA expression to the deletion of SSA1/2 in comparison with the mild heat-shocked wild-type using cDNA microarray.
Results
Yeast cDNA microarray analysis revealed that genes involved in the stress response, including molecular chaperones, were up-regulated in a similar manner in both the ssa1/2 deletion mutant and the mild heat-shocked wild-type. Genes involved in protein synthesis were up-regulated in the ssa1/2 deletion mutant, but were markedly suppressed in the mild heat-shocked wild-type. The genes involved in ubiquitin-proteasome protein degradation were also up-regulated in the ssa1/2 deletion mutant, whereas the unfolded protein response (UPR) genes were highly expressed in the mild heat-shocked wild-type. RT-PCR confirmed that the genes regulating protein synthesis and cytosolic protein degradation were up-regulated in the ssa1/2 deletion mutant. At the translational level, more ubiquitinated proteins and proteasomes were detected in the ssa1/2 deletion mutant, than in the wild-type, confirming that ubiquitin-proteasome protein degradation was up-regulated by the deletion of SSA1/2.
Conclusion
These results suggest that the mechanism for rescue of denatured proteins in the ssa1/2 deletion mutant is different from that in the mild heat-shocked wild-type: Activated protein synthesis in the ssa1/2 deletion mutant supplies a deficiency of proteins by their degradation, whereas mild heat-shock induces UPR.
Similar content being viewed by others
Background
Exposure to certain kinds of environmental stress factors, such as chemical, heat, osmotic, etc., induces living organisms to express stress proteins, thereby enabling the organism to acquire stress tolerance. This phenomenon is called the "stress response". Especially, the heat-inducible proteins termed "heat-shock proteins (Hsps)" constitute an important part of the stress-responsive proteins [1]. HSP70s (70 kDa HSPs) were discovered in Drosophila melanogaster, and their homologs have been found in various organisms including yeast [2, 3]. HSP70s also function as molecular chaperones [2, 3]. In the Saccharomyces cerevisiae genome, there are ca. 14 HSP70-like genes. The SSA, SSB and SSE families are cytosolic HSP70 [4–6], whereas the SSC1 is localized to the mitochondria [7, 8]. In addition, KAR2 (BiP) is localized to the endoplasmic reticulum [9–12]. The SSA family contains 4 genes, SSA1, SSA2, SSA3 and SSA4 [13]. Not only are the SSA1 and SSA2 genes constitutively expressed, they are also 96% identical at the nucleotide level [2]. Moreover, there is no change in the phenotype of deletion in either of the SSA1 and SSA2 genes compared with the wild-type. In addition, they do not show thermotolerance without pre-heat treatment at 37°C [14]. However, the ssa1/2 double deletion mutant acquires thermotolerance even at 23°C, and shows a slow growth rate [14]. A suppressor, EXA3-1 which is an allele of HSF1 encoding a heat shock factor [15, 16] recovers its growth rate. This phenomenon in the ssa1/2 deletion mutant is speculated to result from the overexpression of certain Hsps [17]. HSP104 and SSA4 are found to be highly expressed in the ssa1/2 deletion mutant [4, 18].
SSA1 is involved in protein transport and the rescue of denatured proteins [19–22], and possesses ATPase activity [23]. Sti1p activates ATPase activity of Ssa1p [24]. In addition, Hsp70 is a co-chaperone with Hsp104 and Hsp40 in both S. cerevisiae and E.coli [25, 26]. The relationship between these chaperones and human misfolding disease has been shown [27, 28]. On the other hand, SSA2 is involved in protein transport into the vacuole [29, 30]. Thus, SSA1 is multi-functional, and the ssa1/2 double deletion mutant shows drastic changes needed to acquire thermotolerance, which is similar to the mild heat-shocked wild-type. As Ssa1p and Ssa2p are cytosolic molecular chaperones, it is hypothesized that unfolded proteins appear by the double deletion of SSA1/2.
However, genome-wide expression analysis of the ssa1/2 deletion mutant using cDNA microarray has not been carried out. We believe that gene expression profiling of the ssa1/2 deletion mutant is necessary not only to describe the genomic response developed by yeast to the deletions, but also to reveal the mechanism of the response to denatured proteins. To support the cDNA microarray data, we also performed RT-PCR, and immunoblot analysis of several yeast proteins separated by two-dimensional gel electrophoresis (2-DGE). We demonstrate that the deletion of SSA1/2 genes induces up-regulation of the genes involved in both protein degradation and synthesis, whereas mild heat shock induces UPR.
Results
Comparison of the mRNA expression profiles between the ssa1/2 deletion mutant and the mild heat-shocked wild-type
To investigate the mechanism of the response to denatured proteins comprehensively, the mRNA expression profiling of the ssa1/2 deletion mutant was carried out using yeast cDNA microarray, in comparison with the mild heat-shocked wild-type. The number of up-regulated genes in the ssa1/2 deletion mutant was 144, while that in the mild heat-shocked wild-type by exposure for 30 and 60 min at 43°C was 274 and 400, respectively. The functionally categorized up-regulated genes are shown in Figs. 1A and 2A. The most highly up-regulated genes were categorized into "Cell rescue, defense, and virulence (stress-inducible proteins)" in both ssa1/2 and mild heat-shocked wild-type, of which the rates were 8% and 10%, respectively. On the other hand, the number of down-regulated genes in ssa1/2 was 94, while that in the mild heat-shocked wild-type by exposure for 30 and 60 min at 43°C was 610 and 643, respectively. The functionally categorized down-regulated genes are shown in Figs. 1B and 2B.
The overview of expressed genes in the ssa1/2 deletion mutant. S. cerevisiae JN14 (ssa1/2) and JN54 (wild-type) cells were incubated at 30°C to a logarithmic phase (OD660 = 1). The up-regulated genes (over 2- fold expressed) and down-regulated genes (over 2-fold suppressed) in the ssa1/2 deletion mutant were determined by twice induction of three individual experiments. These genes were functionally categorized using Comprehensive Yeast Genome Database (CYGD) in Munich International Center of Protein Sequence (MIPS) [52]. A, up-regulated genes; B, down-regulated genes.
The overview of expressed genes in the mild heat-shocked wild-type. S. cerevisiae JN54 (wild-type) cells were incubated at 30°C to a logarithmic phase (OD660 = 1), and were then treated with mild heat-shock at 43°C for 30 or 60 min. The up-regulated genes (over 2- fold expressed) and down-regulated genes (over 2-fold suppressed) in mild heat-shocked wild-type were determined by twice induction of three individual experiments. These genes were functionally categorized as in Figure 1. A, up-regulated genes. B, down-regulated genes.
In the ssa1/2 deletion mutant, the percentages of up-regulated genes categorized in "Cell rescue defense and virulence", "Transport facilitation" and "Protein fate" was approximately 2–6 times larger than those of the down-regulated genes (Fig. 3A), and the opposite results were found in "Cellular communication/signal transduction mechanism" category (Fig. 3B). In the mild heat-shocked wild-type, there were no categories in which the percentages of up-regulated genes were over 2-times larger than those of the down-regulated genes (Fig. 4A). However, the percentage of down-regulated genes in "Protein synthesis" was particularly larger (ca. 170-times) than that of the up-regulated genes (Fig. 4B). Thus, the number of up-regulated genes in "Protein synthesis" was remarkably smaller than that of the down-regulated genes in the heat-shocked wild type. Conversely, in the ssa1/2 deletion mutant, the number of up-regulated genes in "Protein synthesis" was larger than that of the down-regulated genes. Therefore, we focused on protein synthesis and correlated protein fate as well as "Cell rescue, defense and virulence" in the ssa1/2 deletion mutant.
The comparison between up-regulated genes and down-regulated genes in the ssa1/2 deletion mutant. Functional categories were the same as in Figure 1. The ratios of up/down-regulated genes or down/up-regulated genes were calculated using the percentages of each category in Fig. 1. A, up/down-regulated genes; B, down/up-regulated genes.
The comparison between up-regulated genes and down-regulated genes in the mild heat-shocked wild-type. Functional categories are as given in Figure 1. The ratios of up/down-regulated genes or down/up-regulated genes were calculated using the percentages of each category in Fig. 2. A, up/down-regulated genes; B, down/up-regulated genes.
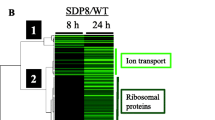
Figure 5 shows a detailed comparison of these categorized genes up-regulated in the ssa1/2 deletion mutant and in the mild heat-shocked wild-type. Figure 5A shows the comparison of all the genes up-regulated in the ssa1/2 deletion mutant and the mild heat-shocked wild-type. In the categories of "Cell rescue, defense and virulence", several Hsps, including molecular chaperones, were commonly up-regulated in the ssa1/2 deletion mutant and the mild heat-shocked wild-type (Fig. 5B). Although genes related to protein synthesis were greatly suppressed in the mild heat-shocked wild-type, ribosomal protein genes were found to be up-regulated only in the ssa1/2 deletion mutant (Fig. 5C). Table 1 shows the expression level of these genes: RPL37A, RPL25, MRP8, RPS15, MRPL10, RSM18, RPL8B and RSM10.
On the other hand, in the category of "Protein fate", the PRE1, RPN4, RPN12 and SCL1 genes that encode for cytosolic proteasome subunits, were found to be up-regulated in ssa1/2 deletion mutant (Fig. 5D). In addition, the UBC4 (ubiquitin conjugating enzyme) gene was also up-regulated. Table 2 shows the expression level of the genes involved in protein degradation. These results suggest that the ubiquitin-proteasome protein degradation pathway is activated in the ssa1/2 deletion mutant. Although a few ubiquitin-proteasome genes (UBI4, UBC5 and UBP9) were up-regulated in the mild heat-shocked wild-type, they were not in common with those up-regulated in the ssa1/2 deletion mutant (Fig. 5D). The proteasome genes up-regulated in the mild heat-shocked wild-type included vacuolar protein genes (AUT7, LAD4 and APG17), and unfolded protein response (UPR) genes (DER1, PDI1 and ERO1) (Fig. 5D).
Confirmatory RT-PCR for proteolytic degradation- and ribosomal biogenesis-related genes
To verify that both protein synthesis and degradation are activated in the ssa1/2 deletion mutant, RT-PCR analysis of several proteolytic degradation genes and cytosolic ribosomal protein genes was carried out. Proteasome subunit genes (PRE1, RPN4, RPN12, and SCL1), an ubiquitin conjugating enzyme gene (UBC4), and cytosolic ribosomal protein genes were found to be up-regulated in the ssa1/2 deletion mutant compared with the wild-type (Fig. 6A and Fig. 6B). This result supports the cDNA microarray data showing that both ubiquitin-proteasome protein degradation and protein synthesis were activated by deletion of the SSA1/2 genes. Only KAR2 was highly expressed among the UPR genes (Fig. 6A).
RT-PCR analysis of ribosomal protein and proteolytic degradation genes in the ssa1/2 deletion mutant and wild-type. RT-PCR was carried out as described in "Materials and Methods", and primers, product size and numbers of PCR cycle are described on Table 3. The RT-PCR products were run on a 4% Nu-Sieve 3:1 Plus agarose gel. A, genes for proteolytic degradation. B, ribosomal protein genes.
Immunoblot analysis of proteolytic degradation-related gene products
To confirm that ubiquitin-proteasome protein degradation is activated at the translational level in the ssa1/2 deletion mutant, immunoblot analysis was performed. Pre1p (20 S proteasome subunit) and Rpn4p (Ubiquitin-mediated 26 S proteasome subunit) increased in the ssa1/2 deletion mutant compared with the wild-type (Fig. 7). An anti-multi ubiquitin mouse monoclonal antibody (FK2) [31, 32] detects only ubiquitin that is covalently bound to substrate proteins, i.e. ubiquitinated proteins, and not free ubiquitin [31, 32]. Ubiquitinated proteins especially with molecular weights less than 30 kDa increased in the ssa1/2 deletion mutant in comparison with the wild-type (Fig. 8).
Discussion
In the present study, we reveal global differences in gene expression between yeast cells lacking two cytosolic HSP70 s, SSA1 and SSA2, and the mild-heat-shocked wild-type using cDNA microarray technologoly.
Results from cDNA microarray analysis reveal that the stress-inducible protein genes, including molecular chaperones, were up-regulated in the ssa1/2 deletion mutant in a similar fashion as seen in the mild heat-shocked wild-type (Figs. 1A, 2A, and 5B). It is clear that thermotolerance is due to expression of these stress-inducible proteins. In the ssa1/2 deletion mutant, HSF1 suppressing growth rate of the ssa1/2 [15, 16] was expressed normally and its expression level was unchanged (data not shown). Several genes involved in the ubiquitin-proteasome protein degradation pathway were up-regulated in the ssa1/2 deletion mutant (Fig. 5D and Table 2). UBC4 [33, 34] was also up-regulated in the ssa1/2 deletion mutant, which is consistent with a previous report [35]. UBC4/5 is necessary for binding between the substrates and Lys48 of ubiquitin that is a target of the 26 S proteasome [34], and for binding between the substrates and Lys63 of ubiquitin, that is not a target of the 26 S proteasome. In addition to UBC4, we found up-regulation of several proteasome genes (PRE1, RPN4, RPN12 and SCL1) in the ssa1/2 deletion mutant. PRE1 and SCL1 encode 20 S proteasome, and RPN4 and RPN12 encode 26 S proteasome [36]. RPN4 (SON1) is a factor involved in ERAD (endoplasmic reticulum associated degradation) [37]. All these genes are essential for degradation of the ubiquitinated proteins [36, 38]. RT-PCR data support the up-regulation of these proteasome genes by the deletion of SSA1/2 (Fig. 6A). Moreover, we confirmed that Pre1p and Rpn4p were up-regulated in the ssa1/2 deletion mutant at the translational level by immunoblotting (Fig. 7). This result provided further evidence that proteolytic degradation by proteasomes was stimulated by the deletion of SSA1/2. As shown in Fig. 8, more ubiquitinated proteins, especially with molecular weights less than 30 kDa, were detected in the ssa1/2 deletion mutant than in the wild-type. The deletion of UBP3 in ssa1/2 has been reported to lead to a significant increase in the number of the ubiquitinated proteins, mainly with molecular weights of more than 30 kDa [35]. The expression level of UBP3 did not change in the ssa1/2 deletion mutant compared with the wild-type (data not shown). Therefore, the increase of ubiquitinated proteins in ssa1/2 is not caused by the deletion of UBP3. There are two possibilities for the increase of ubiquitinated proteins in the ssa1/2 deletion mutant. First, insufficiency of the UPR in the ssa1/2 deletion mutant may lead to activation of the ubiquitin-proteasome protein degradation system. Thus, ubiquitination of the target proteins increases and the expression of proteasome genes is induced. Second, some defect of deubiquitination occurs in the ssa1/2 deletion mutant, consequently leading to the accumulation of ubiquitinated proteins in the cell followed by cell death. Furthermore, proteolytic degradation by proteasomes is facilitated. On the other hand, we found that several genes encoding ribosomal proteins were up-regulated in the ssa1/2 deletion mutant (Figs. 5C, 6A, and Table 1), implying that protein synthesis is activated by the deletion of SSA1/2.
In case of the mild heat-shocked wild-type, genes involved in protein synthesis were significantly suppressed (Figs. 2B and 4B), and the proteasome genes up-regulated in the ssa1/2 deletion mutant did not show any change in their expression levels (Fig. 5D). Instead, some UPR genes (PDI1, DER1, ERO1 and KAR2) were up-regulated (Fig. 5D), implying that UPR occurs during mild heat-shock. The mechanism of UPR is known to induce the up-regulation of ER chaperones for refolding when unfolded proteins accumulate in the ER [39]. In the ssa1/2 deletion mutant, the expression of three UPR genes (PDI1, DER1 and ERO1) remained unchanged (data not shown), and only KAR2 was up-regulated (Fig. 3).
Gasch et al. has reported the genome-wide expression analysis of yeast cells exposed to environmental changes [40]. We compared our data on the mild heat shocked wild-type yeast cells with their data on the wild-type cells shifted to 37°C from 25°C. The stress-inducible protein genes up-regulated in the mild-heat shocked wild type (SSA3, SSA4, SSE2, CTT1, HSP26, HSP78, and HSP104) (Fig. 5B) are in common with the results obtained by Gasch et al. [40]. In the category of "Protein fate", more than 70% of the up-regulated genes in our experiments are also in common with their results [40], even though there is a time lag with their experiments. However, DER1, one of the UPR genes, was not found to be up-regulated in their study during the entire the heat-shock treatment period [40]. In contrast, DER1 was up-regulated in our experiments (Fig. 5D). This may be due to the difference in the temperature and time of heat-shock treatment. On the other hand, in the category of "Protein synthesis", ribosomal protein genes are significantly suppressed in their experiments [40], which is consistent with our data. From these comparisons, it can be said that our data on the mild heat-shocked wild-type is similar to that reported by Gasch et al. [40] in the categories of "Cell rescue, defense, and virulence", "Protein fate" and "Protein synthesis".
It is reasonable that UPR is activated and protein synthesis is suppressed in the mild heat-shocked wild-type. We speculate on the reasons as to why the genes involved in both protein degradation and protein synthesis are up-regulated in ssa1/2 deletion mutant. In the normal state, proteins are synthesized on the ribosome, followed by post-translational modifications in the ER or the Golgi apparatus to finally become mature and functional entitles. Schubert et al [41] showed that 30% of the de novo synthesized proteins are degraded before coming to maturity. Therefore, it can be reasoned that post-translational protein denaturation occurs moderately even under normal conditions. However, organisms have developed several mechanisms in their response to the denatured proteins. UPR is one of the ER quality control mechanisms [39]. In addition, the refolding of denatured proteins is carried out by cytosolic chaperones [25, 42, 43], including SSA1/2 [20, 21]. It can be hypothesized that the deletion of SSA1/2 leads to the suppression of refolding, which is then followed by an accumulation of the denatured proteins in cells. The genes involved in proteolytic degradation may be up-regulated to remove such denatured proteins. However, if the ubiquitin-proteasome system keeps on degrading proteins, the depletion of the proteins essential for growth and development will occur. It is suggested that protein synthesis is activated to supply the proteins deleted by proteolytic degradation in the ssa1/2 deletion mutant. In the ssa1/2 deletion mutant, several hexose transporter genes (HXT2, HXT4, HXT6, HXT7), and the genes that belong to early part of glycolysis (GLK1, HXK1) were up-regulated (data not shown). The expression of these genes, involved in energy generation, may be required for sustaining the increased protein synthesis in the ssa1/2 deletion mutant. HXT genes up-regulated in the ssa1/2 deletion mutant are low-glucose dependent [44–46]. It is possible that the uptake of glucose is activated to generate energy, because energy is consumed by protein synthesis that is induced by the deletion of SSA1/2.
These results indicate that different mechanisms of the response to denatured proteins are employed between the ssa1/2 deletion mutant and the mild heat-shocked wild-type even though several up-regulated Hsps (molecular chaperones) are common between the ssa1/2 deletion mutant and the mild heat-shocked wild-type (Fig. 5B). When Hsp104p, Ydj1p (yeast Hsp40p), and Ssa1p exist together, their chaperone activities increase significantly [25]. From this, it is suggested that the deletion of SSA1/2 induces the suppression of their chaperone activities. Recently, the cooperation of Hsp26p wih Hsp104p/Hsp70p/Hsp40p chaperone system on protein disaggregation in yeast was reported [47, 48]. Hsp26p co-aggregated with substrate is suggested to be a target of the Hsp104p/Hsp70p/Hsp40p chaperone system [47, 48]. Although Ssa1p is able to disaggregate the early Hsp26p-substrate complex (small soluble aggregates), Hsp104p is essential in refolding the late Hsp26p-substrate complex (big insoluble aggregates) [47, 48]. Moreover, excess or stoichiometric Hsp26p against denatured substrates is essential for effective refolding [47]. In the ssa1/2 deletion mutant, an increase in the mRNA expression levels of HSP104 and HSP26 was seen (Fig. 5B). Although the refolding of denatured proteins is sure to succeed if HSP104/Hsp104p and HSP26/Hsp26p are highly expressed, it is a fact that the ubiquitin-proteasome degradation system is facilitated in the ssa1/2 deletion mutant. It can be speculated that as constitutive protein denaturation occurs, the ubiquitin-proteasome degradation system is required in addition to the chaperone refolding system in the ssa1/2 deletion mutant. Furthermore, there is a possibility that protein refolding by molecular chaperones and ubiquitin-proteasome protein degradation are related. In mammalian cells, the following model has been reported; denatured proteins are refolded by Hsp70-HSP40 chaperone-mediated maturation pathway under the treatment of Hsp90 inhibitor, and then denatured proteins are degraded by ubiquitin-proteasome [49]. It is interesting to note that the ubiquitin-proteasome protein degradation system in yeast is induced when the chaperone function is inhibited by the deletion of SSA1/2. However, at present, our data are not sufficient to propose a similar model in yeast, and this remains a topic for future study.
Conclusion
The protein synthesis and ubiquitin-proteasome degradation system were up-regulated in the ssa1/2 deletion mutant, whereas UPR genes were up-regulated but protein synthesis was strongly suppressed in the mild heat-shocked wild-type. These results suggest that the mechanism for rescue of denatured proteins in the ssa1/2 deletion mutant differs from that in the mild heat-shocked wild-type, although the phenomena on acquisition of thermotolerance are similar.
Methods
Strains and growth condition
S. cerevisiae JN14 is the ssa1/2 deletion mutant strain (MATa his3-11, 3-15 leu2-3, 2-112 ura3-52 trp1-?1 lys2? Ssa1-3::HIS3 ssa2-2::URA3) [15]. S. cerevisiae JN54 (MATa his3-11, 3-15 leu2-3, 2-112 ura3-52 trp1-?1 lys2?) is the parent strain (wild-type) of the ssa1/2 double mutant [15]. Yeast cells were incubated in 100 ml of YPD (1% yeast extract, 2% polypeptone and 2% glucose) medium at 30°C to a logarithmic phase (OD660 = 1) using 500 ml Erlenmeyer flasks, and collected by centrifugation (2,800 × g). Cells were washed with distilled water three times, and stocked in a -80°C deep freezer until used for total RNA extraction.
Heat-shock treatment
S. cerevisiae JN54 (wild-type) cells were incubated in YPD medium at 30°C to a logarithmic phase (OD660 = 1), followed by treatment with mild heat-shock at 43°C for 30 or 60 min in pre-warmed (43°C) 100 ml of YPD medium using 500 ml Erlenmeyer flasks. Heat-shocked cells were collected by centrifugation. Cells were washed with distilled water three times, and stocked in a -80°C deep freezer until used for total RNA extraction.
RNA extraction and hybridization to a cDNA microarray
Total RNA was extracted by the hot-phenol method [50]. The extraction of mRNA and reverse-transcription to cDNA was done according to Momose and Iwahashi [51]. Poly (A) +RNA was purified from total RNA with an Oligotex-dT30 mRNA purification kit (TaKaRa, Otsu, Shiga, Japan). Fluorescence-labeled cDNA was synthesized with a Cyscribe cDNA Labeling Kit (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) and 0.5 mM Cy3-UTP (Amersham Biosciences) or 0.5 mM Cy5-UTP. Cy3-UTP was used for the wild-type (as control), and Cy5-UTP was used for the ssa1/2 deletion mutant (as sample). For the heat-shock experiment, Cy3-UTP was used for control cells (30°C), and Cy5-UTP was used for mild heat-shocked cells (43°C). Synthesized cDNA were hybridized to a Kuhara DNA chip (DNA Chip Research, Yokohama, Kanagawa, Japan) at 65°C for 48 h.
cDNA microarray analysis
Hybridized cDNA microarray were washed, dried, and scanned using Scanarray 4000 (GSI Lumonics, Billerica, MA, USA). Quantification of gene expression was done using the Genepix ver. 4.0 quantitative microarray analysis application program (Axon Instruments, Union City, CA, USA). The ratio of intensity Cy5/Cy3 was calculated and normalized with negative control spots. All the calculations and normalizations were done using "Chip Cleanser" program [52]. The functional categorization of genes was performed using GeneSpring (Silicon Genetics, Redwood City, CA, USA), and Comprehensive Yeast Genome Database (CYGD) at the Munich International Center of Protein Sequence (MIPS) database [52]. The over 2- fold expressed genes by the deletion of SSA1/2 or the mild heat-shocked treatment in the wild-type were selected as up-regulated genes and determined by at least twice- induced, out of three individual experiments. Similarly, the over 2- fold suppressed genes by the deletion of SSA1/2 or the mild heat-shocked treatment in the wild-type were selected as down-regulated genes, and determined by twice suppressed, out of three individual experiments [51, 53]. The values for up- or down-regulated genes were the average ratio from three independent experiments. The data obtained in this experiment are available with the accession numbers GSE3315 (ssa1/2 deletion mutant) and GSE3316 (mild heat-shocked wild-type) in the Gene Expression Omnibus Database (GEO) [54].
RT-PCR analysis
Total RNA extraction was carried out as described above. RT-PCR was performed using the One Step RNA PCR Kit (AMV) (TaKaRa), according to the instructions provided by the manufacturer. The primers used for RT-PCR are described in Table 3, and 0.1 μg of total RNA were used for RT-PCR. After reverse transcription, samples were subjected to a cycling regime of 20–25 cycles (details are mentioned in Table 3). Five μl of RT-PCR products were loaded into the wells of a 4% Nu-Sieve 3:1 Plus agarose (Cambrex Bio Science Rockland, Inc. Rockland, ME, USA) gel, and electrophoresis was carried out for 50 min at 100 V. The gels were stained using 10 μg/ml ethidium bromide followed by visualization of the stained bands with an UV-transilluminator (ATTO, Tokyo, Japan).
Antibodies
Anti-Pre1p peptide (Res. No., 17-38) and anti-Rpn4p peptide (Res. No., 499-509) rabbit polyclonal antibodies were ordered to Sigma Genosys (Tokyo, Japan). The anti-multi ubiquitin mouse monoclonal antibody (FK2, Cat. No. SPA-205) was purchased from Stressgen Bioreagents Ltd. Partnership (Victoria, B.C., Canada) [31, 32].
Two-dimensional gel electrophoresis (2-DGE) and Immunoblot analysis
Yeast cells were washed with distilled water three times. Total protein was extracted from cells homogenized with lysis buffer [(7 M urea (ICN Biomedicals, Aurora, OH, USA), 2 M thiourea (Sigma, St. Louis, MO, USA), 4% CHAPS (Sigma), 1% carrier ampholyte (pH 3.5-10, Amersham Biosciences), 18 mM Tris-HCl, pH 7.5, 14 mM Trizma base (Sigma), EDTA-free Proteinase Inhibitor (Roche Diagnostics, Manheim, Germany), 0.2 % Triton X-100, reduced (Sigma), 14.4 mM DTT (Sigma)]. Resuspended cells were broken with glass beads at 4°C for 10 min, and centrifuged at 20,000 × g for 10 min. Cell lysate was centrifuged again at 20,000 × g for 7 min. Equal amounts (350 μg) of protein were subjected to 2-DGE, following O'Farrell's method [55]. Electrophoresis [IEF, carried out in a glass capillary tube of 13 cm length and 3 mm diameter (Nihon Eido, Tokyo, Japan) and SDS-PAGE (12.5% or 15% polyacrylamide gel, 5% stacking and 12.5% or 15% separation gel; using standard glass gels plates obtained from Nihon Eido) in the second dimension] was carried out at a constant current of 35 mA for 2-1/2 h or until the dye (250 μL BPB; 0.1% (w/v) in 10% (v/v) glycerol in MQ) reached the bottom of the gel [56]. Ten μL of the commercially available "ready-to-use" molecular mass standards (Precision Plus Protein Standards, Dual Color, Bio-Rad, Hercules, CA, USA) were loaded next to the acidic end of the IEF tube gel. Reproducibility of 2-DGE protein profiles was confirmed by running at least 3 independent protein samples extracted from the cells of wild-type and the ssa1/2 deletion mutant. Electrotransfer of proteins on gel to a PVDF (NT-31, Nihon Eido) membrane was carried out at 1 mA/cm2 with a semi-dry blotter (Nihon Eido) as described previously [57], followed by immunostaining using antibodies (described above). The anti-Pre1p and anti-Rpn4p rabbit polyclonal antibodies were diluted to 1:50,000, and anti-multi ubiquitin mouse monoclonal antibody (FK2) was diluted to 1:60,000. The ECL plus Western Blotting Detection System protocol for blocking, primary and secondary antibody (anti-Rabbit IgG, Horseradish peroxidase linked whole antibody; from donkey) incubation was followed exactly as described (Amersham Biosciences). Immunoassayed proteins were visualized on an X-ray film (X-OMAT AR, Kodak, Tokyo, Japan) using an enhanced chemiluminescence protocol according to the manufacturer's directions (Amersham Biosciences).
Abbreviations
- SSA:
-
stress seventy family A
- RT-PCR:
-
reverse transcription polymerase chain reaction
- HSP:
-
heat-shock protein
- 2-DGE:
-
two-dimensional gel electrophoresis
- CHAPS:
-
3- [(3-cholamidopropyl) dimethylaminol]-1-propanesulfonate
- EDTA:
-
ethylenediaminetetraacetic acid
- DTT:
-
dithiothreitol
- IEF:
-
isoelectric focusing
- SDS-PAGE:
-
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PVDF:
-
polyvinylidene difluoride
- CYGD:
-
Comprehensive Yeast Genome Database
- MIPS:
-
Munich International Center of Protein Sequence
- GEO:
-
Gene Expression Omnibus Database
- UPR:
-
unfolded protein response
- ERAD:
-
endoplasmic reticulum associated degradation
References
Nover L: Inducers of HSP synthesis: heat shock and chemical stressors. Heat Shock Response. Edited by: Nover L. 1991, Florida: CRC Press, 5-40.
Craig EA: The heat shock response. Crit Rev Biochem. 1985, 18: 239-280.
Lindquist S: The heat shock response. Annu Rev Biochem. 1986, 55: 1151-1191. 10.1146/annurev.bi.55.070186.005443.
Werner-Washburne M, Stone DE, Craig EA: Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987, 7: 2568-2577.
Lopez-Buesa P, Pfund C, Craig EA: The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proc Natl Acad Sci USA. 1998, 95: 15253-15258. 10.1073/pnas.95.26.15253.
Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA: The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J Biol Chem. 2004, 279: 21992-22001. 10.1074/jbc.M313739200.
Craig EA, Kramer J, Shilling J, Werner-Washburne M, Holmes S, Kosic-Smithers J, Nicolet CM: SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol. 1989, 9: 3000-3008.
Liu Q, Krzewska K, Liberek K, Craig EA: Mitochondrial Hsp70 Ssc1: role in protein folding. J Biol Chem. 2001, 276: 6112-6118. 10.1074/jbc.M009519200.
Rose MD, Misra LM, Vogel JP: KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989, 57: 1211-1221. 10.1016/0092-8674(89)90058-5.
Tokunaga M, Kawamura A, Kohno K: Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J Biol Chem. 1992, 267: 17553-17559.
Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K: Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biophys Biochem Res Commun. 2000, 279: 445-450. 10.1006/bbrc.2000.3987.
Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ: Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001, 20: 262-271. 10.1093/emboj/20.1.262.
Ingolia TD, Slater MR, Craig EA: Saccharomyces cerevisiae contains a complex multigene family related to the major heat shock-inducible gene of Drosophila. Mol Cell Biol. 1982, 2: 1388-1398.
Craig EA, Jacobsen K: Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell. 1984, 38: 841-849. 10.1016/0092-8674(84)90279-4.
Nelson RJ, Heschl MF, Craig EA: Isolation and characterization of extragenic suppressors of mutations in the SSA hsp70 genes of Saccharomyces cerevisiae. Genetics. 1992, 131: 277-285.
Halladay JT, Craig EA: A heat shock transcription factor with reduced activity suprpresses a yeast HSP70 mutant. Mol Cell Biol. 1995, 15: 4890-4897.
Werner-Washburne M, Becker J, Kosic-Smithers J, Craig EA: Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J Bacteriol. 1989, 171: 2680-2688.
Sanchez Y, Parsell DA, Taulien J, Vogel JL, Craig EA, Lindquist S: Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol. 1993, 175: 6484-6491.
Becker J, Walter W, Yan W, Craig EA: Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996, 16: 4378-4386.
Bush GL, Meyer DI: The refolding activity of the yeast heat shock proteins Ssa1 and Ssa2 defines their role in protein translocation. J Cell Biol. 1996, 135: 1229-1237. 10.1083/jcb.135.5.1229.
Kim S, Schilke B, Craig EA, Horwich AL: Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc Natl Acad Sci USA. 1998, 95: 12860-12865. 10.1073/pnas.95.22.12860.
Shulga N, James P, Craig EA, Goldfarb DS: A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J Biol Chem. 1999, 274: 16501-16507. 10.1074/jbc.274.23.16501.
Ziegelhoffer T, Lopez-Buesa P, Craig EA: The dissociation of ATP from hsp70 of Saccharomyces cerevisiae is stimulated by both Ydj1p and peptide substrates. J Biol Chem. 1995, 270: 10412-10419. 10.1074/jbc.270.18.10412.
Wegele H, Haslbeck M, Reinstein J, Buchner J: Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003, 278: 25970-25976. 10.1074/jbc.M301548200.
Glover JR, Lindquist S: Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998, 94: 73-82. 10.1016/S0092-8674(00)81223-4.
Zolkiewski M: ClpB cooperates with DnaK, DnaJ and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999, 274: 28083-28086. 10.1074/jbc.274.40.28083.
Krobitsch S, Lindquist S: Aggregation of Huntingtin in yeast varies with the length of polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000, 97: 1589-1594. 10.1073/pnas.97.4.1589.
Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY: Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002, 157: 997-1004. 10.1083/jcb.200112104.
Brown CR, McCann JA, Chiang HL: The heat shock protein Ssa2p is required for import of Fructose-1,6-Bisphosphatase into Vid vesicles. J Cell Biol. 2000, 150: 65-76. 10.1083/jcb.150.1.65.
Satyanarayana C, Schröder-Köhne S, Craig EA, Schu PV, Horst M: Cytosolic Hsp70s are involved in the transport of aminopeptidase 1 from the cytoplasm into the vacuole. FEBS Lett. 2000, 470: 232-238. 10.1016/S0014-5793(00)01324-7.
Fujimuro M, Sawada H, Yokosawa H: Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994, 349: 173-180. 10.1016/0014-5793(94)00647-4.
Takada K, Hirakawa T, Yokosawa H, Okawa Y, Taguchi H, Ohkawa K: Isolation of ubiquitin-E2 (ubiquitin conjugating enzyme) complexes from erythroloukaemia cells using immunoaffinity techniques. Biochem J. 2001, 356: 199-206. 10.1042/0264-6021:3560199.
Seufert W, Jentsch S: Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990, 9: 543-550.
Arnason T, Ellison MJ: Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994, 14: 7876-7883.
Baxter BK, Craig EA: Isolation of UBP3, encoding a de-ubiquitinating enzyme, as a multicopy suppressor of a heat-shock mutant strain of Saccharomyces cerevisiae. Curr Genet. 1998, 33: 412-419. 10.1007/s002940050354.
Hochstrasser M: Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996, 30: 405-439. 10.1146/annurev.genet.30.1.405.
Ng DT, Spear ED, Walter P: Unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol. 2000, 150: 77-88. 10.1083/jcb.150.1.77.
Johnson ES, Ma PC, Ota IM, Varshavsky A: A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995, 270: 17442-17456. 10.1074/jbc.270.29.17442.
Patil C, Walter P: Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol. 2001, 13: 349-356. 10.1016/S0955-0674(00)00219-2.
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO: Genomic Expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000, 11: 4241-4257.
Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR: Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000, 404: 770-774. 10.1038/35004754.
Parsell DA, Kowal AS, Singer MA, Lindquist S: Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994, 372: 475-478. 10.1038/372475a0.
Kawai R, Fujita K, Iwahashi H, Komatsu Y: Direct evidence for the intracellular localization of HSP104 in Saccharomyces cerevisiae by immunoelectron microscopy. Cell Stress Chaperones. 1999, 4: 46-53.
Özcan S, Johnston M: Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995, 15: 1564-1572.
Özcan S, Johnston M: Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999, 63: 554-569.
Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E: Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexose in Saccharomyces cerevisiae. FEBS Lett. 1999, 464: 123-128. 10.1016/S0014-5793(99)01698-1.
Haslbeck M, Meiss A, Stromer T, Walter S, Buchner J: Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem. 2005, 280: 23861-23868. 10.1074/jbc.M502697200.
Cashikar AG, Duennwald M, Lindquist SL: A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005, 280: 23869-23875. 10.1074/jbc.M502854200.
Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L: Chaperone-depenedent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA. 2002, 99: 12847-12852. 10.1073/pnas.202365899.
Kohrer K, Domdey H: Guide to Yeast Genetics and Molecular Biology. Methods Enzymol. Edited by: Guthrie C, Funk GR. 1990, San Diego: Academic Press, 194: 398-401.
Momose Y, Iwahashi H: Bioassay of cadmium using a DNA microarray: genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Environ Toxicol Chem. 2001, 20: 2353-2360. 10.1897/1551-5028(2001)020<2353:BOCUAD>2.0.CO;2.
MIPS: Munich International Center of Protein Sequence. http://mips.gsf.de/
Kitagawa E, Momose Y, Iwahashi H: Correlation of the structures of agricultural fungicides to gene expression in Saccharomyces cerevisiae upon exposure to toxic doses. Environ Sci Technol. 2003, 37: 2788-2793. 10.1021/es026156b.
GEO: Gene Expression Omnibus Database. http://www.ncbi.nlm.nih.gov/geo/
O'Farrell PH: High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975, 250: 4007-4021.
Hajduch M, Rakwal R, Agrawal GK, Yonekura M, Pretova A: High-resolution two-dimentional electrophoresis separation of proteins from metal-stressed rice (Oryza sativa L.) leaves: drastic reductions/ fragmentations of ribulose-1, 5-bisphosphate carbosylate/ oxygenase and induction of stress-related proteins. Electrophoresis. 2001, 22: 2824-2831. 10.1002/1522-2683(200108)22:13<2824::AID-ELPS2824>3.0.CO;2-C.
Rakwal R, Agrawal GK, Kubo A, Yonekura M, Tamogami S, Saji H, Iwahashi H: Defense/ stress responses elicited in rice seedlings exposed to the gaseous air pollutant sulfur dioxide. Environ Exp Bot. 2003, 49: 222-235. 10.1016/S0098-8472(02)00072-2.
Acknowledgements
The authors appreciate the helpful comments from the anonymous referees which helped improve the manuscript. We thank Dr Elizabeth A Craig for providing the JN14 and the JN54 strains. We also thank Dr. Yuko Momose, Ms. Emiko Kitagawa, Dr. Yoshinori Murata, Mr. Mine Odani and Dr. Satomi Murata-Mizukami for advice relating to cDNA microarray analysis, and Ms. Junko Shibato for supporting the protein experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
RM planned and designed the study, performed the experiments and the data analysis, wrote the main draft of the paper, and generated the figures. KA organized all the research, and provided advice for preparing the manuscript. RR designed the protein experiments and RT-PCR analysis, and contributed in figure making and in editing the manuscript. HI planned all the research and designed the experiments, and suggested the draft of the paper. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Matsumoto, R., Akama, K., Rakwal, R. et al. The stress response against denatured proteins in the deletion of cytosolic chaperones SSA1/2 is different from heat-shock response in Saccharomyces cerevisiae. BMC Genomics 6, 141 (2005). https://doi.org/10.1186/1471-2164-6-141
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-6-141